Abstract
We have generated extreme ionizing radiation resistance in a relatively sensitive bacterial species, Escherichia coli, by directed evolution. Four populations of Escherichia coli K-12 were derived independently from strain MG1655, with each specifically adapted to survive exposure to high doses of ionizing radiation. D37 values for strains isolated from two of the populations approached that exhibited by Deinococcus radiodurans. Complete genomic sequencing was carried out on nine purified strains derived from these populations. Clear mutational patterns were observed that both pointed to key underlying mechanisms and guided further characterization of the strains. In these evolved populations, passive genomic protection is not in evidence. Instead, enhanced recombinational DNA repair makes a prominent but probably not exclusive contribution to genome reconstitution. Multiple genes, multiple alleles of some genes, multiple mechanisms, and multiple evolutionary pathways all play a role in the evolutionary acquisition of extreme radiation resistance. Several mutations in the recA gene and a deletion of the e14 prophage both demonstrably contribute to and partially explain the new phenotype. Mutations in additional components of the bacterial recombinational repair system and the replication restart primosome are also prominent, as are mutations in genes involved in cell division, protein turnover, and glutamate transport. At least some evolutionary pathways to extreme radiation resistance are constrained by the temporally ordered appearance of specific alleles.
A survey of bacteria and archaea identifies 11 phyla that contain species with unusually high resistance to the lethal effects of ionizing radiation (IR). These phyla are not closely related to each other and do not share a common lineage, and all include genera that are considered radiosensitive (9). The existence of so many unrelated and isolated radioresistant species in the phylogenetic tree argues that the molecular mechanisms that protect against IR-induced damage evolved independently in these organisms, suggesting that at least some species have the capacity to acquire radioresistance through evolutionary processes if they are subjected to appropriate selective pressure.
The first of these species to be discovered, and the best studied to date, is the bacterium Deinococcus radiodurans. The molecular basis of the extraordinary radioresistance of Deinococcus has not been elucidated, but well-constructed proposals abound. Radioresistance has variously been attributed to the condensed structure of the nucleoid (29, 40, 56), the elevated levels of Mn ion present in the cytosol as a mechanism to control protein oxidation (11, 12), a specialized RecA-independent DNA repair process (54), and other species attributes (9). Radioresistance in Deinococcus is probably mechanistically related to desiccation resistance derived from evolution in arid environments (37, 45), although this may not be the origin of the phenotype in all relevant species (9).
An understanding of the genetic underpinnings of bacterial radiation resistance holds promise for yielding insights into the mechanistic basis of radiation toxicity, along with the potential for new approaches to facilitate recovery from radiation injury in other organisms, including humans. To better define the genetic, biochemical, and physiological characteristics most important for radioresistance, we employed a strategy to allow the cells to inform us. In brief, we generated radioresistant variants of radiosensitive bacteria and defined the genetic changes underlying the new phenotype.
In 1946, Evelyn Witkin established that it was possible to increase the resistance of Escherichia coli B to DNA damage (50). She exposed cultures to high doses of UV light, killing most of the population and selecting for variants better able to tolerate UV. In the 6 decades since the Witkin report, additional investigators have repeated this result, demonstrating that iterative cycles of high-dose exposure to a DNA damaging agent can heritably enhance a culture's ability to tolerate that DNA damaging agent. Increases in IR resistance have been reported for E. coli (17), Salmonella enterica serovar Typhimurium (14), and Bacillus pumulis (44), organisms that are otherwise considered radiosensitive. Davies and Sinskey (14) showed that for S. enterica serovar Typhimurium LT2, the number of cycles of exposure and recovery correlates with the level of radioresistance achieved. After 84 cycles, they generated a strain displaying inactivation kinetics similar to that of Deinococcus radiodurans, with a D10 value (the dose needed to inactivate 90% of the population) 200-fold higher than that of the parental strain.
For this study, we expanded on these earlier studies by independently generating four IR-resistant populations of Escherichia coli K-12 MG1655 (4). Our effort included an important innovation relative to the earlier studies—we characterized the evolved populations with an experimental program that included the complete genomic resequencing of multiple strains purified from three of the populations, taking advantage of new sequencing technologies. The result is an increasingly detailed data set—based on a single robust model system—that allows us to (i) explore the molecular basis of radiation resistance in bacteria and (ii) test current hypotheses and search for novel mechanisms of radiation resistance.
MATERIALS AND METHODS
Irradiation-outgrowth protocol.
Ten-milliliter cultures of E. coli strain MG1655 were grown to stationary phase in LB broth with aeration at 37°C. Two 1-milliliter samples were removed, one was archived at −80°C, and the other was transferred to a 1.5-ml plastic tube before exposure to IR (60Co source from a Shepherd model 484 irradiator; 19 Gy/min). After irradiation, an aliquot was removed and serially diluted in physiological 0.9% saline, and dilutions were spotted onto a plate to estimate survival. The balance of the irradiated culture was used to inoculate fresh LB broth. Survivors were allowed to grow to stationary phase (12 to 18 h), and the protocol was repeated. The dose of radiation administered was adjusted to allow approximately 1% survival after 1 day at 37°C, with the dose increasing as radioresistance increased.
Survival curves.
Cultures grown to stationary phase in LB broth (at 37°C) were harvested by centrifugation, and cells were resuspended in physiological saline. The cell suspensions were exposed to the 60Co source until the desired dose was achieved. Irradiated cultures were diluted in physiological saline and plated immediately on LB agar. The surviving fraction was calculated by determining the titer of the surviving population from the number of CFU that appeared 1 day after irradiation and dividing that number by the titer of the unirradiated culture.
Comparative genome sequencing (CGS).
