Abstract
Bacterially derived exotoxins kill eukaryotic cells by inactivating factors and/or pathways that are universally conserved among eukaryotic organisms. The genes that encode these exotoxins are commonly found in bacterial viruses (bacteriophages). In the context of mammals, these toxins cause diseases ranging from cholera to diphtheria to enterohemorrhagic diarrhea. Phage-carried exotoxin genes are widespread in the environment and are found with unexpectedly high frequency in regions lacking the presumed mammalian “targets,” suggesting that mammals are not the primary targets of these exotoxins. We suggest that such exotoxins may have evolved for the purpose of bacterial antipredator defense. We show here that Tetrahymena thermophila, a bacterivorous predator, is killed when cocultured with bacteria bearing a Shiga toxin (Stx)-encoding temperate bacteriophage. In cocultures with Tetrahymena, the Stx-encoding bacteria display a growth advantage over those that do not produce Stx. Tetrahymena is also killed by purified Stx. Disruption of the gene encoding the StxB subunit or addition of an excess of the nontoxic StxB subunit substantially reduced Stx holotoxin toxicity, suggesting that this subunit mediates intake and/or trafficking of Stx by Tetrahymena. Bacterially mediated Tetrahymena killing was blocked by mutations that prevented the bacterial SOS response (recA mutations) or by enzymes that breakdown H2O2 (catalase), suggesting that the production of H2O2 by Tetrahymena signals its presence to the bacteria, leading to bacteriophage induction and production of Stx.
Genes encoding bacterial exotoxins are frequently carried in bacteria by bacteriophages (4, 19). Phage-encoded exotoxins such as cholera toxin, diphtheria toxin, botulinium toxin, and Shiga toxin cause disease in mammals (4). Although these toxins do affect humans and other mammals, these phage-carried exotoxin genes are found at high frequencies in free phages and lysogenic bacteria that are isolated from environments where the presumed corresponding targets are not prevalent (6). These exotoxins kill eukaryotic cells by means of receptors and pathways that are generally conserved among eukaryotic organisms. These observations have led to the hypothesis that humans and other susceptible mammals are neither the original nor primary “targets” of these toxins (31).
If not mammals, then what are the true targets of these ubiquitous exotoxins? One clue may come from a consideration of the bacterial ecology and evolutionary biology. A major source of bacterial mortality is consumption by single-cell eukaryotic predators, such as ciliates and other protozoa (13). The coordinated release of exotoxins, at either the pre- or postingestional state (31), could comprise one of the bacterium's major antipredator defense strategies. Hence, humans may be innocent bystanders in the evolutionary battle between protozoans and their bacterial prey.
A family of phages strongly related to the well-characterized coliphage λ genes carry genes encoding Shiga-like exotoxin (Stx). Stx-encoding bacteriophages are associated with a broad range of hosts, including several serotypes of Escherichia coli, Enterobacter cloacae, Shigella flexneri, and Citrobacter freundii. One well-studied Stx-encoding strain is Escherichia coli O157:H7 strain EDL933. This strain was responsible for the first reported multistate U.S. outbreak of Shiga toxin-caused hemorrhagic colitis in 1982 (40). Related strains continue to cause serious food- and waterborne infections worldwide.
Analysis of the genome sequence of the EDL933 bacterial strain and the lambdoid bacteriophages liberated from it revealed that the disease-causing Shiga toxin is encoded by each of two lysogenic lambdoid prophages, 933W and 933J (32, 34, 36, 38). Lambdoid bacteriophages all share a common developmental program. Upon infection of a bacterial cell, the lambdoid phages choose between two developmental fates. The phage can grow lytically, thereby killing the host. Alternatively, in lysogenic growth, the phage chromosome inserts into the host chromosome and is replicated along with it until a signal that induces lytic growth is perceived by the lysogenized phages. Since transcription of the stx genes is under the control of a promoter that is active only during lytic growth (53), Stx is not produced when the toxin-encoding bacteriophages are in the lysogenic state. Inactivation of the respective bacteriophage repressors (53) causes lytic induction of the lysogenic phages, and exotoxin production increases substantially upon phage lysogen induction (27, 32, 34, 54). Stx is apparently not exported through any bacterial secretory machinery. Its release from the bacteria depends on phage genes that cause bacterial cell lysis (52, 53). These genes are also expressed only during phage lytic growth. Therefore, the Shiga toxin gene can reside harmlessly within the bacterial host until phage induction causes production and release of Shiga toxin.
Stx is a compound A-B5 toxin, consisting of a 32-kDa A subunit and a pentameric 9.7-kDa B subunit. The five B subunits form a disulfide-bonded ring, into which is inserted the C-terminal end of the A subunit (41-43, 45). Stx toxicity in mammalian cells is mediated by the binding of the B subunit of the holotoxin to its receptor, globotriaosylceramide (Gb3), on the surface of mammalian cells. Holotoxin enters mammalian cells by clathrin-dependent, receptor-mediated endocytosis (42). The B subunit directs the retrograde transport of the A subunit from endosomes to the Golgi apparatus and finally to the endoplasmic reticulum (22, 44), where the catalytic A subunit is cleaved from the holotoxin by furin and released into the cytoplasm (56). Once inside cells, the StxA subunit causes cell death by selectively removing the adenine from position 4224 on 22S rRNA, thereby inhibiting protein synthesis.
