Abstract
Haemophilus influenzae is a human-specific gram-negative coccobacillus that causes a variety of human infections ranging from localized respiratory infections to invasive diseases. Hsf is the major nonpilus adhesin in encapsulated strains of H. influenzae and belongs to the trimeric autotransporter family of proteins. The Hsf protein contains two highly homologous binding domains, designated HsfBD1 and HsfBD2. In this study we characterized the differential binding properties of HsfBD1 and HsfBD2. In assays using HeLa cells, we found that bacteria expressing either full-length Hsf or HsfBD1 by itself adhered at high levels, while bacteria expressing HsfBD2 by itself adhered at low levels. Immunofluorescence microscopy and a cellular enzyme-linked immunosorbent assay using purified proteins revealed that the binding affinity was significantly higher for HsfBD1 than for HsfBD2. Purified HsfBD1 was able to completely block adherence by bacteria expressing either HsfBD1 or HsfBD2, while purified HsfBD2 was able to block adherence by bacteria expressing HsfBD2 but had minimal activity against bacteria expressing HsfBD1. Conversion of the residue at position 1935 in the HsfBD1 binding pocket from Asp to Glu resulted in HsfBD2-like binding properties, and conversion of the residue at position 569 in the HsfBD2 binding pocket from Glu to Asp resulted in HsfBD1-like binding properties, as assessed by adherence assays with recombinant bacteria and by immunofluorescence microscopy with purified proteins. This work demonstrates the critical role of a single amino acid in the core of the binding pocket in determining the relative affinities of the HsfBD1 and HsfBD2 binding domains.
Haemophilus influenzae is a gram-negative coccobacillus that causes both serious invasive diseases and localized respiratory tract infections in humans (10, 17, 19). Isolates of H. influenzae can be separated into encapsulated and nonencapsulated or so-called nontypeable strains (12). Most strains recovered from patients with invasive disease are encapsulated and express the type b capsule, while the majority of strains associated with respiratory tract infections are nontypeable (19).
The pathogenesis of disease due to H. influenzae type b begins with colonization of the upper respiratory tract (4, 8, 11, 13, 16, 19). Most type b strains are capable of expressing hemagglutinating pili, which mediate bacterial attachment to oropharyngeal epithelial cells, extracellular matrix proteins, and mucin and promote colonization. Mutant strains that lack hemagglutinating pili are also capable of adherence and colonization, highlighting the fact that nonpilus adhesive factors also exist (4, 5, 8, 20). In recent work, we have demonstrated that the major nonpilus adhesin in H. influenzae type b is a large protein called Hsf, which forms short fibers visible by electron microscopy (15).
The Hsf adhesin is encoded by the hsf locus and is a trimeric autotransporter protein that shares significant homology with Hia, a trimeric autotransporter adhesin that is present in ∼25% of nontypeable H. influenzae strains. Hsf contains an N-terminal signal sequence, an internal passenger domain with two binding domains, and a C-terminal outer membrane pore-forming domain, analogous to Hia (3, 6). The binding domains in Hsf are called HsfBD1 and HsfBD2 and share high-level homology with each other and with the two binding domains in Hia (2, 14). HsfBD1 and HsfBD2 interact with the same host cell receptor structure on Chang epithelial cells, although with different affinities (3). Based on in vitro experiments using purified proteins and Chang epithelial cells, HsfBD1 has a dissociation constant (Kd) of ∼0.2 nM and HsfBD2 has a Kd of ∼2.5 nM.
In previous work using X-ray crystallography and site-directed mutagenesis, we established that both HiaBD1 and HiaBD2 are trimeric structures with acidic binding pockets formed by contiguous IsNeck and Trp-ring domains (9, 21). Using structural modeling and site-directed mutagenesis, we determined that HsfBD1 and HsfBD2 possess the same fold and trimeric assembly as HiaBD1 and HiaBD2, with conservation of the residues that are essential for HiaBD1 adhesive activity (3).
