Abstract
The death of cells harboring defects in two distinct pathways implicates these pathways in the control of an essential process. Here we report that cells lacking OmpR and harboring constitutively active MalT undergo premature death that involves increased expression of the outer membrane porin LamB.
To survive, a cell must coordinate its response to diverse stimuli simultaneously. These stimuli can originate in the extracellular environment, the envelope, or the cytosol. The coordination requires effective interaction of different, often apparently unrelated, pathways. A powerful tool to identify interactions between seemingly unrelated pathways is a genetic phenomenon called synthetic lethality (SL). SL is defined as the loss of viability that occurs from the combination of two mutations that separately do not cause death (14, 19, 22, 31). Here we used SL to demonstrate that the osmoregulator OmpR and MalT, the transcriptional regulator of the maltose uptake and metabolism system, interact to control a critical process. Loss of control by disruption of ompR in cells that also carry the malT(Con)(T949A) allele (initially termed SLompR) leads to cell death.
Construction and characterization of a conditional synthetic lethal mutant.
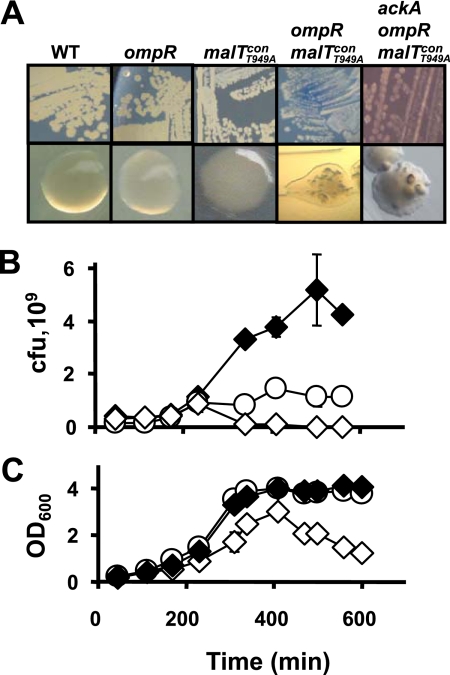
While examining the effect of the small phosphoryl donor acetyl phosphate on the function of the EnvZ-OmpR two-component signal transduction pathway, we attempted to construct an ackA ompR double mutant. We used P1 transduction (30) to introduce an ompR::Tn10 allele (from strain JB100) (Table 1) (2, 6) into an ackA null mutant (strain AJW1939) (18) and observed that approximately 50% of the resulting transductants (designated strain AJW2051) exhibited a set of unique phenotypes. During growth at 37°C on solid tryptone-based media (tryptone broth or LB), colonies became translucent and papillae arose. This behavior contrasted with that of the parental strains and their wild-type (WT) parent (Fig. 1A). Similarly, following growth in liquid tryptone broth or LB, these transductants were unable to form viable colonies (Fig. 1B), a behavior that explains the loss of turbidity as the cultures entered late exponential phase (Fig. 1C). The use of a live-dead stain in combination with light microscopic observation showing that cells grown in liquid culture lyse just prior to entry into stationary phase confirmed the bactericidal nature of the phenotype (data not shown).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Reference |
|---|---|---|
| AJW678 | thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136 lacX74 | 17 |
| AJW1939 | AJW678 ackA::Km | 17 |
| AJW2050 | AJW678 ompR::Tn10 | 2 |
| AJW2051 | AJW678 ackA::Km ompR::Tn10 malT(Con)(T949A) | This study |
| AJW3098 | AJW678 ompR::Tn10 malT(Con)(T949A) | This study |
| AJW3308 | AJW678 ompR::Tn10 malT(Con)(T949A) ΔmalK::Km | This study |
| AJW3499 | AJW678 malT con(T949A) | This study |
| AJW3592 | AJW678 ompR::Tn10 malT(Con)(T949A) ΔlamB::Km | This study |
| JB100 | ompR::Tn10 malTc-1 | 6 |
All other alleles were derived from the KEIO collection (16).