DNA samples were analyzed using the Comparative Genome Sequencing Service of NimbleGen Systems, Inc., as described earlier (1, 32). Briefly, custom “mapping” microarrays were designed to tile the genome of E. coli MG1655 with 29-base oligonucleotides, with a 23-base overlap between sequential oligonucleotides and specific to each DNA strand. Genomic DNA from the radiation-sensitive E. coli “reference” strain was labeled with Cy5, and DNA from each radiation-resistant “test” strain was labeled with Cy3. Five micrograms of each test sample was mixed with an equal amount of reference and competitively hybridized to microarrays. Ratios of hybridization intensities (reference/test intensities) were calculated and plotted versus genome position to give a high-resolution map of possible mutation sites that localized each mutation to a window of 29 bases (length of reporting probe). Likely sites of mutation were identified using a custom algorithm based on hybridization intensity ratios significantly above background and with good concordance of results from corresponding probes from both strands. Possible sites of mutation were chosen for resequencing by hybridization to a custom microarray designed specifically in response to data from the initial mapping microarrays. Array-based resequencing exploits differential hybridization of genomic DNA to short perfect-match (PM) versus mismatch oligonucleotides. Each nucleotide to be queried is located near the central position of a short oligonucleotide probe. For each PM oligonucleotide, probes with the three possible mismatch nucleotides at the same central position are also synthesized on the array. Probe lengths and mismatch positions are varied based on calculated melting temperatures. The differences in hybridization signal intensities between sequences that bind strongly to the PM oligonucleotides and those that bind poorly to the corresponding mismatch oligonucleotides allow the correct base at a given sequence position to be discerned accurately and reliably. The arrays designed to resequence each resistant E. coli line contained 392,000 probes, with 4 probes per base position for each strand. Probes for both forward and reverse strands were included, and 48,600 total bases were resequenced.
Direct DNA sequencing.
Sequencing was performed with Big Dye technology (ABI), using the protocols of the University of Wisconsin Biotechnology Center, after PCR amplification of the target genomic DNA sequence.
High-throughput sequencing using an Illumina instrument.
Evolved strains CB1000 and CB2000 were sequenced using an Illumina genome analyzer. CB1000 was sequenced in a single run (lane 1) that produced ∼7.6 million 36-bp reads. CB2000 was sequenced in two independent runs (lanes 2 and 3), which produced ∼10.3 and ∼6.1 million reads, respectively. Estimation of the level of sequencing error using Illumina's Gerald software revealed that the raw sequence data had an extremely high quality, and therefore assemblies were performed using raw reads without quality scores.
Two different approaches were used to generate assemblies of each template, as described below. One approach used the SeqMan whole-genome assembler (DNASTAR, Inc.), which performs templated assembly whereby sequence reads are first mapped to the reference genome and then assembled into contigs by looking for overlaps between neighbors. In a second round, an attempt was made to include unmapped reads by looking for overlaps to contigs created in the first stage, with no regard to the reference genome. After assembly of the genome, the program SeqManPro was used to generate consensus sequences from the contigs and to identify sequence differences between the consensus and the reference genome. For strain CB2000, which was sequenced in duplicate lanes, the SeqMan approach was used only for the lane that produced the smallest number of reads. The second approach employed the de novo assembler Velvet (55), which generated contigs that were then mapped to the reference genome by use of the contig-reordering functionality of the MAUVE genome alignment program (13). In the case of CB2000, pooling of the duplicate data sets produced a higher level of coverage, with fewer and generally longer contigs, than that obtained by treating either of the two lanes separately with the same assembler. After reordering of the Velvet-based assemblies, they were combined together with the corresponding assembly produced by SeqMan and the reference genome to construct three-way multiple genome alignments with MAUVE. This provided a way to assess whether a reported change might be an artifact of either assembly process.
Although both approaches resulted in assemblies that covered over 95% of the genome, the SeqMan assemblies covered slightly more of the genome (see Results). It should be noted that neither of the programs used for genome assembly is able to reliably include repeated regions, such as insertion elements and rRNA genes, and the differences in coverage may arise from the way that the two programs deal with repeated sequences within the genome. Velvet, which has the advantage of being extremely quick, produced 10-fold more contigs than those generated using the SeqMan assembler. This may be due in part to the difference between a de novo assembly and one that is based on comparison to a reference genome. However, after mapping of the Velvet contigs to the reference sequence and examination of the sequences at the junctions between contigs within MAUVE, it was found that the 3′ ends of contigs were typically duplicated at the 5′ ends of the downstream neighbors, with the duplicated sequence terminated by a single mismatch. This suggests that the greater number of contigs may have been caused by Velvet's attempt to stringently deal with ambiguous base calls in areas of low coverage. After reordering of the Velvet-based assemblies, custom-written Perl scripts were used in an attempt to reduce the number of contigs by merging adjacent and overlapping contigs that remained unjoined by Velvet, presumably due to low coverage or ambiguity. This process led to a >2-fold reduction in the number of contigs in each of the two assemblies.
CB1012, CB1013, CB1014, CB1015, CB1024, and CB1025 were similarly sequenced with Illumina genome analyzers at the DOE Joint Genome Institute, Walnut Creek, CA.
Preparation of cell cultures for protein and nucleotide determinations.
Overnight cultures of the E. coli MG1655 founder strain and strains CB1000, CB2000, and CB3000 were grown in 10 ml of LB at 37°C. The cultures were diluted 1:100 in 350 ml fresh LB in 2-liter flasks and grown to early log phase at 37°C (∼1 × 108 CFU/ml, or an optical density at 600 nm [OD600] of ∼0.2). The cultures were placed on ice, and 1 ml was removed to determine the cell count. The rest of the culture was harvested in 100-ml aliquots by centrifugation at 8,000 rpm for 10 min at 4°C, using a Beckman JA16.250 rotor and an Avanti J-20I centrifuge. The LB was removed from the cell pellets, the cells were washed with 6 ml of ice-cold 10 mM Tris-acetate (80%+; pH 7.5)-10 mM MgSO4, and centrifugation was repeated. Collected and washed cell pellets were frozen in liquid N2 and stored at −80°C.