In the current study we examined the structural basis for the different binding affinities of HsfBD1 and HsfBD2. In initial experiments, we found that the differences between HsfBD1 and HsfBD2 were easier to observe with HeLa cells than with Chang cells, reflecting the fact that the receptor density is lower on HeLa cells. Our results demonstrated the critical role of a single amino acid in the core of the binding pocket in determining the relative affinities of HsfBD1 and HsfBD2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. Escherichia coli strains were grown on Luria-Bertani (LB) agar or in LB broth and were stored at −80°C in LB broth with 30% glycerol. H. influenzae strains were grown on chocolate agar or in brain heart infusion broth supplemented with hemin and NAD (1) and were stored at −80°C in brain heart infusion broth with 30% glycerol. Selection for plasmids in E. coli strains was performed using ampicillin at a concentration of 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80dlacZΔM15 ΔlacU169 deoR recA endA1 | Life Technologies |
| BL21(DE3) | hsdS gal (λ cI ts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 18 |
| H. influenzae strains | ||
| C54b−p−Hsf+ | Capsule-deficient, nonpiliated mutant of type b strain C54 | 5 |
| C54b−p−Hsf− | Derivative of C54b−p−Hsf+ containing a mini-Tn10 kan element in the hsf locus | 5 |
| Plasmids | ||
| pT7-7 | Cloning vector, Ampr | New England Biolabs |
| pDC601 | pT7-7 containing hsf from H. influenzae strain C54 | 15 |
| pDC601(E569A) | pDC601 encoding E-to-A mutation at residue 569 in Hsf | 3 |
| pDC601(D1935A) | pDC601 encoding D-to-A mutation at residue 1935 in Hsf | 3 |
| pDC601(E569A/D1935A) | pDC601 encoding E-to-A and D-to-A mutations at residues 569 and 1935 in Hsf | 3 |
| pNS1 | Derivative of pHAT10 that contains hia upstream sequence and coding sequence for the Hia signal peptide, the HAT epitope, and coding sequence for the Hia translocator domain | 3 |
| pNS1-HsfBD1 | pNS1 containing coding sequence for HsfBD1 from H. influenzae strain C54 | This study |
| pNS1-HsfBD2 | pNS1 containing coding sequence for HsfBD2 from H. influenzae strain C54 | This study |
| pNS1-HsfBD1(Y1934V) | pNS1/HsfBD1 encoding Y-to-V mutations at residue 1934 in Hsf | This study |
| pNS1-HsfBD1(D1935E) | pNS1/HsfBD1 encoding D-to-E mutation at residue 1935 in Hsf | This study |
| pNS1-HsfBD2(V568Y) | pNS1/HsfBD2 encoding V-to-Y mutation at residue 568 in Hsf | This study |
| pNS1-HsfBD2(E569D) | pNS1/HsfBD2 encoding E-to-D mutation at residue 569 in Hsf | This study |
| pGEX-6-P-1/HsfBD1 | pGEX-6-P-1 containing coding sequence for HsfBD1 from H. influenzae strain C54 | 3 |
| pGEX-6-P-1/HsfBD2 | pGEX-6-P-1 containing coding sequence for HsfBD2 from H. influenzae strain C54 | 3 |
| pGEX-6-P-1/HsfBD1(Y1934) | Derivative of pGEX-6-P-1/HsfBD1 encoding Y-to-V mutation at residue 1934 in Hsf | This study |
| pGEX-6-P-1/HsfBD1(D1935E) | Derivative of pGEX-6-P-1/HsfBD1 encoding D-to-E mutation at residue 1935 in Hsf | This study |
| pGEX-6-P-1/HsfBD2(V568Y) | Derivative of pGEX-6-P-1/HsfBD2 encoding V-to-Y mutation at residue 568 in Hsf | This study |
| pGEX-6-P-1/HsfBD2(E569D) | Derivative of pGEX-6-P-1/HsfBD2 encoding E-to-D mutation at residue 569 in Hsf | This study |
In order to generate pNS1/HsfBD1 and pNS1/HsfBD2, DNA corresponding to Hsf residues 1899 to 2031 (HsfBD1) and 532 to 661 (HsfBD2) was amplified by PCR from pDC601, engineering BamHI and KpnI sites at the 5′ and 3′ ends, respectively. The resulting fragments were digested with BamHI and KpnI and then ligated into BamHI-KpnI-digested pNS1. Plasmids with point mutations in HsfBD1 and HsfBD2 were generated by performing site-directed mutagenesis on pNS1/HsfBD1 or pNS1/HsfBD2, as appropriate, using a QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions. Similarly, glutathione S-transferase (GST) expression constructs with mutations in HsfBD1 and HsfBD2 were generated by performing site-directed mutagenesis on pGEX-6-P-1/HsfBD1 or pGEX-6-P-1/HsfBD2, as appropriate. Mutagenized plasmids were confirmed to have the intended mutations by nucleotide sequencing.
Purification of GST fusion proteins.
GST fusion proteins were purified as described previously (3). Briefly, E. coli BL21(DE3) strains harboring pGEX-6P-1 derivatives were grown at 37°C in LB medium containing 100 μg/ml ampicillin to an optical density at 600 nm of ∼0.4 to 0.5. Cultures were then induced with isopropryl-β-d-thiogalactopyranoside (IPTG) at a concentration of 0.1 mM for 3 h at 30°C. Bacteria were harvested by centrifugation at 6,600 × g, resuspended in lysis buffer (phosphate-buffered saline [PBS], 5 mM EDTA, 1:100 Complete Mini protease inhibitor cocktail [Roche]), and lysed by sonication. Cell fragments were removed by centrifugation at 10,000 × g, and GST fusion proteins were isolated by affinity chromatography using immobilized glutathione (Pierce) according to the manufacturer's directions.