A major source of bacterial mortality is consumption by single-cell protozoan predators. Ciliates, including Tetrahymena thermophila, control bacterial densities in many ecosystems (3). Protozoan bacterivory has been shown to reduce bacterial numbers (47) and is therefore accepted as a pivotal process in microbial “population control.” Earlier investigations have demonstrated the utility of using Tetrahymena species and E. coli as a model predator-prey interaction for studying food web dynamics (23). Tetrahymena cells can be grown using only E. coli as a food source (23). Methods to quantify predation of bacterial cultures by Tetrahymena have been described (11, 49).
We propose that the Stx-encoding bacteriophages evolved to reside within the bacteria to function as part of their antipredator arsenal and that the presence of the toxin gene may confer an evolutionary advantage on the growth and survivability of this bacterial population by killing the predator. We tested this hypothesis by exploring how the presence of an exotoxin-encoding bacteriophage resident within bacteria influences the growth and survival of the bacterial population and a model unicellular eukaryotic predator, Tetrahymena thermophila.
A recent study compared the relative survival of Stx+ and Stx− bacteria in cocultures with Tetrahymena pyriformis (49). Consistent with a role of exotoxins in augmenting the fitness of bacterial populations that carry them, these investigators found that the ratio of Stx+ to Stx− bacteria increased under these conditions. However, the mechanism by which Stx enhances survivability was not clear from this study. Under the conditions used in those experiments, the presence of Stx-encoding phages increased the survivability of bacteria in food vacuoles inside T. pyriformis. However, the results also indicated that increased bacterial survival was largely independent of Stx expression. That study did not examine the effect of bacterial Stx production on the growth and survivability of the Tetrahymena predator, a factor that could profoundly influence bacterial survival. The focus of the work presented here is the effect of Stx on Tetrahymena viability.
We find that when cocultured with a model predator, Tetrahymena thermophila, Stx-encoding bacteria kill this predator. Stx-encoding strains are less efficiently predated than are strains that do not encode this exotoxin. We also found that Tetrahymena appears to release a factor that signals the bacteria that a predator is present and stimulates the production of toxin by the bacteria. The results are consistent with a role for bacterial exotoxins in the bacterial antipredator arsenal.
MATERIALS AND METHODS
Strains and chemicals.
EDL933 was obtained from the ATCC, and 933r, an isogenic recA mutant of EDL933 (16), was obtained from Jörg Hacker, Universität Würzburg. EDL933WΔstx, an EDL933 variant bearing a complete deletion of all stx genes (18), was obtained from Christine Miller, Institut National de la Recherche Argonomique. The W3110::λ lysogen was constructed using wild-type λ bacteriophage as described previously (2). Tetrahymena thermophila (CU427.4) was obtained from the Tetrahymena Stock Center (Cornell University).
A plasmid encoding Stx2 holotoxin (pStxAB) was constructed by amplifying DNA carrying the entire stxAB region of bacteriophage 933W (38) from genomic DNA of the EDL933 and inserting this DNA into the plasmid pET17b (EMB Biosciences). A plasmid encoding only Stx2A (pStx2A) was constructed by disrupting the stx2B gene in pStx2AB. This disruption was accomplished by inserting the coding sequence of the chloramphenicol resistance (Cmr) gene, isolated from pACYC184 by PCR, into the unique PflMI restriction site in the stxB gene. As assessed by immunodot analysis (50), both these plasmids direct the low-level expression of holotoxin or Stx2A subunit when transformed into bacterial cells lacking the T7 RNA polymerase gene.
Catalase and superoxide dismutase were obtained from Worthington Biochemicals. Mitomycin C was obtained from Sigma. Purified Stx2 holotoxin and anti-Shiga toxin antibodies were purchased from Toxin Technologies.
Effect of coculture on Tetrahymena and bacterial viability.
Tetrahymena and bacterial cells needed for the coculture experiments were prepared as follows. Cultures of the specified bacteria were to grown saturation at 37°C in M9 plus 0.08% glucose. Cells in these cultures were harvested, washed twice with M9 plus 0.08% sodium citrate, and suspended in the same medium. Tetrahymena cells were diluted fivefold from saturated liquid cultures and grown for 2 days at 30°C in protease peptone plus FeCl2. These cells were harvested, washed twice with 10 mM Tris-HCl (pH 7.4), and suspended in M9 plus 0.08% sodium citrate in a volume sufficient to give 104 cells/ml. To each washed Tetrahymena culture or control cultures containing no Tetrahymena, either 108 washed bacterial cells/ml or an equal volume of medium were added. The cocultures were maintained at 30°C. Where indicated, 4 μg/ml of either superoxide dismutase or catalase was added to each culture.
At various times after initiating coculture, two aliquots were removed from each. One aliquot was used to determine the amount of Tetrahymena present in the culture. Tetrahymena counts were obtained by counting the number of Lugol-stained cells or by trypan blue exclusion, as visualized in a hemocytometer. Tetrahymena cells that are killed by exposure to Stx or Stx-expressing bacteria are not visible by either method, presumably because they have lysed. The number of bacteria was determined plating the culture and counting the number of CFU. Each measurement was performed in triplicate and the data averaged. Each experiment was repeated a minimum of three times. The data presented represent the average for the three (or more) replicates.