Cells were counted by serially diluting 1 ml of culture with 10 mM Tris-acetate (80%+; pH 7.5)-10 mM MgSO4 and plating 100 μl on 100- by 15-mm LB-plus-1.5% agar plates. The plates were incubated at 37°C, and colonies were counted the next day. For each dilution, three plates were spread and counted to ensure accuracy.
Preparation of cell extracts for total protein concentration determination.
The total protein concentrations of E. coli MG1655 (founder) and of strains CB1000, CB2000, and CB3000 were determined using the microplate procedure of a BCA protein assay kit from Pierce. Cell pellets were prepared as described above. The cell pellets were removed from storage at −80°C and placed on ice. Pellets were quickly resuspended in 20 mM Tris-Cl, pH 6.8-5% (wt/vol) glycerol-200 mM NaCl-2% sodium dodecyl sulfate (SDS) to a final volume of 2 ml. Samples were incubated on ice for 1 h with occasional mixing by pipetting. Samples were then allowed to reach room temperature, at which point the solutions were mixed and became clear.
Total protein concentration determination.
Standards and samples were analyzed for protein content in the same manner. A standard curve was generated using an albumin standard (Pierce) as directed. Samples were diluted to an appropriate concentration giving an absorbance at 570 nm that fell within the standard curve measurements. Both standards and samples were diluted using 5% (wt/vol) glycerol-200 mM NaCl-2% SDS. The reaction was initiated by mixing the standard or sample with working reagent (prepared as directed) at a ratio of 1:8 to a final volume of 225 μl in a 96-well flat-bottomed microplate (Nunc). Reactions with standards and samples were repeated eight times. The microplates were covered and incubated at 37°C for 30 min and then allowed to cool to room temperature for 5 min. The absorbance at 570 nm was read by a Bio-Tek EXx800 microplate reader. Total protein content in the E. coli strains was determined by using a quadratic line equation fit to the inverse standard curve data and solving for protein content based on the absorbance at 570 nm. The total protein concentration was determined by combining the protein content and cell count data.
Preparation of cell extracts for dNTP concentration determination.
Nucleotides were extracted from the cell pellets described above by a modified dual-extraction procedure (46, 49). The cell pellets were removed from storage at −80°C and placed on ice. The cell pellets were quickly resuspended with 3 ml of cold 60% methanol-1% toluene-0.1 mM Desferal and incubated at −20°C for 4 h, followed by drying under vacuum (using a Jouan RC10.10 SpeedVac). Nucleotides were reextracted in 2 ml of 5% trichloroacetic acid-0.1 mM Desferal by rocking at 4°C for 1 h and then spun at 10,000 × g for 30 min at 4°C. The supernatant was removed, and the acid was neutralized by adding 3 ml of tri-n-octylamine-Freon R11 at a ratio of 215:785 and mixing the sample by inversion. The layers were allowed to separate, and the aqueous top layer was then taken to dryness by lyophilization and the residue was dissolved in 0.25 ml of high-performance liquid chromatography buffer A-buffer B (50-50) (see below). Samples were filtered through a 0.45-μm polyvinylidene difluoride membrane and stored at −80°C for up to 1 week. Known amounts of deoxynucleoside triphosphates (dNTPs) were subjected to the same conditions and compared to a standard curve to determine recovery efficiency. In the case of dCTP, dGTP, dTTP, and dATP, there was 50% recovery.
High-performance liquid chromatography nucleotide analysis.
The chromatographic system consisted of Waters 515 pumps and a Waters 996 photodiode array detector. Standard dNTPs (Amersham) and prepared samples were observed and quantitated at 267 nm. Chromatographic separation of the nucleotides was achieved by using a Varian C18 column (Microsorb-MV 100-5 C18; 250 × 4.6 mm) with a Varian guard column (Metaguard Pursuit 4.6-mm-by-5-μm C18 column). The mobile phase consisted of buffer A (10 mM tetrabutylammonium hydroxide-10 mM KH2PO4-0.25% methanol at pH 7.0) and buffer B (2.8 mM tetrabutylammonium hydroxide-100 mM KH2PO4-30% methanol at pH 5.5). All runs were carried out at a flow rate of 1 ml/min. Separation was achieved with a 60-min linear gradient from 50% buffer A and 50% buffer B to 30% buffer A and 70% buffer B, followed by a wash with 100% buffer B. The column was reequilibrated in 50% buffer A and 50% buffer B for 45 min to achieve reproducible runs. Injections (200 μl each) of bacterial extract or standards were carried out by a Waters 717 autosampler and were performed in duplicate. Peak heights and retention times of unprocessed standards were used to identify dNTP peaks in bacterial extracts for quantitation or in processed standards for determining percent recovery.
Measurements of UV sensitivity and mitomycin C sensitivity.
Measurements of sensitivity to UV and mitomycin C were carried out as described previously (31).
Growth of E. coli cells for metal analysis.