FIG. 1.
ompR malT(Con)(T949A) and ackA ompR malT(Con)(T949A) mutants are synthetic lethal mutants. (A) Colony morphology after growth on LB plates at 37°C for 48 h. Note the translucent appearance of colonies formed by ompR malT(Con)(T949A) and ackA ompR malT(Con)(T949A) mutants (strains AJW3098 and AJW2051, respectively). Also note the presence within the colonies of papillae, which arise from spontaneous suppressor mutants. (B and C) Numbers of CFU and growth curves (C) for ackA (strain AJW1939) (filled diamonds), ompR (strain AJW2050) (open circles), and ackA ompR malT(Con)(T949A) (strain AJW2051) (open diamonds) mutants. Cells were grown at 37°C in LB, and samples were taken at regular intervals to determine the optical density at 600 nm (OD600) and numbers of CFU. The error bars indicate the standard deviations for triplicate determinations and are shown only if they are larger than the symbol. The lower numbers of CFU for ompR mutants can be attributed to the observation that ompR mutants grow at the same rate as WT cells (based on optical density measurements) but divide at a lower rate (based on cell number measurements) (25, 26).
The observed phenotypes depended on the medium composition and temperature. When grown on glucose-supplemented M63 minimal media (either solid or liquid), the transductants exhibited growth phenotypes similar to those of the parental strains and their WT parent (data not shown). Similarly, growth on tryptone-based media at 22°C resulted in healthy, mucoid colonies (data not shown). In contrast, incubation at 28°C or a higher temperature resulted in translucent colonies and the accumulation of papillae, a behavior that became more pronounced as the temperature increased.
The lack of OmpR causes cell death.
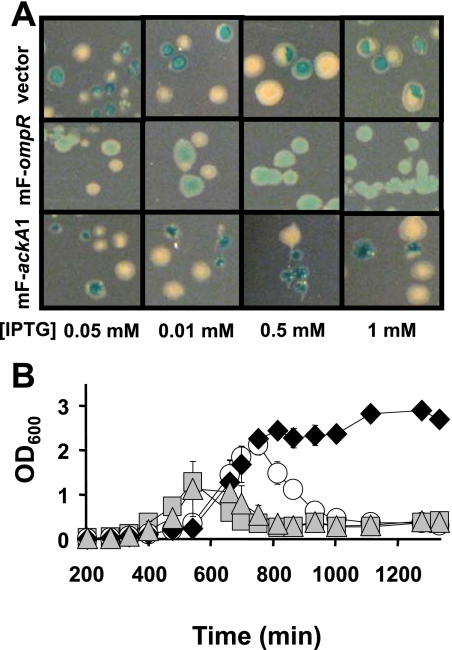
To determine if disruption of ompR contributed to cell death, we employed pRC7, an unstable single-copy mini-F (mF) plasmid carrying ompR and lacZ under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac promoter (3, 12). The mF plasmid lacks the natural F-factor stabilization systems; therefore, it is unstable and thus easily lost when no antibiotic selection is applied. If the vector carries an essential gene, cells have to maintain the plasmid even without antibiotic selection, and all colonies are blue. If the plasmid does not carry an essential gene, it can be lost; thus, colonies with blue and white sectors and pure white colonies can form.
When cells of strain AJW2051 were transformed with mF-ompR, all surviving colonies were blue (Fig. 2A, middle panels). Similarly, in broth, single-copy mF-ompR (Fig. 2B) complemented the defect. Expression of WT OmpR from the multicopy pFR29 vector also complemented the growth defect. In contrast, expression of the mutant proteins OmpR(T83I) and OmpR(V203M) (20, 21) or of WT EnvZ did not complement the growth defect (data not shown). These results provide positive evidence that disruption of ompR contributes to cell death.
FIG. 2.