Quantitative adherence assays.
Chang epithelial cells (Wong-Kilbourne derivative, clone 1-5c-4 [human conjunctiva], ATCC CCL 20.2) and HeLa cells (human cervical epidermoid carcinoma, ATCC HTB 33) were maintained in minimum essential medium supplemented with nonessential amino acids and 10% (vol/vol) fetal bovine serum. Cells were cultivated at 37°C in 5% CO2. Adherence assays were performed as described previously (3, 16, 17). For adherence inhibition assays, monolayers were preincubated with purified GST fusion proteins for 1.5 h at 37°C in 5% CO2 prior to inoculation of bacteria. The level of adherence was calculated by dividing the number of adherent CFU per monolayer by the number of inoculated CFU per monolayer. All assays were performed in triplicate, which allowed calculation of means and standard errors. Values were compared to determine statistically significant differences using the two-tailed Student t test.
Detection of protein binding by immunofluorescence microscopy.
Immunofluorescence microscopy was performed as described previously (7). Briefly, Chang and HeLa cells were seeded at a density of 8 × 104 cells per well onto glass coverslips in 24-well plates, and the plates were incubated overnight. Cell monolayers were washed once with PBS and fixed for 15 min with 2.5% (vol/vol) paraformaldehyde-0.2% (vol/vol) glutaraldehyde in PBS. Monolayers were washed again with PBS, and potentially reactive sites were quenched for 10 min with 20 mM ethanolamine in PBS. Following quenching, nonspecific binding sites were blocked for 30 min with 3% bovine serum albumin in PBS, and cell monolayers were then incubated for 1 h with the relevant purified GST fusion protein at a concentration of 100 nM. Protein binding was detected using an anti-GST antibody (2 μg/ml; GE Healthcare) and a Cy2-conjugated secondary antibody (1:200; Jackson Immuno Research). Samples were mounted and examined by confocal scanning microscopy.
Quantitation of protein binding by cellular ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (7). Briefly, Chang and HeLa cells were seeded into 96-well tissue culture plates at a density of 1.8 × 105 cells per well, and the plates were incubated overnight. Cell monolayers were washed once with PBS and fixed for 15 min with 2.5% (vol/vol) paraformaldehyde-0.2% (vol/vol) glutaraldehyde in PBS. The monolayers were washed again with PBS, and potentially reactive sites were quenched for 15 min with 20 mM ethanolamine in PBS. Subsequently, nonspecific binding sites were blocked for 30 min with 3% bovine serum albumin in PBS, the relevant purified GST fusion protein was added, and the cells were incubated for 2 h at 37°C in 5% CO2. The monolayers were washed four times with PBS and incubated with an anti-GST antibody (1:4,000; GE Healthcare) and then with a horseradish peroxidase-conjugated secondary antibody (1:4,000; Sigma). Protein binding was quantitated by adding a 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) (ABTS) peroxidase substrate solution (Roche) and measuring the absorbance at 405 nm with a microplate reader (Bio-Tek Instruments). All experiments were performed in triplicate on multiple occasions. Kd values were determined using the GraphPad Prism software.
Flow cytometry.
Binding of purified GST-HsfBD1 to Chang cells and binding of purified GST-HsfBD1 to HeLa cells were compared using flow cytometry, employing GST-HsfBD1 as a probe for receptor density and GST alone as a measure of background fluorescence. Confluent monolayers of Chang and HeLa cells in tissue culture flasks were washed once with PBS, and cells were detached from the flasks using Cellstripper (MediaTech, Inc.). Following collection, cells were washed again with PBS and fixed with 4% formaldehyde in PBS for 15 min and then were washed with 2% fetal calf serum (FCS) in PBS and resuspended in 2% FCS in PBS at a density of 2 × 106 cells/ml. Subsequently, 0.5-ml aliquots of cells were incubated with 100 μM purified GST-HsfBD1 or 100 μM purified GST at 4°C for 1 h. Samples were washed twice with 2% FCS in PBS and then resuspended in 2% FCS in PBS and incubated with a 1:250 dilution of an Alexa fluor 488-conjugated anti-GST antibody at 4°C for 1 h. Samples were washed with PBS and analyzed by flow cytometry using a Becton Dickinson FACSCalibur running on Becton Dickinson CellQuest software. The ratio of the geometric mean of the fluorescence associated with binding of GST-HiaBD1 to the geometric mean of the fluorescence associated with GST alone was calculated separately for Chang cells and HeLa cells, allowing comparison of GST-HsfBD1 binding values.
Sequence alignment.
Amino acid sequence alignment was performed using BLAST at NCBI (http://www.ncbi.nlm.nih.gov/blast/) and ClustalW at EBI (http://www.ebi.ac.uk/clustalw/).
RESULTS
HsfBD1 and HsfBD2 mediate different levels of adherence to HeLa cells.