LB medium was prepared in glassware that was treated to minimize metal contamination in the following manner. The flask was soaked in 2 N nitric acid for 12 h, followed by soaking in 1% (vol/vol) nitric acid for 24 h. This was followed by rinsing in high-purity water prior to use. For growth of cells, 2-ml aliquots of LB medium were transferred to sterile 15-ml polypropylene disposable centrifuge tubes (Corning) and inoculated with scrapings from freezer stocks of the founder strain, CB1000, CB2000, CB3000, and CB4000. The cultures were incubated overnight at 37°C with shaking. The next day, 25-ml aliquots of LB medium were transferred to sterile 50-ml polypropylene disposable centrifuge tubes (Corning) and inoculated in duplicate with 0.25 ml of each overnight culture. The cultures were incubated at 37°C with shaking to an OD600 of >0.2. The cultures were chilled for 10 min on ice. Cells (20 ml) were spun down in a clinical centrifuge, and the cell pellets were resuspended in 2 ml fresh LB. Aliquots of 1.2 ml of cells were transferred to treated microcentrifuge tubes prepared by the metal analysis facility and then spun down. The supernatants were removed, and the cell pellets were stored at −20°C. In order to determine the number of cells in each cell pellet, serial 1:10 dilutions were made in LB, using the chilled cultures remaining after 20 ml of cells was removed for centrifugation. Fifty-microliter samples of the 10−5 dilution were spread on LB plates and incubated overnight at 37°C. Colonies were counted, and numbers were adjusted for dilution.
Growth of D. radiodurans cells for metal analysis.
TGYM medium was prepared in glassware that was treated to minimize metal contamination in the following manner. The flask was soaked in 2 N nitric acid for 12 h, followed by soaking in 1% (vol/vol) nitric acid for 24 h. This was followed by rinsing in high-purity water prior to use. For growth of cells, 25-ml aliquots of TGYM medium were transferred to sterile 50-ml polypropylene disposable centrifuge tubes (Corning) and inoculated in duplicate with scrapings from freezer stocks of D. radiodurans. The cultures were incubated for 24 h at 30°C with shaking to an OD600 of 0.2. The cultures were chilled for 10 min on ice. Cells (20 ml) were spun down in a clinical centrifuge, and the cell pellets were resuspended in 2 ml fresh TGYM. Aliquots of 1.2 ml of cells were transferred to treated microcentrifuge tubes prepared by the metal analysis facility and then spun down. The supernatants were removed, and the cell pellets were stored at −20°C. In order to determine the number of cells in each cell pellet, serial 1:10 dilutions were made in TGYM, using the chilled cultures remaining after 20 ml of cells was removed for centrifugation. Fifty-microliter samples of the 10−5 dilution were spread on LB plates and incubated overnight at 37°C. Colonies were counted, and numbers were adjusted for dilution.
Metal analysis.
The bacterial pellets were submitted for analysis in acid-cleaned polypropylene vials and treated at 40°C with ultrapure HNO3. The samples were subsequently diluted to volume for analysis with 2% HNO3. Samples were analyzed in the Trace Element Clean Laboratory at the Wisconsin State Laboratory of Hygiene, using high-resolution inductively coupled plasma mass spectrometry. Each sample was measured twice, and the metal content is reported as μg per pellet of bacteria submitted.
PFGE.
Pulsed-field gel electrophoresis (PFGE) of digested genomic DNAs from the E. coli founder strain and E. coli radioresistant strains CB1000, CB2000, and CB3000 was carried out using a slightly modified protocol from the Centers for Disease Control and Prevention (CDC) for subtyping of E. coli O157:H7 (http://www.cdc.gov/pulsenet/protocols.htm).
Cell growth for PFGE.
Cultures were grown overnight at 37°C with shaking. For each overnight culture, two 125-ml flasks with 50 ml fresh LB were inoculated with a 1:1,000 dilution and grown to early log phase at 37°C (∼1 × 108 CFU/ml, or an OD600 of ∼0.2). An appropriate volume was harvested by centrifugation in a microcentrifuge to pellet 1.35 × 108 cells. The medium was removed from the pellet, and pellets were frozen with liquid N2 and then stored at −80°C. Additionally, 1 ml was removed to determine the cell count.
Irradiation of cultures to determine effects of radiation prior to PFGE.
Both flasks were brought to the McArdle Lab (UW-Madison) Mark I 137Cs irradiator from J. L. Shepherd and Associates. One flask, labeled “mock,” was placed outside the irradiator. The second flask was irradiated. The flux was 7.82 Gy/min. Samples were taken at 60- or 120-min intervals for 4 h. At each time point, 1.5 ml of culture was removed from the irradiated flask. The OD600 was used to determine the appropriate volume to harvest by centrifugation to pellet 1.35 × 108 cells. At the end of 4 h, both flasks were placed on ice, and samples were removed from both the mock and irradiated flasks. Additionally, 1 ml was removed to determine cell survival. When indicated, cells were counted by serially diluting 1 ml of culture with 10 mM Tris-acetate (80%+; pH 7.5)-10 mM MgSO4 and plating 100 μl on 100- by 15-mm LB-plus-1.5% agar plates. The plates were incubated at 37°C, and colonies were counted the next day. For each dilution, three plates were spread and counted to ensure accuracy.
Treatment of samples to follow genome reconstitution.
Aliquots (40 μl) of overnight liquid cultures of the founder strain, CB1000, and CB2000 were used to inoculate 20 ml of LB in 125-ml flasks. Cells were grown at 37°C with shaking at 200 rpm until the culture reached an OD600 of 0.2. Samples (1 ml) were collected prior to irradiation and stored. The remainder of each culture was moved to a 15-ml conical tube and irradiated to 5,000 Gy. The cells were spun down, resuspended in 20 ml fresh LB medium in a 125-ml flask, and allowed to recover at 37°C with shaking at 200 rpm. At 1-hour intervals, a 1-ml sample of each culture was removed, and the OD600 was measured. Based on that measurement, a volume of cells was removed from the culture equivalent to 1 ml of cells with an OD600 of 0.2 (with volumes adjusted up or down depending on the actual OD600 to equate cell masses for each sample), spun down, and frozen at −20°C for subsequent processing.