OmpR, but not AckA, rescues the lethal phenotype of ackA ompR malT(Con)(T949A) mutants. An ackA ompR malT(Con)(T949A) mutant (strain AJW2051) was transformed with the empty mF vector, the mF vector harboring a WT ompR allele (mF-ompR), or two distinct WT ackA alleles (mF-ackA1 and mF-ackA2). (A) Transformants were grown on LB plates at 37°C for 48 h. Expression of ackA and ompR was induced with IPTG. Note the mixture of blue, white, and sectored colonies obtained with 0.5 and 1 mM IPTG in the upper and lower panels, in contrast to the uniform blue colonies in the middle panels. (B) Growth curves for transformants grown in LB supplemented with 0.5 mM IPTG at 37°C. Circles, empty vector; diamonds, mF-ompR; squares, mF-ackA1; triangles, mF-ackA2. The error bars indicate the standard deviations of triplicate cultures. OD600, optical density at 600 nm.
The lack of AckA does not cause cell death.
We also tested if disruption of ackA contributed to cell death. In contrast to ompR, two distinct mF-ackA plasmids, one that included the native ackA ribosomal binding site (mF-ackA1) and one that included an optimized ribosomal binding site (mF-ackA2), failed to restore growth (Fig. 2A and B). Both plasmids exhibited AckA activity and complemented several ackA phenotypes, including mucoidy and poor motility (data not shown). Expression of WT ackA from a multicopy plasmid also failed to sustain growth of the mutant cells, and colonies transformed with ackA plasmids formed papillae (Fig. 2A, lower panels; data not shown). These observations indicated that disruption of ackA does not contribute to death. More importantly, they led us to hypothesize that strain AJW2051 carries an additional mutation [initially designated SLompR and later identified as malT(Con)(T949A)] that is responsible for causing cell death in the absence of OmpR. To test this hypothesis, we introduced the ompR::Tn10 allele from strain AJW2051 into cells of WT (ackA+) strain AJW678. Approximately 50% of the resulting transductants (strain AJW3098) died, and the dying colonies accumulated papillae. Taken together, these results led us to hypothesize that SLompR is linked to the ompR::Tn10 allele.
Characterization of the ompR SLompR mutant.
We characterized the ompR SLompR double mutant (strain AJW3098) by comparing its phenotypes to those of an ackA ompR SLompR triple mutant (strain AJW2051). The two mutants displayed the same translucent phenotype, and both of them accumulated papillae (Fig. 1A). The phenotype was universal; it could be observed in several different strain backgrounds (BW25113, MG1655, MC4100) and with different ompR null alleles (data not shown). Like the triple ackA ompR SLompR mutant, the double ompR SLompR mutant could be complemented with mF-ompR upon induction with IPTG. Subsequent depletion by back-dilution of the double mutant twice in fresh LB again led to a loss of turbidity, as observed with the complemented triple mutant (data not shown).
In contrast to the double mutant, the triple mutant grew more slowly, formed mucoid colonies, and migrated poorly in semisolid motility agar. These differences in growth, mucoidy, and motility are indicative of the ackA mutant allele (13). Furthermore, the triple mutant survived longer than the double mutant (data not shown) (Fig. 2C), most likely due to the slower growth attributed to the ackA mutation.
Mapping of SLompR.
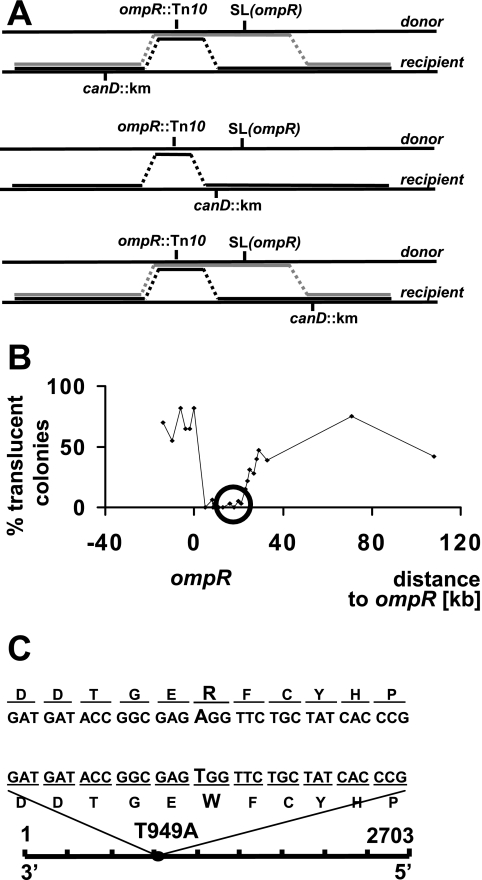