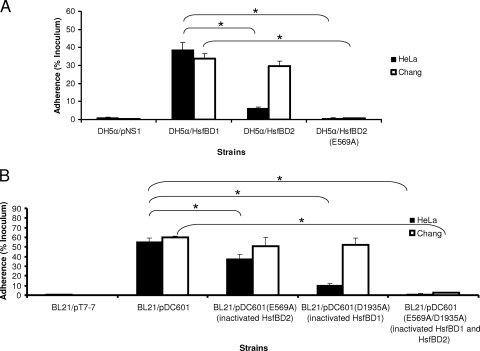
In previous in vitro studies using purified GST fusion proteins and Chang epithelial cells, we observed that the binding affinity of GST-HsfBD1 was significantly higher than the binding affinity of GST-HsfBD2 when the affinity was assessed by cellular ELISA and immunofluorescence microscopy (3). Despite the difference in binding affinity, E. coli DH5α expressing HsfBD1 by itself (in a presentation vector called pNS1) and DH5α expressing HsfBD2 by itself adhered to Chang cells at similar levels (3). In the current study, we set out to further characterize the binding properties of HsfBD1 and HsfBD2 and began by examining HsfBD1- and HsfBD2-mediated bacterial adherence to HeLa cells. As shown in Fig. 1A, E. coli DH5α expressing HsfBD1 by itself demonstrated high-level adherence, while E. coli DH5α expressing HsfBD2 by itself demonstrated only low-level adherence; thus, these results differed from the results obtained with Chang cells. The low-level adherence associated with HsfBD2 was completely eliminated when the HsfBD2 binding pocket was disrupted via mutagenesis of the residue at position 569 that changed this residue from glutamic acid to alanine (HsfBD2-E569A), paralleling observations with Chang cells and suggesting that the same binding pocket is involved in interactions between HsfBD2 and both HeLa cells and Chang cells (3).
FIG. 1.
Adherence of Hsf-expressing bacteria to HeLa cells and Chang cells. (A) Adherence by E. coli derivatives expressing HsfBD1 by itself, HsfBD2 by itself, or HsfBD2(E569A) (containing a point mutation that disrupts the binding pocket) in the presentation vector pNS1. (B) Adherence by E. coli derivatives expressing full-length Hsf (pDC601), full-length Hsf with a mutation that disrupts HsfBD2 [pDC60(E569A)], full-length Hsf with a mutation that disrupts HsfBD1 [pDC601(D1935A)], and full-length Hsf with mutations that disrupt both HsfBD2 and HsfBD1 [pDC601(E569A/D1935A)]. For both panel A and panel B, monolayers were incubated with bacteria for 30 min at 37°C and were then rinsed to remove nonadherent bacteria. Dilutions were plated on agar to determine the number of adherent CFU per monolayer, and adherence was calculated by dividing the number of adherent CFU by the number of inoculated CFU per monolayer. The bars and error bars indicate the means and standard errors of three measurements from representative experiments. The asterisks indicate statistically significant differences (P < 0.05) between the values indicated by the brackets.
To determine whether HsfBD1 and HsfBD2 mediate different levels of adherence to HeLa cells in the context of the native Hsf protein, we performed quantitative adherence assays using DH5α expressing full-length Hsf with inactivating mutations in the binding pocket of either HsfBD1 or HsfBD2. As shown in Fig. 1B, the adherence levels were consistent with those observed when HsfBD1 and HsfBD2 were expressed by themselves. In particular, the level of adherence remained high when only HsfBD1 was functional (HsfB2 was disrupted) and was low when only HsfBD2 was functional (HsfBD1 was disrupted). Adherence was completely abolished when both HsfBD1 and HsfBD2 were disrupted.
Purified GST-HsfBD1 and GST-HsfBD2 bind to the same host cell receptor on HeLa cells, but the binding affinities are different.
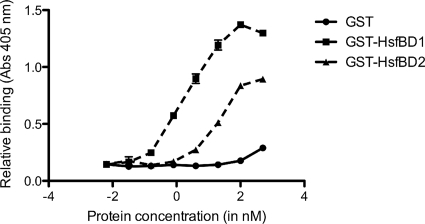
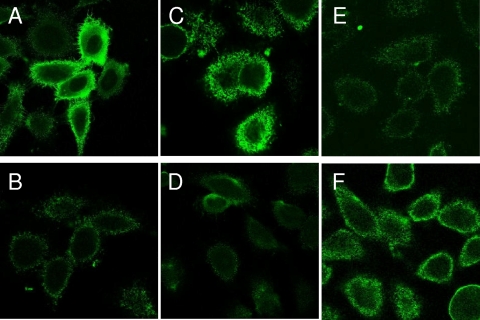
To further analyze the interaction of HsfBD1 and HsfBD2 with HeLa cells, we performed cellular ELISAs with purified GST-HsfBD1 and purified GST-HsfBD2. As shown in Fig. 2, we observed high-affinity binding with GST-HsfBD1 and lower-affinity binding with GST-HsfBD2. As determined with the GraphPad Prism program, the Kd for HsfBD1 was ∼0.2 nM and the Kd for HsfBD2 was ∼3.0 nM, observations similar to our previous observations with Chang cells (3). Consistent with these results, immunofluorescence microscopy revealed that GST-HsfBD1 was associated with strong punctuate fluorescence, while GST-HsfBD2 was associated with only weak fluorescence (Fig. 3A and 3B).