Preparation of plugs for PFGE.
Beyond the initial collection of cell pellets, the CDC protocol was followed exactly. Briefly, cell pellets (described above) were suspended in 200 μl cell suspension buffer (100 mM Tris-Cl-100 mM EDTA, pH 8.0) and 10 μl of 20-mg/ml predigested proteinase K (Sigma). Immediately thereafter, an equal volume of 60°C prewarmed 1% SDS-1% SeaKem Gold agarose (Cambrex) was added and mixed. Aliquots were quickly dispersed into disposable PFGE plug molds (Bio-Rad) and allowed to solidify at 4°C for 10 min (four plugs per sample). The plugs were removed and placed in separate 50-ml conical tubes. Each set of plugs was incubated at 54°C, with agitation at 150 rpm, for 2 h in 5 ml of cell lysis buffer (50 mM Tris-Cl-50 mM EDTA, pH 8.0, + 1% Sarkosyl) and 25 μl of 20-mg/ml predigested proteinase K. The plugs were washed by decanting the lysis buffer and adding 15 ml of 50°C distilled H2O, followed by incubation at 50°C, with shaking at 150 rpm, for 15 min. The process was repeated once more with pure H2O and then four times with Tris-EDTA. The plugs were stored in 15 ml of fresh Tris-EDTA at 4°C.
Digestion of genomic DNA in plugs.
Each plug (prepared as described above) was preequilibrated in a separate 1.65-ml Eppendorf tube by being covered with 200 μl 1× buffer from the enzyme manufacturer. After 15 min, the buffer was removed by pipette, and 200 μl of a restriction digestion mix (20 μl 10× buffer, 173 μl distilled H2O, 2 μl 10 mg ml−1 bovine serum albumin, and 5 μl of 10 U μl−1 enzyme) was added. Samples were incubated for at least 2 h at the appropriate enzyme temperature. Afterwards, plugs were washed twice with 0.5× Tris-borate-EDTA (TBE) before being loaded into the gel.
Running PFGE.
We used a Bio-Rad CHEF-DRII apparatus with a pump and chiller. For each lane, the plug was cut in half and placed directly on the 15-well comb. The samples were allowed to air dry for 5 min before pouring of 55°C precooled 1% SeaKem Gold agarose (in 0.5× TBE). The gel was allowed to solidify for 45 min, and then the comb was removed and the wells were filled with additional 1% SeaKem Gold agarose. After 10 min, the gel was removed from the casting stand and placed into a 14°C prechilled 0.5× TBE-filled PFGE chamber. The gel was run at 14°C and 6 V for 19 h, with an initial switch time of 2 s and a final switch time of 60 s. DNA was visualized using ethidium bromide.
RESULTS
Directed evolution of IR resistance in Escherichia coli.
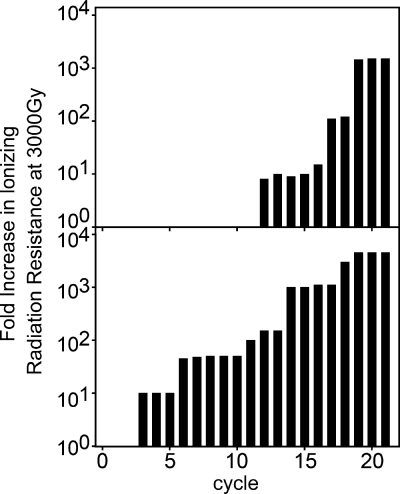
IR-resistant populations of MG1655 were selected by subjecting cultures derived from a single colony isolate (referred to as the founder) to 20 iterative cycles of irradiation and outgrowth. The length of each exposure was adjusted to kill >99% of the population, with this dose increasing from 2,000 Gy for the first cycle to 10,000 Gy on the last cycle. Surviving cells were harvested postirradiation and allowed to recover in rich medium until reaching stationary phase; this population then served as the source of the next culture to be irradiated. As predicted by earlier studies of bacterial evolution (16, 19, 28), the radioresistance of the successively irradiated population increased in a stepwise manner (Fig. 1), suggesting that acquisition of this phenotype requires several genomic alterations. In the first trial, after 20 cycles, an evolved population with enhanced radiation resistance was generated and designated population IR-1-20. (For convenience, the bacterial populations obtained after each cycle are identified by selective agent, trial, and cycle. IR-1-20 identifies the population recovered during our first trial after 20 cycles of exposure to IR and outgrowth.) A survey of 62 purified strains derived from the population (all were single colony isolates generated using procedures designed to avoid the selection of genetically identical strains) exhibited an increase in survival of 1,000- to 5,000-fold when subjected to irradiation at 5,000 Gy; two colonies remained as sensitive as the founder. A radiation-resistant single colony isolate, designated CB1000, was chosen for further analysis. This culture was grown for 100 generations without selection, and the phenotype was maintained, with no measurable loss of IR resistance.
FIG. 1.
Increase in radiation resistance during selection. Cells were subjected to cycles of 60Co irradiation and outgrowth by a protocol described in Materials and Methods (also see the supplemental material). The relative increase in survival at 3,000 Gy is shown as a function of cycle for the trials leading to the isolation of CB1000 and CB2000.