Purification of StxB.
StxB was purified from JM105:pSBC54 (Ampr) (1) as described previously (5). The concentrated protein was stored at −80°C in 10 mM phosphate-buffered saline supplemented with 15% glycerol.
Effect of holo-Shiga toxin on Tetrahymena viability.
Tetrahymena was grown in protease peptone to a density of 106 cells/ml. These cells were diluted 100-fold in fresh medium and incubated for up to 6 h at 30°C in the absence or presence of (i) 1 to 1,000 ng/ml of purified Stx2A or (ii) 25 ng/ml Stx2A in the presence of increasing concentrations of purified StxB subunit. As controls for the effect of adding protein to the growth medium, Tetrahymena was separately incubated with increasing concentrations of heat-denatured toxin or an equivalent weight of bovine serum albumin (BSA). The number of viable Tetrahymena cells was determined at various times after initiating incubation. Each measurement was performed in triplicate and the data averaged. Each experiment was repeated a minimum of three times. The data presented represent the average of the three (or more) replicates.
Statistical methods.
Error bars presented in the figures represent standard deviations of the means of multiple (>3) replicate experiments. t tests were used to test the significance of differences between the mean of the measured initial amounts and the amounts of bacteria and/or Tetrahymena after treatment in each experiment.
RESULTS
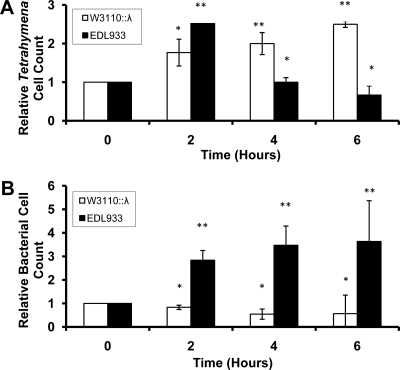
To determine whether unicellular eukaryotes can be potential targets of exotoxins such as Shiga toxin, we measured the growth of Tetrahymena thermophila when these cells were cocultured with several bacterial strains. As a first step in our study, T. thermophila, acclimated to feed on non-phage-bearing W3110 bacteria, was washed and suspended in medium containing Stx+ or Stx− bacteria. Under these culture conditions, T. thermophila growth requires the presence of bacteria. Consistent with the use of bacteria as food by T. thermophila, when a Stx− bacterial strain that is lysogenized with wild-type bacteriophage λ, a phage that does not encode an exotoxin, is grown in the presence of W3110::λ, the number of T. thermophila cells approximately doubles within 6 h (Fig. 1A). In contrast, when T. thermophila is fed the Shiga toxin-encoding strain EDL933, the number of T. thermophila cells decreases to less than half of the input number of cells by 6 h (Fig. 1A). Hence, EDL933 does not support T. thermophila growth; instead, this bacterium causes T. thermophila death.
FIG. 1.
Relative amounts of Tetrahymena (A) and E. coli (B) cells surviving coculture. Cocultures and cell counts were performed as described in Materials and Methods. Cell numbers are expressed as the fold change in the number of cells remaining in culture at increasing lengths of time after beginning incubation. At the start of incubation, bacteria and Tetrahymena cells were present at 108 and 105 cells/ml, respectively. Error bars represent standard deviations from ≥3 independent experiments, with each experiment being comprised of a minimum of three individual measurements *, P < 0.01; **, P < 0.001.
The killing of T. thermophila seen in the presence of the Stx-encoding bacteria (Fig. 1A) is not due to a reduction in the amount of their bacterial food source. Figure 1B shows that in coculture with T. thermophila, the amount of EDL933 increases over 6 h in coculture. In contrast, the number of W3110::λ cells decreases ∼2-fold over the same time period. These observations are consistent with the idea that the presence of Stx-encoding bacteriophages is advantageous to a bacterial population, because when subject to T. thermophila predation, the Stx-encoding bacteria survive better than those that do not encode Stx.
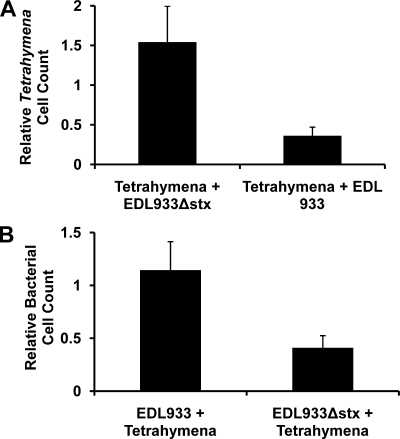
Compared to W3110::λ, EDL933 contains approximately 1 Mb of additional DNA, much of which carries other pathogenicity-related genes in addition to stx. Thus, to more directly test the idea that Shiga toxin encoded by the lysogenized bacteriophage can kill T. thermophila, we determined whether an EDL933 strain bearing complete disruption/deletion of both the stx1 and stx2 gene clusters, EDL933Δstx (18), affects the growth of T. thermophila. Similar to the results presented in Fig. 1, when T. thermophila is fed the Shiga toxin-encoding strain EDL933, the number of T. thermophila cells decreases > 2-fold over a 6 h incubation (Fig. 2A). In contrast when T. thermophila are cocultured with the EDL933Δstx strain, the number of T. thermophila increases by ∼50% in this time period (Fig. 2A). These observations demonstrate that the killing of T. thermophila in cocultures with EDL933 is due to Stx encoded by this bacterial strain. Consistent with the findings in Fig. 1, in coculture with the T. thermophila, the number of EDL933Δstx bacteria decrease >2-fold over the 6-h incubation period, while the number of EDL933 bacteria increases slightly (Fig. 2B). This finding supports the idea that Shiga toxin can function as part of an antipredator defense strategy.