FIG. 2.
Relative binding of HsfBD1 and HsfBD2 to HeLa cells as determined by a cellular ELISA. Monolayers of HeLa epithelial cells were incubated with purified GST-HsfBD1, GST-HsfBD2, or only GST for 2 h. Unbound proteins were removed by rinsing, and monolayers were incubated with an anti-GST antibody and then with a horseradish peroxidase-conjugated secondary antibody. Protein binding was quantified by adding ABTS substrate solution and measuring the absorbance at 405 nm with a microplate reader.
FIG. 3.
Binding to HeLa cells by purified GST-Hsf proteins as assessed by immunofluorescence microscopy. HeLa cells were incubated with 100 nM purified GST fusion proteins for 2 h. Unbound proteins were removed by rinsing, and protein binding was detected by incubation with an anti-GST antibody and a Cy2-conjugated secondary antibody. Samples were analyzed by confocal scanning microscopy. (A) GST-HsfBD1; (B) GST-HsfBD2; (C) GST-HsfBD1(Y1934V); (D) GST-HsfBD2(V568Y); (E) GST-HsfBD1(D1935E); (F) GST-HsfBD2(E569D).
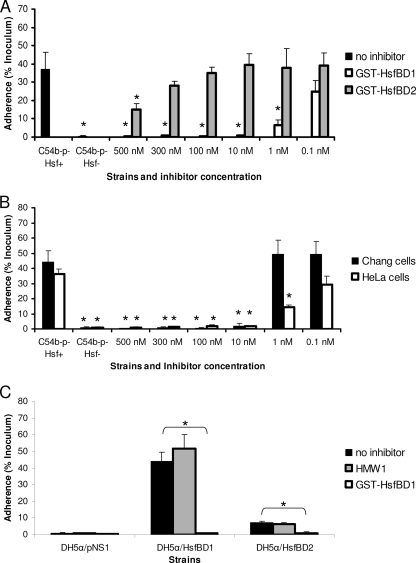
To extend our findings, we assessed the abilities of purified GST-HsfBD1 and purified GST-HsfBD2 to inhibit Hsf-mediated bacterial adherence to HeLa cells. As shown in Fig. 4A, preincubation of monolayers with GST-HsfBD1 at concentrations as low as 10 nM resulted in complete inhibition of adherence by H. influenzae strain C54b−p−Hsf+, while preincubation with purified GST-HsfBD2 at concentrations of 500 and 300 nM resulted in only partial inhibition of adherence by strain C54b−p−Hsf+, further confirming the different affinities of HsfBD1 and HsfBD2. As shown in Fig. 4B, the concentration of purified GST-HsfBD1 required to inhibit adherence by H. influenzae strain C54b−p−Hsf+ was lower for HeLa cells than for Chang cells, suggesting that there are different densities of the Hsf host cell receptor in these two cell lines.
FIG. 4.
Inhibition of Hsf-mediated bacterial adherence by purified GST-HsfBD1 and GST-HsfBD2. (A) HeLa cell monolayers were preincubated with 0.1 to 500 nM purified GST-HsfBD1 or GST-HsfBD2 for 90 min. Subsequently, H. influenzae C54b−p−Hsf+ was inoculated onto monolayers, and adherence was measured with quantitative adherence assays. (B) Chang and HeLa cell monolayers were preincubated with 0.1 to 500 nM purified GST-HsfBD1 for 90 min. Subsequently, H. influenzae C54b−p−Hsf+ was inoculated onto monolayers, and adherence was measured with quantitative adherence assays. (C) HeLa cell monolayers were preincubated with 100 nM purified GST-HsfBD1 or purified HMW1 for 90 min. Subsequently, DH5α expressing the presentation vector alone, HsfBD1, or HsfBD2 was inoculated onto monolayers, and adherence was measured with quantitative adherence assays. In all panels, adherence is expressed as a percentage of the bacterial inoculum that bound to the epithelial cell monolayers. The bars and error bars indicate the means and standard errors of three measurements from representative experiments. In panel A, the asterisks indicate statistically significant differences (P < 0.05) for comparisons with H. influenzae C54b−p−Hsf+ with no inhibitor. In panel B, the asterisks indicate statistically significant differences (P < 0.05) for comparisons with H. influenzae C54b−p−Hsf+ with no inhibitor with Chang and HeLa cells. In panel C, the asterisks indicate statistically significant differences (P > 0.05) between the values indicated by the brackets (e.g., adherence by DH5α/HsfBD1 was statistically significantly different when monolayers were preincubated with no inhibitor versus GST-HsfBD1).