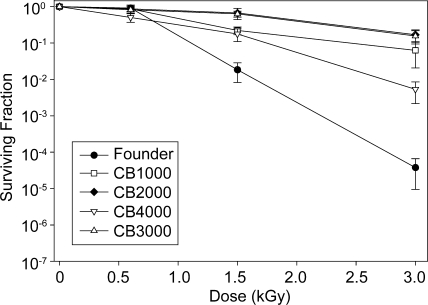
This selection protocol was repeated independently three more times. Each replicate was initiated using an isolated colony derived from a frozen stock of the founder. The outcomes of these additional trials were the same, resulting in the isolation of three more radioresistant populations. These were designated populations IR-2-20, IR-3-20, and IR-4-20. One single colony isolate was taken from each of these populations, generating purified strains designated CB2000, CB3000, and CB4000. The founder, like other E. coli K-12 strains, is quite sensitive to IR; exposure to 3,000 Gy gamma radiation (60Co; 19 Gy/min) reduced the viability 4 orders of magnitude compared to that of the unirradiated culture (Fig. 2). The D37 value for CB1000 was 1,100 Gy, whereas the D37 value for CB2000 and CB3000 was 2,000 Gy—approximately threefold less than the D37 value measured for actively growing cultures of Deinococcus radiodurans R1 (41). The D37 value for the founder was 730 Gy. Higher doses of IR revealed a major improvement in resistance. CB1000, CB2000, and CB3000 exhibited 1,500- to 4,500-fold increases in survival relative to the founder after exposure to 3,000 Gy (Fig. 2). CB4000 was approximately 10-fold less radioresistant than the other isolates.
FIG. 2.
Survival curves for evolved E. coli strains. Survival curves were generated as described in Materials and Methods (also see the supplemental material) for the founder strain and the evolved strains. Results were obtained using a 60Co irradiator (19 Gy/min).
Characterization of evolved strains.
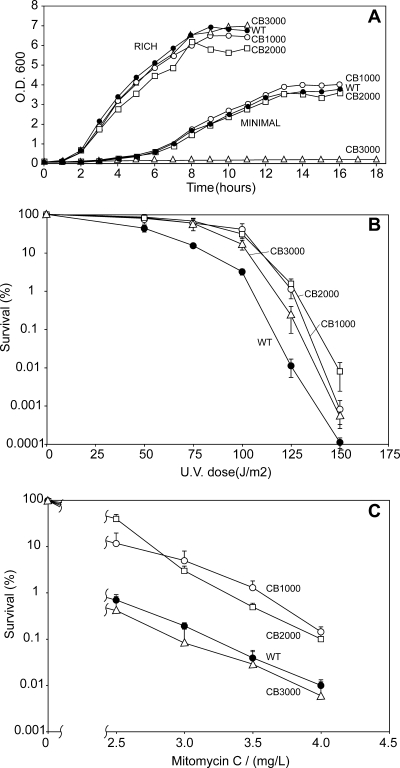
CB1000, CB2000, and CB3000 were studied further. The growth rates of these isolates in rich medium were unaffected by the changes that conferred radiation resistance relative to growth of the founder (Fig. 3A). In minimal medium, CB1000 and CB2000 exhibited growth curves identical to that of the founder (Fig. 3A); CB3000 did not grow in minimal medium due to the presence of a mutation affecting arginine biosynthesis (see below). All three strains exhibited an approximately 10-fold increase in survival after UV irradiation (Fig. 3B) relative to that of the founder. There were modest increases in resistance to mitomycin C for CB1000 and CB2000, while CB3000 exhibited no change in mitomycin C resistance (Fig. 3C).
FIG. 3.
Characterization of radioresistant E. coli. (A) Growth curves. Growth of E. coli strains in rich LB medium and minimal M9 medium is shown. (B) UV survival. Survival curves are shown for E. coli strains exposed to UV radiation. (C) Mitomycin C survival. Survival of E. coli strains exposed to mitomycin C is shown.
Microscopy of DAPI (4′,6-diamidino-2-phenylindole)-stained cells revealed no obvious alterations in morphology, and nucleoid shape did not change (see Fig. S1 in the supplemental material). Cells from Deinococcus radiodurans, stained and visualized using identical methods, reveal a highly condensed, toroid-like nucleoid structure (56).
Daly and colleagues proposed that accumulation of Mn ions in radiation-resistant species facilitates recovery from radiation injury (12), specifically moderating damage to cellular protein components (11). In our trials, there were no significant changes in measured metal content in any of the four radioresistant isolates relative to the founder (see Table S1 in the supplemental material). The measured Mn/Fe ratio was approximately 0.07 for all unirradiated strains. For CB1000 and the founder, there were no significant changes in metal content when the cells were subjected to irradiation at 600 Gy (CB2000, CB3000, and CB4000 were not tested). When cells were examined during exponential growth in rich medium, the DNA concentration per cell, approximately 5 nM, did not differ from that of the founder. The total cellular content of deoxyribonucleotides and protein also did not vary in the three isolates tested (see Table S2 in the supplemental material).
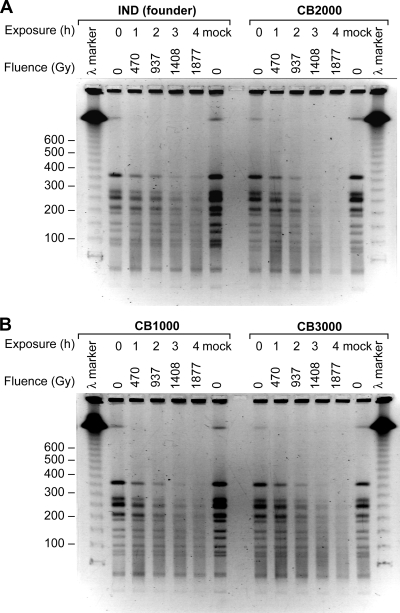
The radiation resistance and genome recovery observed in our isolates could have arisen from passive protection of cellular DNA or from active DNA repair processes. We directly examined the rate of DNA degradation. Strains CB1000, CB2000, and CB3000—with the founder strain used as a control—were irradiated (137Cs [7.8 Gy/min]), and samples were taken after exposure to increasing doses of radiation. The genomic DNA was subjected to digestion with NotI, followed by PFGE. All three of the evolved strains derived from the founder exhibited genomic degradation that was at least as great as that observed for the founder (Fig. 4), indicating the presence of potential new enzymatic processes contributing to genomic degradation in the evolved strains. There are no apparent new mechanisms for the passive protection of genomic DNA in the evolved strains. We examined protein oxidation directly by assaying for protein carbonylation spectrophotometrically and by immunodetection of 2,4-dinitrophenylhydrazine-derivatized protein carbonyls. There were no detectable differences noted between the founder strain, CB1000, CB2000, and CB3000 (data not shown).