FIG. 2.
Role of Stx in Shiga toxin-encoding bacterially mediated killing of Tetrahymena (A) and inhibition of Tetrahymena predation of bacteria (B). Tetrahymena was cocultured with EDL933 or EDL933Δstx (18), a strain bearing complete disruptions in both the stx1 and stx2 gene clusters. Cocultures and cell counts were performed as described in Materials and Methods. Cell numbers are expressed as the fold change in the number of cells remaining in culture at 6 h relative to the number of cells present at the start of the coculture. Input cell numbers, experimental design, and error analysis were as described in the legend to Fig. 1. Differences between Stx+ and Stx− are significant at a P value of <0.005 or greater.
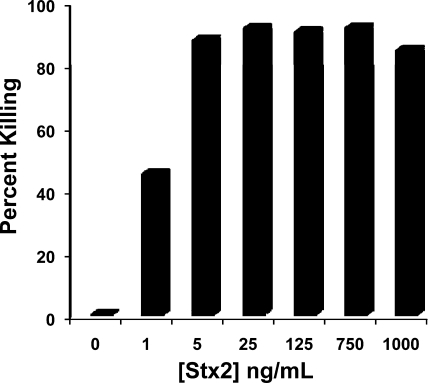
To directly demonstrate that Stx can kill T. thermophila, we determined whether addition of purified Stx2 could kill T. thermophila in axenic cultures. We found that addition of increasing concentrations of Stx2 progressively decreases the number of T. thermophila cells that survive after a 6-hour incubation with toxin (Fig. 3). Under our conditions, addition of 25 ng/ml of purified Stx kills ≥90% of the cells in culture. Similarly, partially purified Stx-containing extracts (33) obtained from mitomycin C-induced EDL933 cells also kill T. thermophila (not shown). These findings confirm that Shiga toxin is responsible for EDL933-mediated killing of T. thermophila.
FIG. 3.
Effect of purified Stx2 on survival of axenically growing Tetrahymena. The indicated amount of purified Stx2 holotoxin was added to 105 cells/ml of axenically growing Tetrahymena. The number of live Tetrahymena cells was determined after 6 h. Data are presented as percent of input Tetrahymena cells killed during a 6-h incubation with indicated amount of Stx2.
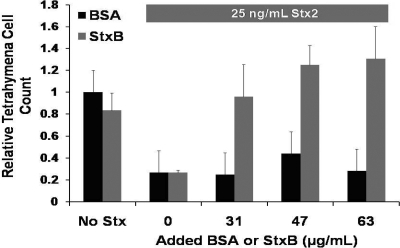
We wished to delineate the factors that regulate the cytotoxic effect of Shiga toxin on T. thermophila. Stx entry into mammalian cells requires the binding of the B subunit of the holotoxin to its Gb3 receptor on the surface of mammalian cells. We reasoned that if the B subunit has a role in Stx-dependent T. thermophila killing, addition of excess StxB subunit should compete with holotoxin's cytotoxic effects. Added partially purified B subunit alone does not exhibit any cytolethal effect on T. thermophila cultures (Fig. 4), a finding that is similar to that obtained with mammalian cells (20, 29). Addition of an increasing weight of the partially purified StxB subunit in the presence of intact Stx progressively inhibits killing of T. thermophila caused by the added purified Stx. The inhibitory effect of added B subunit in Stx-mediated killing saturates at between 47 and 63 μg/ml. This finding implies that the added B subunit competes with binding of holo-Stx to specific receptor/trafficking sites in T. thermophila. The StxB blockade of holotoxin-mediated killing is specific; addition of similar quantities of BSA (Fig. 4) or boiled B subunit (not shown) does not inhibit Stx-mediated T. thermophila killing.
FIG. 4.
StxB-mediated inhibition of Stx2A holotoxin-dependent Tetrahymena killing. Tetrahymena (105 cells/ml) was axenically grown in the presence of 65 μg/ml of purified StxB (gray bars) subunit or BSA (black bars) alone or with 25 ng/ml of Stx2 holotoxin and increasing concentrations of either StxB or BSA. The number of live Tetrahymena cells was determined after 6 h. Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena at the start of incubation. Error bars represent standard deviations from ≥3 independent experiments, with each experiment being comprised of a minimum of three individual measurements.
Figure 4 indicates that in order to interfere with Stx-mediated bacterial killing, an excess of StxB over the amount of Stx holotoxin must be added to the cultures. We do not yet know the nature or number of Stx holotoxin receptor/trafficking sites within T. thermophila. The requirement for “excess” StxB to block killing indicates that there many such sites, although the observation that the StxB inhibitory effect is saturable suggests that the number of sites is not infinite. Consistent with our findings, excess B subunit is also needed to block Stx holotoxin-mediated killing of mammalian cells (20, 30). Addition of substoichiometric ratios of Stx holotoxin to Gb3 receptor is sufficient to kill mammalian cells (35), and thus excess StxB subunit is needed to saturate the receptors to prevent killing. We speculate that a similar situation occurs with T. thermophila. Nonetheless, regardless of the precise mechanism by which StxB blocks Stx-mediated T. thermophila killing, the data in Fig. 4 indicate that the B subunit has a role in mediating cytotoxicity by Stx holotoxin.