To further map the location of SLompR, we performed a series of tripartite crosses. We hypothesized that donation of the ompR SLompR region to recipient strains using P1 transduction should yield a mixture of two different colony types. Type 1 colonies carrying both ompR and SLompR should be nonviable and translucent when they are grown on or in tryptone-based media at 37°C. In contrast, type 2 colonies carrying ompR but not SLompR should be viable and nontranslucent. We thus transferred the ompR::Tn10 SLompR region from the donor strain AJW2051 to recipient cells carrying null mutations in candidate genes (single-gene deletions, each designated canD and marked with kanamycin resistance) (1). canD genes were chosen to span the region that includes and flanks the ompR and SLompR region (Fig. 3A). Transductants were selected for both tetracycline and kanamycin resistance (and hence both the ompR and canD alleles) on M63 plates supplemented with 0. 4% glucose and incubated at room temperature. Transductants were subsequently screened on LB plates at 37°C for translucence. Depending on the relative locations of ompR, SLompR, and canD, the frequency of translucent colonies varied. Plotting the frequency of translucent offspring versus canD location in the genome generated a distinct pattern, which allowed us to map SLompR to the mal-rtc locus (Fig. 3B).
FIG. 3.
Mapping of malT(Con)(T949A) by P1 transduction. (A) Mapping strategy. The ompR allele was transduced into strains carrying mutant genes (designated canD) spanning the ompR SLompR region. Depending on the relative locations of ompR, SLompR, and canD, the frequency of translucent colonies varies. Black lines indicate crossover events that do not yield translucent colonies (i.e., result in an ompR mutant). Gray lines indicate crossover events that give rise to translucent colonies (i.e., result in an ompR SLompR double mutant). (B) Frequency of translucent colonies after transduction of ompR into canD mutants. After transduction, translucent and nontranslucent colonies were counted, and the percentage of translucent colonies was calculated and plotted. The x axis shows the distance of canD genes and the mal-rtc locus (circled) relative to ompR. At least 80 colonies were tested for each transduction. (C) Location of SLompR. The 2,703-nucleotide DNA sequence of malT and the approximate position of the malT(Con)(T949A) mutation (circle) are shown in the lower part of the panel. The enlargement in the upper part of the panel shows the nucleotide and amino acid changes corresponding to the malT(Con)(T949A) mutation.
Mapping also allowed us to make a statement about the nature of the SLompR mutation. If SLompR was a loss-of-function allele, then its transduction into the corresponding canD deletion mutant (i.e., the specific case where SLompR is canD) would have resulted in 100% translucent colonies during mapping. However, Fig. 3B shows no such peak in the mal-rtc locus. On this basis, we concluded that SLompR is unlikely to be a loss-of-function allele and thus tested the alternative possibility that the SLompR mutation is a gain-of-function mutation. To test this hypothesis, we overexpressed each of the genes in the mal-rtc region in either the WT parent (strain AJW678) or the ompR mutant (strain AJW2050) with the expectation that, compared to the WT parent, the ompR mutant would display higher sensitivity to overexpression of at least one of these genes. We found that overexpression of gntT, rtcA, rtcB, rtcR, or malP had no significant effect on either the WT parent or the ompR mutant. In contrast, the ompR mutant was more sensitive than its WT parent to overexpression of malT or malQ (data not shown). At present, we cannot explain why an ompR mutant is more sensitive to malQ overexpression.
Subsequent sequencing revealed that the SLompR mutation is a point mutation located within the first one-third of the malT open reading frame. Replacement of thymine with adenine at position 949 (T949A) in the nucleotide sequence had led to replacement of a tryptophan residue (W) with an arginine residue (R) (Fig. 3C). We hypothesized that this amino acid change caused the production of a gain-of-function MalT [MalT(Con)] protein. Below, the SLompR mutation is referred to as malT(Con)(T949A) and the mutant protein is referred to as MalT(Con)(W317R).