To determine whether HsfBD1 and HsfBD2 interact with the same cellular receptor on HeLa cells, we preincubated cell monolayers with purified GST-HsfBD1 and then inoculated monolayers with DH5α expressing either HsfBD1 or HsfBD2. As a control, we preincubated monolayers with purified HMW1, a separate H. influenzae adhesin with a different cell binding specificity. As shown in Fig. 4C, preincubation with 100 nM purified GST-HsfBD1 resulted in complete inhibition of both HsfBD1- and HsfBD2-mediated adherence, while preincubation with purified HMW1 had no effect on adherence. Preincubation with 100 nM purified GST-HsfBD2 completely inhibited HsfBD2-mediated adherence but had no significant effect on HsfBD1-mediated adherence (data not shown). These results suggest that HsfBD1 and HsfBD2 recognize the same cellular receptor on HeLa cells, an observation similar to earlier observations for Chang cells (3).
To confirm that HeLa cells and Chang cells have different densities of the Hsf receptor, we performed flow cytometry using purified GST-HsfBD1 as a probe for receptor molecules and purified GST as a negative control and for measuring background fluorescence. The ratio of the geometric mean fluorescence with GST-HsfBD1 to the geometric mean fluorescence with GST alone was 2.5 for HeLa cells and 8.9 for Chang cells, indicating that there were fewer bound HsfBD1 molecules and hence ∼70% fewer receptor molecules on HeLa cells than on Chang cells.
Homology models of HsfBD1 and HsfBD2 and site-directed mutagenesis.
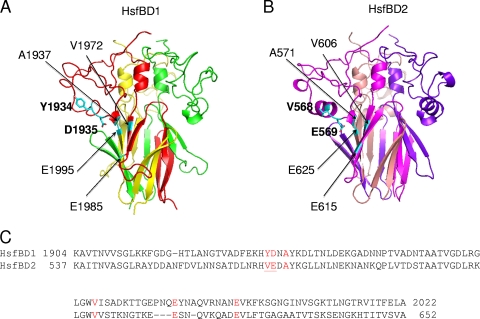
Previous homology models of HsfBD1 and HsfBD2 based on the HiaBD1 structure suggested that there are binding pockets very similar to the binding pocket in HiaBD1 (3). To refine the homology models of HsfBD1 and HsfBD2, we used both the HiaBD1 structure and the recently solved HiaBD2 structure (9). As shown in Fig. 5A and 5B, the refined models of HsfBD1 and HsfBD2 share a highly intertwined β-meander fold in the Trp-ring domain with HiaBD1 and HiaBD2. Examination of the refined models and alignment of the HsfBD1, HsfBD2, HiaBD1, and HiaBD2 sequences revealed that the binding pocket in HsfBBD1 is formed by residues Y1934, D1935, A1937, V1972, E1985, and E1995 and the binding pocket in HsfBD2 is formed by residues V568, E569, A571, V606, E615, and E625 (Fig. 5C). Closer inspection revealed two notable differences between the HsfBD1 and HsfBD2 binding pockets; namely, the Y1934 and D1935 residues in HsfBD1 corresponded to V568 and E569 in HsfBD2 (Fig. 5C). Based on the homology models, both Y1934 and V568 lie in the outermost helix of the structure perpendicular to the threefold axis of the trimer, and both D1935 and E569 lie in a pivotal position initiating a conserved helix-helix kink in the IsNeck domain. While the side chains of Y1934 and V568 point outward away from the trimer, the acidic side chains of D1935 and E569 point inward and appear to be at the center of the binding pocket (Fig. 5A and 5B).
FIG. 5.
Homology modeling and sequence alignment of HsfBD1 and HsfBD2. Homology model structures of HsfBD1 and HsfBD2 were generated based on the crystal structures of HiaBD1 and HiaBD2. (A) Overall homology model structure of HsfBD1, showing the residues that form the binding pocket and highlighting the side chains of the tyrosine and aspartic acid residues in the binding pocket. (B) Overall homology model structure of HsfBD2, showing the residues that form the binding pocket and highlighting the side chains of the valine and glutamic acid residues in the binding pocket. (C) Alignment of HsfBD1 and HsfBD2 sequences. The residues that make up the acidic binding pocket for each binding domain are red, and the residues in the binding pocket that were mutated by site-directed mutagenesis are underlined.