The results in Fig. 4 indicate that the B subunit mediates the cytotoxicity of added purified Stx. To determine whether the B subunit also plays a role in the cytolethal effect of bacteria bearing Stx-encoding bacteriophages (Fig. 1 and 2), we cocultured T. thermophila with EDL933Δstx bearing either of two plasmids, one encoding Stx2 holotoxin and the other encoding only the Stx2A subunit, and measured the growth kinetics of T. thermophila and bacteria. When cocultured with EDL933Δstx bearing the Stx holotoxin-encoding plasmid (EDL933Δstx/pStxAB), the number T. thermophila cells decreases ∼3-fold over a 6-h incubation. The efficiency of T. thermophila killing is slightly larger than that seen in cocultures with EDL933, presumably due to the increased production of Stx holotoxin from the plasmid. Consistent with the results presented in Fig. 1 and 2 and our proposal that Stx acts as an antipredator agent, the number of bacterial cells bearing pStxAB increases in the presence of T. thermophila.
In contrast, T. thermophila are not killed when cocultured with EDL933Δstx bearing a plasmid that encodes only the Stx2A subunit (EDL933Δstx/pStxA). Instead, this stxA+ strain supports T. thermophila growth to a similar extent as do non-Stx encoding bacteria (Fig. 1). Also, the number of EDL933Δstx/pStxA cells decreases in the presence of T. thermophila, showing that these cells are more efficiently predated than are the stx+ EDL933Δstx/pStxAB cells. Control immunoblots indicate that identical amounts of StxA subunit are produced by the EDL933Δstx/pStxA and EDL933Δstx/pStxAB strains. These findings indicate that the B subunit is involved in killing of T. thermophila mediated by Stx-encoding bacteria and is essential to its potential role as an antipredator defense molecule.
Shiga toxin is not produced by EDL933 as long as the resident toxin-encoding bacteriophages remain in their lysogenic state. Lytic induction of lysogenic bacteriophages occurs upon inactivation of the phage repressor (53). The best-studied mechanism of repressor inactivation involves RecA-stimulated repressor autoproteolysis (9, 37, 46) that occurs during the host's SOS response to DNA damage. Fuchs et al. (16) showed that 933r, an otherwise isogenic recA mutant of EDL933, has severely reduced virulence in mice. Hence, we used 933r to examine whether the bacterial SOS response plays a similar role in mediating T. thermophila killing. In contrast to the results with wild-type EDL933, when T. thermophila is fed 933r, the number of T. thermophila cells remains unchanged. This finding indicates that recA mutation blocks the bacterially mediated killing of T. thermophila.
The form of RecA that stimulates repressor autocleavage arises as a consequence of DNA damage (15). Such damage can be caused by numerous agents, including natural products (e.g., microbially produced antibiotics) and reactive oxygen species (ROS). We hypothesized that ROS released by T. thermophila (14, 39) may indicate the presence of this predator to EDL933 cells. The ROS would be expected to activate the bacterial SOS response, causing induction of the Stx-encoding prophages in these cells, leading to Stx production and release.
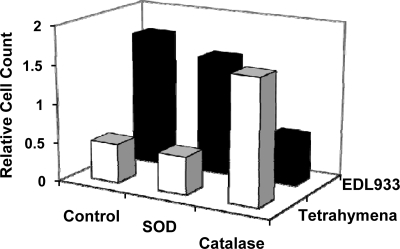
We tested this idea by coculturing EDL933 and T. thermophila for 2 h in the absence or presence of either superoxide dismutase or catalase, enzymes that degrade superoxide or H2O2, respectively. As found previously, the number of T. thermophila cells decreases over time in the absence of any antioxidant enzymes, while the number of EDL933 cells increases (Fig. 5). We obtained identical results with cocultures containing superoxide dismutase, suggesting that this enzyme does not influence toxin production by EDL933. In contrast, the number of T. thermophila cells increased when these cells were cocultured in the presence of catalase, while the number of EDL933 cells decreased (Fig. 5). Therefore, the EDL933/T. thermophila cocultures with catalase behaved identically to cocultures consisting of T. thermophila and the control bacterium, W3110::λ. This finding suggests that catalase blocks the release of Shiga toxin by EDL933 by removing H2O2. This observation is consistent with the finding that the bacterial SOS response mediates enhanced Shiga toxin production by EDL933, leading to increased T. thermophila killing.
FIG. 5.
Effect of antioxidant enzymes on the survival of EDL933 and Tetrahymena in cocultures. Cocultures containing 105 Tetrahymena cells/ml and 108 bacterial cells/ml were grown in the absence or presence of 4 μg/ml of either superoxide dismutase (SOD) or catalase, as indicated. Tetrahymena cell counts are given as the number of live cells remaining at end of the 6-h incubation relative to number of cells present at start of incubation.