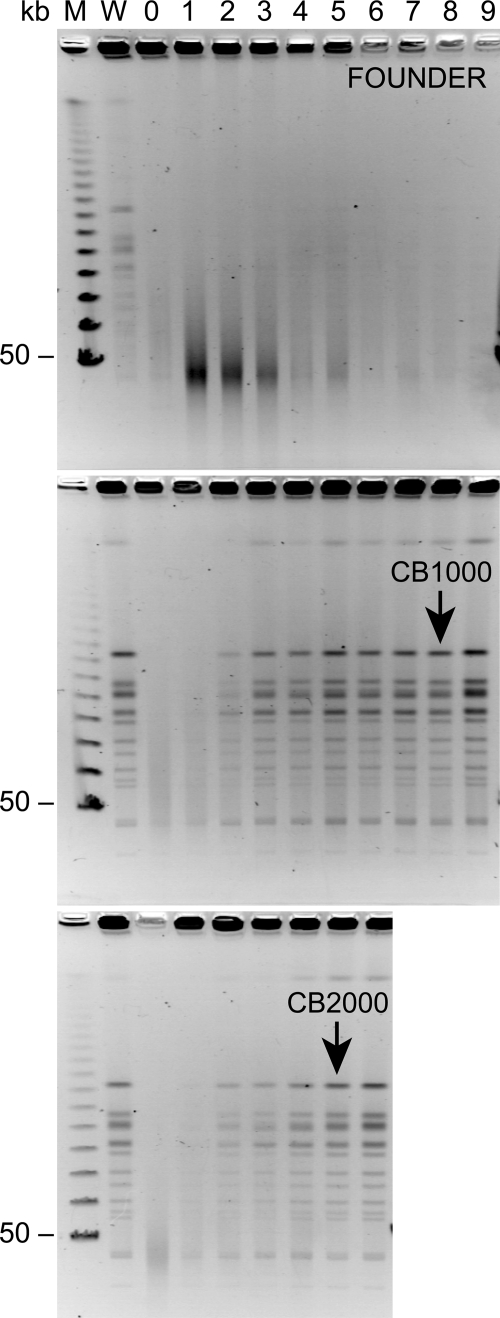
FIG. 4.
DNA degradation due to irradiation. PFGE was performed on DNAs from evolved strains after irradiation at the doses indicated. DNAs were digested with the enzyme NotI prior to electrophoresis.
We also examined the recovery of genomic DNA from CB1000 and CB2000 after exposure to 5,000 Gy of 137Cs (7.8 Gy/min) and compared this with the fate of genomic DNA in the founder strain. The acquired phenotype was evident in this experiment (Fig. 5). The DNA from the founder did not recover after this dose of radiation over a 9-hour time course. Even the fragmented DNA appeared to disappear with time, probably reflecting nuclease degradation. No growth of the irradiated cell culture was evident over a period of 9 h. In contrast, the genomic DNAs from CB1000 and CB2000 were repaired, with the normal NotI banding pattern clearly visible in the pulsed-field gel after 2 hours in both cases. Visible genome restoration appeared to peak after 3 to 4 h. The increase in genomic DNA was not due to growth of undamaged survivors. No increase in bacterial cell mass was evident in the cultures until 8 h and 5 h for CB1000 and CB2000, respectively. These results indicate that the genomic DNA was repaired well before the initiation of normal genome replication and cell division.
FIG. 5.
Genome reconstitution in evolved strains of E. coli. PFGE was used to follow genome reconstitution after irradiation at 5,000 Gy. Samples collected at 1-hour intervals are shown. DNAs were digested with NotI prior to electrophoresis. The black arrows indicate the approximate times when the CB1000 and CB2000 cultures resumed normal growth, as determined by OD measurements. Sample collection was halted 1 hour after the resumption of growth in both cases.
We tested several of the isolated strains for a mutator phenotype. Previous studies with E. coli point to two potentially significant contributors. (i) IR has the potential to introduce mutations through DNA damage, with mutations arising during DNA synthesis on a damaged template by the replicative polymerase or one of several specialized translesion polymerases (5, 20, 51). (ii) E. coli populations harbor individuals with a mutator phenotype, i.e., cells that lack one or more of the components necessary for high-fidelity DNA replication (33, 39). The high spontaneous mutation frequency associated with mutators serves as a source of genetic variation that is acted on during selection. The mutator phenotype eventually reverts, and if the resulting strain carries an adaptive mutation, it survives. CB1000, CB2000, CB3000, and CB4000 were all tested to determine their status as mutators by measuring spontaneous rates of mutation to rifampin (rifampicin) resistance (39). Relative to the founder, no increase in the frequency of Rifr mutations was noted for any of the strains, and the observed rates were well within reported ranges for wild-type (nonmutator) strains of E. coli (E. Wood, unpublished data).
Genetic changes in evolved strains.