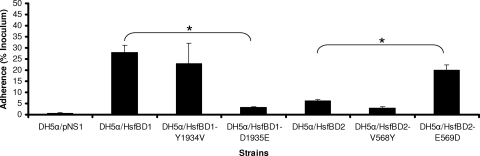
To assess the role of the Y1934/V568 and D1935/E569 residues in the different binding affinities of HsfBD1 and HsfBD2, we generated individual Y1934V and D1935E mutations in HsfBD1 (inserting the HsfBD2 residues) and individual V568Y and E569D mutations in HsfBD2 (inserting the HsfBD1 residues). As shown in Fig. 6, DH5α expressing HsfBD1 containing the D1935E mutation by itself demonstrated low-level HsfBD2-like adherence, and DH5α expressing HsfBD2 containing the E569D mutation by itself demonstrated high-level HsfBD1-like adherence. In contrast, the Y1934V mutation by itself in HsfBD1and the V568Y mutation by itself in HsfBD2 had no effect on adherence.
FIG. 6.
Effect of site-directed mutagenesis on Hsf-mediated adherence to HeLa cells: adherence to HeLa cells by E. coli DH5α harboring the presentation vector pNS1 or expressing HsfBD1, HsfBD1(Y1934V), HsfBD1(D1935E), HsfBD2, HsfBD2(V568Y), or HsfBD2(E569D) in pNS1. The mutations in HsfBD1 change amino acids to the corresponding amino acids in HsfBD2, and the mutations in HsfBD2 change amino acids to the corresponding amino acids in HsfBD1. Bacteria were inoculated onto monolayers, and adherence was measured with quantitative adherence assays. Adherence is expressed as a percentage of the bacterial inoculum that bound to the epithelial cell monolayers. The bars and error bars indicate the means and standard errors of three measurements from a representative experiment. The asterisks indicate statistically significant differences (P < 0.05) between the values indicated by the brackets.
To confirm the results of the bacterial adherence assays, we generated GST fusion proteins containing individual point mutations in HsfBD1 and HsfBD2 and examined the binding of these proteins by immunofluorescence microscopy. The D1935E mutation alone in HsfBD1 resulted in decreased binding to HeLa cells, similar to binding by GST-HsfBD2 (Fig. 3C), and the E569D mutation alone in HsfBD2 resulted in increased binding to HeLa cells, similar to binding by GST-HsfBD1 (Fig. 3F). In contrast, the Y1934V mutation by itself in HsfBD1 and the V568Y mutation by itself in HsfBD2 had no effect on binding (Fig. 3E and 3D, respectively).
DISCUSSION
In this study we examined the binding properties of the HsfBD1 and HsfBD2 binding domains in the H. influenzae Hsf adhesin. Using HeLa cells as a model system for quantifying binding, we found that the adherence by recombinant bacteria expressing HsfBD1 alone was greater than the adherence by recombinant bacteria expressing HsfBD2 alone. Consistent with these results, we observed that the binding affinity of purified HsfBD1 was higher than the binding affinity of HsfBD2, as assessed by cellular ELISA and immunofluorescence microscopy. Modeling of the HsfBD1 and HsfBD2 structures based on the crystal structures of HiaBD1 and HiaBD2 and alignment of the amino acid sequences of HsfBD1 and HsfBD2 revealed differences between the HsfBD1 and HsfBD2 binding pockets at two residues. Further analysis established the critical role of a single residue in the center of the binding pocket in determining the relative affinities of HsfBD1 and HsfBD2.
In earlier crystallography studies, we found that the binding pocket in both HiaBD1 and HiaBD2 is an acidic cleft formed by protruding helices of the IsNeck domain and the β-sheet of the Trp-ring domain on each face of the trimeric structure (9, 21). Modeling of the HsfBD1 and HsfBD2 structures reveals the same IsNeck domain and Trp-ring domain. The three residues in the core of the binding pocket in HsfBD1 are D1935, A1937, and V1972, and the three residues in the core of the binding pocket in HsfBD2 are E569, A571, and V606. In our mutagenesis studies, we converted D1935 to glutamic acid in HsfBD1 and observed a decrease in binding affinity, and we converted E569 to aspartic acid in HsfBD2 and observed an increase in binding affinity. This conversion of HsfBD1 to an HsfBD2-like structure and of HsfBD2 to an HsfBD1-like structure suggests that the slightly shorter side chain of aspartic acid is essential for optimal interaction with the host cell receptor, perhaps to avoid a steric effect associated with glutamic acid, which contains an extra CH2 group.
In considering our findings with HsfBD1 and HsfBD2, it is interesting to compare the binding pockets of HiaBD1 and HiaBD2 in the Hia trimeric autotransporter expressed in nontypeable H. influenzae. The three residues in the core of the binding pocket in HiaBD1 are D618, A620, and V656 and are associated with a Kd of 0.05 to 0.1 nM, and the three residues in the core of the binding pocket in HiaBD2 are Q82, A84, and V120 and are associated with a Kd of 1 to 2 nM (7, 21). Thus, similar to our observations with HsfBD1 and HsfBD2, the key determinants of the relative binding affinities of HiaBD1 and HiaBD2 appear to be the aspartic acid in HiaBD1 and the glutamine in HiaBD2, underscoring the importance of the length of the side chain at this position and raising the question of whether an acidic side chain is essential for optimal interaction.