The malT(Con)(T949A) mutant exhibits increased LamB levels.
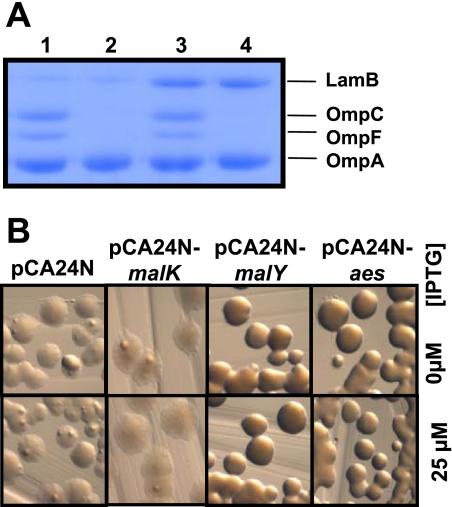
The transcription factor MalT positively regulates 10 genes organized in five operons (4, 5, 27). Constitutively active malT(Con) mutants are known to cause increased expression of all regulon members, including the outer membrane porin LamB (9, 11). To test if MalT(Con)(W317R) increases MalT regulon expression, we made outer membrane preparations and monitored LamB levels. We indeed found that the malT(Con)(T949A) (strain AJW3445) and ompR malT(Con)(T949A) (strain AJW3098) mutants exhibited increased LamB levels (Fig. 4A, lanes 3 and 4) compared to WT cells and the ompR mutant (Fig. 4A, lanes 1 and 2). This result provides further evidence that the SLompR mutation is located in malT. We hypothesize that MalT(Con)(W317R) is a constitutively active protein.
FIG. 4.
MalT(Con)(W317R) is constitutively active. (A) malT(Con)(T949A) mutants exhibit elevated LamB levels, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of outer membrane preparations. Cells were grown in LB at 37°C and harvested during late exponential phase. Gels were stained with Coomassie brilliant blue. Lane 1, WT strain (AJW678); lane 2, ompR::Tn10 strain (AJW2050); lane 3, malT(Con)(T949A) strain (AJW3445); lane 4, ompR::Tn10 malT(Con)(T949A) strain (AJW3098). (B) Inhibition of MalT(Con)(W317R) with MalY and Aes. The ompR malT(Con)(T949A) mutant (strain AJW3098) was transformed with plasmids carrying genes coding for MalT inhibitors (MalK, MalY, Aes) or the empty vector. The resulting transformants were streaked on LB plates supplemented with IPTG to induce gene expression, and growth was observed after incubation at 37°C for 48 h.
SL of the ompR malT(Con)(T949A) mutant is subject to catabolite repression.
Transcription of malT is regulated by catabolite repression; i.e., exposure to glucose represses malT transcription (7, 8). If malT(Con)(T949A) was responsible for SL in the ompR mutant background, then exposure to glucose should permit the ompR malT(Con)(T949A) mutant to survive in nonpermissive growth conditions. Indeed, when grown at 37°C in LB medium (either solid or liquid) supplemented with 0.4% glucose, cells of the ompR malT(Con)(T949A) mutant displayed WT-like growth (data not shown). This result is supported by our initial finding that the ompR malT(Con)(T949A) mutant survives on glucose-supplemented M63 minimal medium (data not shown) and supports our hypothesis that malT(Con)(T949A) encodes a constitutively active MalT protein.
Overexpression of MalY and Aes but not MalK permits survival of the ompR malT(Con)(T949A) mutant.
Gain-of-function MalT [MalT(Con)] proteins can be inhibited by one or more of the following inhibitors: MalK, MalY, and Aes (16, 24, 28). MalK provides a feedback loop between maltose transport and MalT activation. Under conditions in which no maltose is transported into the cell, MalK directly interacts with MalT to inhibit activation by maltotriose (16). Like MalK, the inhibitors MalY and Aes also interfere with maltotriose binding (24, 28).