DISCUSSION
The data presented here clearly show that Shiga toxin, either released from an induced lysogenic bacteriophage or added exogenously, is capable of killing the single-cell eukaryote Tetrahymena thermophila. To our knowledge, this is the first direct demonstration of Shiga toxin lethality in any protozoan or other bacterial predator. Our results also show that the presence of a Shiga toxin-encoding gene within bacteria decreases T. thermophila predation efficiency. Our findings are consistent with the results of a previous investigation showing that in the presence of Tetrahymena species, bacterial exotoxins augment the fitness of bacterial populations that carry them (49). Together with these previous findings (49), our results strongly suggest that Stx can function as an antipredator defense and support the hypothesis that this toxin may have originated in bacteria as an antipredator adaptation.
There is a growing body of evidence that predation by bacterivorous protozoa can play a large role in shaping the composition of bacterial populations (24, 25, 48, 55). The cytotoxic effect of a bacterial exotoxin on a protozoan predator indicates that these molecules can have important ecological functions within natural microbial communities. Therefore, future studies with T. thermophila can provide information about the evolution and function of bacteriotoxins in ciliate predators.
Two of our findings suggest that various aspects of the interaction between Shiga toxin-encoding bacteria and mammalian cells have an ancient evolutionary origin in the predator-prey interaction between these bacteria and protozoans. First, the finding that EDL933 bacteria appear to sense the presence of the T. thermophila by detecting the presence of ROS and respond by inducing the synthesis and release of Stx may represent a foreshadowing of the situation found in the mammalian response to bacterial infection. Consistent with the findings of others (14, 39), we have observed that T. thermophila releases H2O2 into the medium (data not shown). Similarly, ROS such as H2O2 or superoxide, generated and released by leukocytes and neutrophils, activate the SOS response in Shiga toxin-encoding E. coli, leading to toxin release (51) and subsequent death of the “attacking” eukaryotic cell. Since H2O2 is present in internal vesicles of Tetrahymena species (14), it is also possible that bacteriophage induction occurs inside the cells after the bacteria are eaten.
Second, similar to its role in mammalian cell killing, we found that the B subunit plays an essential role in Stx-mediated T. thermophila killing. In mammalian systems, Shiga toxin is produced outside of the cell as a consequence of lytic growth of Stx-encoding bacteriophage. Stx entry into mammalian cells occurs by receptor-mediated endocytosis, which requires the binding of the holotoxin's B subunit to the neutral glycolipid Gb3 receptor. Mammalian cells lacking this glycolipid are immune to the toxin (7). Tetrahymena has all of the machinery for clathrin-mediated endocytosis (12, 21). Therefore, Tetrahymena could use a clathrin-dependent receptor-mediated endocytosis mechanism to import Shiga toxin. However, exhaustive studies performed in our labs and in the laboratories of Clifford Lingwood at the University of Toronto (personal communication) failed to identify any Stx binding lipids in the glycolipid-containing fraction. Moreover, inspection of the T. thermophila genome sequence (8, 10) failed to identify any close homologues of the gene that encodes the mammalian Gb3 synthase enzyme (26). These observations are inconsistent with Stx entry into Tetrahymena occurring by “standard” Gb3 receptor-mediated endocytosis. Nonetheless, our findings showing that excess B subunit blocks the killing of T. thermophila by purified Stx2 holotoxin (Fig. 4) and that expression of the B subunit is required for T. thermophila intoxication suggest that alternative Stx receptors could be present in these cells.
Tetrahymena cells can ingest particulate and soluble nutrients through the oral apparatus and funnel them into their food vacuoles. Hence, in the absence of a Shiga toxin receptor, uptake of either released Stx or Stx-encoding bacteria via Tetrahymena's oral cavity might provide an alternative route or the sole route for toxin entry into these cells. This ability to capture bacteria through their oral apparatus could provide a novel route for toxin entry into Tetrahymena cells. If this idea is correct, it suggests that the B subunit affects Shiga toxin cytotoxicity in Tetrahymena by influencing intracellular trafficking.
Considering that the lysogenic bacterial host must die to release the Shiga toxin, the fitness benefits of this antipredator defense mechanism cannot accrue to the individual organism but would be to the overall bacterial population, a population that would include cells that are not lysogenic for the toxin-encoding bacteriophage. At first glance this strategy may seem overly “altruistic.” However, the genes encoding this exotoxin are found on mobile, temperate bacteriophages. Due to the high propensity to grow lytically (28), the phage has limited ability to lysogenize naïve hosts. The propensity of the phage to lytically infect, and thereby kill, the naïve hosts would restrict the benefit of Shiga toxin antipredator activities for the fitness of this segment of the bacterial population. It should be noted that lytic growth in the naïve hosts would increase the amount of Stx produced, amplifying the killing capacity of the original sacrificed cell. The ability of nonlysogens to enhance Stx production by a bacterial population has already been demonstrated (17).
Hence, the presence of toxins genes, in this case Shiga toxin genes, in the temperate phages and linkage of toxin expression to lytic growth may provide substantial advantages to bacterial lysogens that “choose” to harbor these phages. Utilization of an exotoxin encoded on an inducible phage is then a cost-effective defensive strategy for the bacterial population. These lysogenic populations sacrifice a few cells to produce a toxin that kills its major predator and produce infectious phage that have the potential to eliminate bacterial competitors (53).