In previous studies we observed that recombinant bacteria expressing HsfBD1 by itself and recombinant bacteria expressing HsfBD2 by itself adhered at comparable levels to Chang epithelial cells (3); thus, these results differ from the results in the current study with HeLa cells. The fact that adherence to Chang and HeLa cells is mediated by the same HsfBD1 and HsfBD2 binding pockets suggests that the two cell lines express the same Hsf receptor structure. Given that the concentration of purified HsfBD1 required to inhibit Hsf-mediated adherence was lower for HeLa cells than for Chang cells, we propose that the Hsf receptor is present at a lower density on HeLa cells than on Chang cells. Consistent with this conclusion, we found that purified GST-HsfBD1 bound at lower levels to HeLa cells than to Chang cells when binding was assessed by flow cytometry. A lower receptor density may result in lower-avidity adherence by whole bacteria, explaining the difference in the magnitude of adherence between recombinant bacteria expressing HsfBD1 and recombinant bacteria expressing HsfBD2. In ongoing work, we are pursuing the identity of the Hsf receptor.
In summary, the HsfBD1 and HsfBD2 binding domains interact with the same host cell receptor but with different binding affinities. Our results demonstrate that the critical determinant of the relative binding affinities of HsfBD1 and HsfBD2 is the length of the side chain of the acidic amino acid in the core of the binding pocket.
Acknowledgments
This work was supported by NIH grant RO1-AI44167 to J.W.S.G. and by Wellcome Trust grant 08087 to G.M. and G.W.
Footnotes
Published ahead of print on 12 June 2009.
REFERENCES
- 1.Anderson, P., R. B. Johnston, Jr., and D. H. Smith. 1972. Human serum activities against Hemophilus influenzae, type b. J. Clin. Investig. 51:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 3.Cotter, S. E., H. J. Yeo, T. Juehne, and J. W. St. Geme III. 2005. Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 187:4656-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farley, M. M., D. S. Stephens, S. L. Kaplan, and E. O. Mason, Jr. 1990. Pilus- and non-pilus-mediated interactions of Haemophilus influenzae type b with human erythrocytes and human nasopharyngeal mucosa. J. Infect. Dis. 161:274-280. [DOI] [PubMed] [Google Scholar]
- 5.Geme, J. W., III, and D. Cutter. 1995. Evidence that surface fibrils expressed by Haemophilus influenzae type b promote attachment to human epithelial cells. Mol. Microbiol. 15:77-85. [DOI] [PubMed] [Google Scholar]
- 6.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 7.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St. Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46:731-743. [DOI] [PubMed] [Google Scholar]
- 8.Loeb, M. R., E. Connor, and D. Penney. 1988. A comparison of the adherence of fimbriated and nonfimbriated Haemophilus influenzae type b to human adenoids in organ culture. Infect. Immun. 56:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng, G., J. W. St Geme III, and G. Waksman. 2008. Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. J. Mol. Biol. 384:824-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moxon, E., and T. F. Murphy. 2000. Haemophilus influenzae, p. 2369-2376. In G. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, PA.
- 11.Murphy, T. F., J. M. Bernstein, D. M. Dryja, A. A. Campagnari, and M. A. Apicella. 1987. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to nontypable Haemophilus influenzae: pathogenetic and epidemiological observations. J. Infect. Dis. 156:723-731. [DOI] [PubMed] [Google Scholar]
- 12.Pittman, M. 1931. Variation and type specificity in the bacterial species Hamophilus influenzae. J. Exp. Med. 53:471-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St. Geme, J. W., III. 1997. Insights into the mechanism of respiratory tract colonization by nontypable Haemophilus influenzae. Pediatr. Infect. Dis. J. 16:931-935. [DOI] [PubMed] [Google Scholar]
- 14.St. Geme, J. W., III, and D. Cutter. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St. Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St. Geme, J. W., III, and S. Falkow. 1990. Haemophilus influenzae adheres to and enters cultured human epithelial cells. Infect. Immun. 58:4036-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 19.Turk, D. C. 1984. The pathogenicity of Haemophilus influenzae. J. Med. Microbiol. 18:1-16. [DOI] [PubMed] [Google Scholar]
- 20.Weber, A., K. Harris, S. Lohrke, L. Forney, and A. L. Smith. 1991. Inability to express fimbriae results in impaired ability of Haemophilus influenzae b to colonize the nasopharynx. Infect. Immun. 59:4724-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeo, H. J., S. E. Cotter, S. Laarmann, T. Juehne, J. W. St. Geme III, and G. Waksman. 2004. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 23:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]