To determine if any of the inhibitors could inhibit MalT(Con)(W317R) and thus restore growth, we transformed the ompR malT(Con)(T949A) double mutant (strain AJW3098) with the empty pCA24N vector or with this vector carrying malK, malY, or aes (17). The resultant transformants were grown at 37°C on LB plates with increasing IPTG concentrations to induce expression of the inhibitors. We found that even uninduced expression of MalY or Aes permitted growth, likely due to leakiness of the T5-lac promoter (Fig. 4B). In contrast, expression of MalK did not permit survival even after induction with 0.2 mM IPTG (Fig. 4B and data not shown). The functionality of the plasmid was confirmed using outer membrane preparations to monitor the levels of LamB. As expected, expression of MalK in WT cells resulted in reduced LamB levels (data not shown). These experiments, combined with our outer membrane preparation, led us to conclude that malT(Con)(T949A) encodes a constitutively active MalT(Con)(W317R).
Deletion of lamB suppresses lethality.
Since MalT activates transcription of all MalT regulon members, we hypothesized that increased expression of one or more of these proteins could cause lethality in an ompR mutant background. To identify the MalT regulon member that causes cell death in the absence of ompR, we introduced the ompR allele linked to the malT(Con)(T949A) mutation into several mal deletion mutants and screened for translucence. Of all the mal mutants tested (malEGFKMSZ and lamB; malP and malQ were not tested), only the lamB ompR malT(Con)(T949A) and malK ompR malT(Con)(T949A) triple mutants produced 100% viable colonies. The viability of the malK ompR malT(Con)(T949A) triple mutant could be explained by a polar effect exerted by the malK allele on lamB. Indeed, removing the kanamycin resistance cassette using flp recombinase (10, 23) and thus generating a nonpolar deletion of malK produced nonviable colonies (data not shown). These results are consistent with the hypothesis that increased expression of LamB in the absence of OmpR contributes to death.
The observation that deletion of lamB permits survival of ompR malT(Con)(T949A) mutants may in part explain why the SL phenotype has not been reported before. Schlegel et al. (28) constructed several ompR malT(Con) double mutants in a strain background that includes the malB107 allele and thus lacks LamB (15).
SL is not specific to the malT(Con)(T949A) allele.
Over the past four decades, several different malT(Con) alleles have been reported and studied (9, 28, 29). We asked if any of these alleles also cause a synthetic lethal phenotype in ompR mutants. We cotransduced a representative selection of malT(Con) alleles, each linked to an ompR::Tn10 insertion (derived from strains AS42-46, AS48-50, AS54, and AS56) (28) into WT cells (strain AJW678). Transductants were selected as described above and screened for the translucent phenotype on LB plates at 37°C. We found that all of the malT(Con) alleles tested resulted in a translucent phenotype in an ompR mutant background (data not shown). Thus, we concluded that most, if not all, constitutively active MalT proteins cause death in mutants that lack OmpR.
Concluding remarks.
Constitutively active MalT [in the form of MalT(Con)(W317R)] increases expression of LamB (9, 11). Surprisingly, such expression causes cell death in ompR mutants but not in cells that retain OmpR. There is no known overlap in the OmpR and MalT regulons; therefore, it is unlikely that the cause of death is redundancy of some essential function. Instead, we propose that cells coordinate the expression of the OmpR and MalT regulons to guard against the inappropriate insertion of LamB into the outer membrane. Efforts are under way to understand why LamB is detrimental to ompR mutants.
Acknowledgments
We thank Linda Kenney, Tom Silhavy, Tom Bernhardt, Winfried Boos, and the National Institute for Genetics (Japan) for providing strains, plasmids and phage; Mark Goulian for providing advice regarding outer membrane preparations; Karen Visick, David Keating, and Winfried Boos for fruitful discussions; members of the Wolfe and Visick labs for critical reading of the manuscript; and Shivanee Shah for the initial characterization of strain AJW2051.