Acknowledgments
We thank the members of our laboratories for valuable discussions. We also acknowledge Todd Hennessey for invaluable contributions of material, assistance, and advice.
G.B.K. acknowledges the support of the College of Arts and Sciences, University at Buffalo.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Acheson, D. W., S. B. Calderwood, S. A. Boyko, L. L. Lincicome, A. V. Kane, A. Donohue-Rolfe, and G. T. Keusch. 1993. Comparison of Shiga-like toxin I B-subunit expression and localization in Escherichia coli and Vibrio cholerae by using Trc- or iron-regulated promoter systems. Infect. Immun. 61:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber, W., L. Enquist, B. Hohn, N. E. Murray, K. Murray, R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg. 1983. Lambda II, p. 433-466. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 3.Berninger, U., B. J. Finlay, and P. Kuuppo-Leinikk. 1991. Protozoan control of bacterial abundances in freshwater. Limnol. Oceanogr. 36:139-147. [Google Scholar]
- 4.Brussow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterton, J. R., E. T. Ryan, D. W. Acheson, and S. B. Calderwood. 1997. Coexpression of the B subunit of Shiga toxin 1 and EaeA from enterohemorrhagic Escherichia coli in Vibrio cholerae vaccine strains. Infect. Immun. 65:2127-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casas, V., J. Miyake, H. Balsley, J. Roark, S. Telles, S. Leeds, I. Zurita, M. Breitbart, D. Bartlett, F. Azam, and F. Rohwer. 2006. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in Southern California. FEMS Microbiol. Lett. 261:141-149. [DOI] [PubMed] [Google Scholar]
- 7.Cilmi, S. A., B. J. Karalius, W. Choy, R. N. Smith, and J. R. Butterton. 2006. Fabry disease in mice protects against lethal disease caused by Shiga toxin-expressing enterohemorrhagic Escherichia coli. J. Infect. Dis. 194:1135-1140. [DOI] [PubMed] [Google Scholar]
- 8.Coyne, R. S., M. Thiagarajan, K. M. Jones, J. R. Wortman, L. J. Tallon, B. J. Haas, D. M. Cassidy-Hanley, E. A. Wiley, J. J. Smith, K. Collins, S. R. Lee, M. T. Couvillion, Y. Liu, J. Garg, R. E. Pearlman, E. P. Hamilton, E. Orias, J. A. Eisen, and B. A. Methe. 2008. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeAnda, J., A. R. Poteete, and R. T. Sauer. 1983. P22 c2 repressor-domain structure and function. J. Biol. Chem. 258:10536-10542. [PubMed] [Google Scholar]
- 10.Eisen, J. A., R. S. Coyne, M. Wu, D. Wu, M. Thiagarajan, J. R. Wortman, J. H. Badger, Q. Ren, P. Amedeo, K. M. Jones, L. J. Tallon, A. L. Delcher, S. L. Salzberg, J. C. Silva, B. J. Haas, W. H. Majoros, M. Farzad, J. M. Carlton, R. K. Smith, Jr., J. Garg, R. E. Pearlman, K. M. Karrer, L. Sun, G. Manning, N. C. Elde, A. P. Turkewitz, D. J. Asai, D. E. Wilkes, Y. Wang, H. Cai, K. Collins, B. A. Stewart, S. R. Lee, K. Wilamowska, Z. Weinberg, W. L. Ruzzo, D. Wloga, J. Gaertig, J. Frankel, C. C. Tsao, M. A. Gorovsky, P. J. Keeling, R. F. Waller, N. J. Patron, J. M. Cherry, N. A. Stover, C. J. Krieger, C. del Toro, H. F. Ryder, S. C. Williamson, R. A. Barbeau, E. P. Hamilton, and E. Orias. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenmann, H., H. Harms, R. Meckenstock, E. I. Meyer, and A. J. Zehnder. 1998. Grazing of a Tetrahymena sp. on adhered bacteria in percolated columns monitored by in situ hybridization with fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 64:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elde, N. C., G. Morgan, M. Winey, L. Sperling, and A. P. Turkewitz. 2005. Elucidation of clathrin-mediated endocytosis in Tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 1:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenchel, T. 1987. Ecology of protozoa: the biology of free-living phagotrophic protists. Science Tech Publishers, Madison, WI.