We thank the National Institute of General Medical Sciences (grant GM066130) and the Loyola University Potts Foundation (grant LU#11200) for funding.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, M., L. Enquist, and T. J. Silhavy. 1981. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 3.Bernhardt, T. G., and P. A. de Boer. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boos, W., and A. Bohm. 2000. Learning new tricks from an old dog: MalT of the Escherichia coli maltose system is part of a complex regulatory network. Trends Genet. 16:404-409. [DOI] [PubMed] [Google Scholar]
- 5.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brass, J. M., K. Bauer, U. Ehmann, and W. Boos. 1985. Maltose-binding protein does not modulate the activity of maltoporin as a general porin in Escherichia coli. J. Bacteriol. 161:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon, C. 1982. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J. Bacteriol. 150:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapon, C., and A. Kolb. 1983. Action of CAP on the malT promoter in vitro. J. Bacteriol. 156:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dardonville, B., and O. Raibaud. 1990. Characterization of malT mutants that constitutively activate the maltose regulon of Escherichia coli. J. Bacteriol. 172:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debarbouille, M., H. A. Shuman, T. J. Silhavy, and M. Schwartz. 1978. Dominant constitutive mutations in malT, the positive regulator gene of the maltose regulon in Escherichia coli. J. Mol. Biol. 124:359-371. [DOI] [PubMed] [Google Scholar]
- 12.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 13.Fredericks, C. E., S. Shibata, S. Aizawa, S. A. Reimann, and A. J. Wolfe. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734-747. [DOI] [PubMed] [Google Scholar]
- 14.Fujimitsu, K., M. Su'etsugu, Y. Yamaguchi, K. Mazda, N. Fu, H. Kawakami, and T. Katayama. 2008. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 190:5368-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofnung, M., D. Hatfield, and M. Schwartz. 1974. malB region in Escherichia coli K-12: characterization of new mutations. J. Bacteriol. 117:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly, N., A. Bohm, W. Boos, and E. Richet. 2004. MalK, the ATP-binding cassette component of the Escherichia coli maltodextrin transporter, inhibits the transcriptional activator MalT by antagonizing inducer binding. J. Biol. Chem. 279:33123-33130. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 18.Kumari, S., C. M. Beatty, D. F. Browning, S. J. Busby, E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lestini, R., and B. Michel. 2008. UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J. Bacteriol. 190:5995-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattison, K., R. Oropeza, N. Byers, and L. J. Kenney. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315:497-511. [DOI] [PubMed] [Google Scholar]
- 21.Nara, F., S. Matsuyama, T. Mizuno, and S. Mizushima. 1986. Molecular analysis of mutant ompR genes exhibiting different phenotypes as to osmoregulation of the ompF and ompC genes of Escherichia coli. Mol. Gen. Genet. 202:194-199. [DOI] [PubMed] [Google Scholar]
- 22.Ooi, S. L., X. Pan, B. D. Peyser, P. Ye, P. B. Meluh, D. S. Yuan, R. A. Irizarry, J. S. Bader, F. A. Spencer, and J. D. Boeke. 2006. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 22:56-63. [DOI] [PubMed] [Google Scholar]
- 23.Pan, G., K. Luetke, and P. D. Sadowski. 1993. Mechanism of cleavage and ligation by FLP recombinase: classification of mutations in FLP protein by in vitro complementation analysis. Mol. Cell. Biol. 13:3167-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peist, R., A. Koch, P. Bolek, S. Sewitz, T. Kolbus, and W. Boos. 1997. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J. Bacteriol. 179:7679-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruss, B. M. 1998. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch. Microbiol. 170:141-146. [DOI] [PubMed] [Google Scholar]
- 26.Pruss, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 179:5602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlegel, A., A. Bohm, S. J. Lee, R. Peist, K. Decker, and W. Boos. 2002. Network regulation of the Escherichia coli maltose system. J. Mol. Microbiol. Biotechnol. 4:301-307. [PubMed] [Google Scholar]
- 28.Schlegel, A., O. Danot, E. Richet, T. Ferenci, and W. Boos. 2002. The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY. J. Bacteriol. 184:3069-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuman, H. A. 1982. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J. Biol. Chem. 257:5455-5461. [PubMed] [Google Scholar]
- 30.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Ting, H., E. A. Kouzminova, and A. Kuzminov. 2008. Synthetic lethality with the dut defect in Escherichia coli reveals layers of DNA damage of increasing complexity due to uracil incorporation. J. Bacteriol. 190:5841-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]