- 14.Fok, A. K., and R. D. Allen. 1975. Cytochemical localization of peroxisomes in Tetrahymena pyriformis. J. Histochem Cytochem. 23:599-606. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 16.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 17.Gamage, S. D., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobert, A. P., M. Vareille, A.-L. Glasser, T. Hindre, T. de Sablet, and C. Martin. 2007. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-κB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 178:8168-8174. [DOI] [PubMed] [Google Scholar]
- 19.Herold, S., H. Karch, and H. Schmidt. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115-121. [DOI] [PubMed] [Google Scholar]
- 20.Hurley, B. P., M. Jacewicz, C. M. Thorpe, L. L. Lincicome, A. J. King, G. T. Keusch, and D. W. K. Acheson. 1999. Shiga Toxins 1 and 2 translocate differently across polarized intestinal epithelial cells. Infect. Immun. 67:6670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs, M. E., L. V. DeSouza, H. Samaranayake, R. E. Pearlman, K. W. M. Siu, and L. A. Klobutcher. 2006. The Tetrahymena thermophila phagosome proteome. Eukaryot. Cell 5:1990-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannes, L., and D. Decaudin. 2005. Protein toxins: intracellular trafficking for targeted therapy. Gene Ther. 12:1360-1368. [DOI] [PubMed] [Google Scholar]
- 23.Jost, J. L., J. F. Drake, A. G. Fredrickson, and H. M. Tsuchiya. 1973. Interactions of Tetrahymena pyriformis, Escherichia coli, Azotobacter vinelandii, and glucose in a minimal medium. J. Bacteriol. 113:834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurgens, K., and C. Matz. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie van Leeuwenhoek. 81:413-434. [DOI] [PubMed] [Google Scholar]
- 25.Jurgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keusch, J. J., S. M. Manzella, K. A. Nyame, R. D. Cummings, and J. U. Baenziger. 2000. Cloning of Gb3 synthase, the key enzyme in globo-series glycosphingolipid synthesis, predicts a family of alpha 1, 4-glycosyltransferases conserved in plants, insects, and mammals. J. Biol. Chem. 275:25315-25321. [DOI] [PubMed] [Google Scholar]
- 27.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691-1704. [DOI] [PubMed] [Google Scholar]
- 29.Mallard, F., C. Antony, D. Tenza, J. Salamero, B. Goud, and L. Johannes. 1998. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of Shiga toxin B-fragment transport. J. Cell Biol. 143:973-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcato, P., G. Mulvey, and G. D. Armstrong. 2002. Cloned Shiga toxin 2 B subunit induces apoptosis in Ramos Burkitt's lymphoma B cells. Infect. Immun. 70:1279-1286. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 32.Newland, J. W., N. A. Strockbine, S. F. Miller, A. D. O'Brien, and R. K. Holmes. 1985. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science 230:179-181. [DOI] [PubMed] [Google Scholar]
- 33.Noda, M., T. Yutsudo, N. Nakabayashi, T. Hirayama, and Y. Takeda. 1987. Purification and some properties of Shiga-like toxin from Escherichia coli O157:H7 that is immunologically identical to Shiga toxin. Microb. Pathog. 2:339-349. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 35.Obrig, T. G., C. B. Louise, C. A. Lingwood, B. Boyd, L. Barley-Maloney, and T. O. Daniel. 1993. Endothelial heterogeneity in Shiga toxin receptors and responses. J. Biol. Chem. 268:15484-15488. [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Phizicky, E. M., and J. W. Roberts. 1980. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J. Mol. Biol. 139:319-328. [DOI] [PubMed] [Google Scholar]
- 38.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rico, D., A. Martín-González, S. Díaz, P. de Lucas, and J.-C. Gutiérrez. 2009. Heavy metals generate reactive oxygen species in terrestrial and aquatic ciliated protozoa. Comp. Biochem. Physiol. C 149:90-96. [DOI] [PubMed] [Google Scholar]
- 40.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia-Coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 41.Sandvig, K. 2001. Shiga toxins. Toxicon 39:1629-1635. [DOI] [PubMed] [Google Scholar]
- 42.Sandvig, K., S. Grimmer, S. U. Lauvrak, M. L. Torgersen, G. Skretting, B. van Deurs, and T. G. Iversen. 2002. Pathways followed by ricin and Shiga toxin into cells. Histochem. Cell Biol. 117:131-141. [DOI] [PubMed] [Google Scholar]
- 43.Sandvig, K., S. Olsnes, J. E. Brown, O. W. Petersen, and B. van Deurs. 1989. Endocytosis from coated pits of Shiga toxin: a glycolipid-binding protein from Shigella dysenteriae 1. J. Cell Biol. 108:1331-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandvig, K., and B. van Deurs. 2005. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther. 12:865-872. [DOI] [PubMed] [Google Scholar]
- 45.Sandvig, K., and B. van Deurs. 2000. Entry of ricin and Shiga toxin into cells: molecular mechanisms and medical perspectives. EMBO J. 19:5943-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer, R. T., H. C. Nelson, K. Hehir, M. H. Hecht, F. S. Gimble, J. DeAnda, and A. R. Poteete. 1983. The lambda and P22 phage repressors. J. Biomol. Struct. Dyn. 1:1011-1022. [DOI] [PubMed] [Google Scholar]
- 47.Sherr, B. F., and E. B. Sherr. 1984. Role of heterotrophic protozoa in carbon and energy flow in aquatic ecosystems, p. 412-423. In M. J. Klug and C. A. Reddy, (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, DC.
- 48.Simek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinberg, K. M., and B. R. Levin. 2007. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. Biol. Sci. 274:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vilhena-Costa, A. B., R. M. F. Piazza, J. M. Nara, L. R. Trabulsi, and M. B. Martinez. 2006. Slot blot immunoassay as a tool for plasmid-encoded toxin detection in enteroaggregative Escherichia coli culture supernatants. Diagn. Microbiol. Infect. Dis. 55:101-106. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, P. L., D. W. K. Acheson, and M. K. Waldor. 2001. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69:1934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldor, M. K., and D. I. Friedman. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459-465. [DOI] [PubMed] [Google Scholar]
- 55.Wildschutte, H., D. M. Wolfe, A. Tamewitz, and J. G. Lawrence. 2004. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. USA 101:10644-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, M., and D. B. Haslam. 2005. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]