Twenty additional isolates were derived independently from population IR-1-20, designated CB1012 to CB1031, and set aside for further analysis. The genomes of the founder, CB1000, CB2000, CB3000, and six of the additional isolates from population IR-1-20 were resequenced using one or both of two complementary methods. First, CGS, a microarray-based technology developed by NimbleGen Systems, Inc. (1), was used to detect mutations in the founder, CB1000, CB2000, and CB3000. Second, direct sequencing using an Illumina genome analyzer was used to sequence the entire genomes of CB1000, CB2000, and the six additional isolates from IR-1-20. Data acquired by CGS were extensively validated (see the supplemental material). Table S3 in the supplemental material summarizes the number of contigs and the amount of the 4,639,675-bp reference genome that was covered by the various assemblies derived from Illumina sequencing. In general, the two methods agreed well. For the strains sequenced using both technologies (CB1000 and CB2000), Illumina sequencing confirmed all mutations that had previously been found by the microarray-based CGS. Genomic alterations missed using CGS (and detected by Illumina sequencing) were on the order of 5 to 10%, similar to the levels reported in other, similar studies (22). All mutations found uniquely in CB1000 and CB2000 by Illumina sequencing were confirmed by direct Sanger sequencing methods. The combination of approaches provides confidence that we have detected most, if not all, of the genomic changes in the sequenced strains.
Genomic alterations detected in all sequenced strains by all methods are summarized in Tables S4 and S5 in the supplemental material (single nucleotide polymorphisms and small [≤3-bp] insertions/deletions are catalogued in Table S4, and larger deletions and insertions are listed in Table S5). The founder strain was independently subjected to CGS. In addition, we searched the founder for any mutations present in all sequenced strains by direct Sanger sequencing. Seven genomic alterations were discovered in the founder relative to the published MG1655 genomic sequence (see Table S6 in the supplemental material). These are not included in Tables S4 and S5 in the supplemental material.
In the overall data set, the seven complete genomic sequences from population IR-1-20 provide both documentation of the mutational diversity in this population and an opportunity to detect mutations that are especially abundant. The sequences of CB2000 and CB3000 allow a comparison to strains derived from populations that evolved independently from IR-1-20.
The total number of genomic alterations in a given evolved strain ranged from 40 to 71. Mutation classes are summarized in Table 1. Between 50 and 60% of the differences were nonsynonymous mutations affecting the sequences of protein or tRNA products. Synonymous mutations are included in the report to complete the picture of the mutational landscape, since some of these may affect gene regulation or processes such as RNA folding or RNA-protein interactions. Base substitutions predominated; 97.5% of the detected genomic alterations fell into this category. Approximately 83% of the base substitutions were either GC→AT or AT→GC transitions (Table 1), consistent with previous reports that detailed the pattern of mutations generated when E. coli K-12 strain AB1157 was irradiated using 60Co (52). A number of insertions and deletions were detected. All of the characterized strains exhibited a deletion of the e14 prophage. This was the only universal genomic alteration (relative to the MG1655 sequence database) not also detected in the founder. CB1000 contains a 3-bp in-frame deletion in the fruB (b2169; included in Table S4 in the supplemental material) coding sequence, and CB3000 carries a 93-bp deletion within argC (b3958; included in Table S5 in the supplemental material) that renders the cell an arginine auxotroph. Evidence of transposition by the IS1 insertion sequences (43) was detected in multiple isolates from population IR-1-20. In CB1000, novel IS1 insertions were found in yaiP (b3063) and fimE (b4313). The fimE gene was also targeted with an IS5 insertion in CB1031 (see Table S5 in the supplemental material).
TABLE 1.
Summary of mutations in evolved E. coli strainsa
| Mutation | No. of mutations
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CB1000 | CB1012 | CB1013 | CB1014 | CB1015 | CB1024 | CB1025 | CB2000 | CB3000 | Combined | |
| Transitions | ||||||||||
| C→T | 19 | 20 | 5 | 10 | 15 | 6 | 9 | 13 | 14 | 111 |
| G→A | 18 | 9 | 11 | 16 | 11 | 11 | 12 | 17 | 17 | 122 |
| T→C | 7 | 7 | 6 | 4 | 6 | 13 | 4 | 11 | 11 | 69 |
| A→G | 15 | 16 | 7 | 8 | 8 | 14 | 11 | 10 | 6 | 95 |
| Transversions | 1 | 1 | ||||||||
| G→T | 4 | 4 | 1 | 1 | 1 | 3 | 5 | 3 | 5 | 27 |
| C→A | 1 | 1 | 2 | 1 | 1 | 5 | 6 | 5 | 2 | 24 |
| A→C | 1 | 1 | 2 | 1 | 1 | 6 | ||||
| T→G | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||
| G→C | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 10 | ||
| C→G | 1 | 2 | 3 | 6 | ||||||
| A→T | 1 | 2 | 1 | 3 | 7 | |||||
| T→A | 1 | 1 | 1 | 1 | 1 | 5 | ||||
| Insertions | 3 | 1 | 4 | |||||||
| 767-bp IS1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | ||
| Deletions | 1 | 1 | ||||||||
| e14 prophage | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| 93-bp deletion | 1 | 1 | ||||||||
| Totals | 71 | 63 | 40 | 46 | 49 | 56 | 53 | 70 | 64 | 503 |
| In genesa | 56 | 51 | 28 | 38 | 39 | 50 | 40 | 57 | 53 | 412 |
| Synonymous | 13 | 9 | 5 | 17 | 13 | 12 | 14 | 16 | 23 | 122 |
| Nonsynonymous | 43 | 42 | 23 | 21 | 26 | 38 | 26 | 41 | 30 | 290 |
| Intergenic | 11 | 10 | 5 | 3 | 7 | 4 | 8 | 11 | 9 | 68 |
Tabulations in the final four rows include all mutations tabulated in Table S4 in the supplemental material, including short insertions and deletions (≤3 bp).
In Table 1, mutations within genes have also been sorted with respect to their effects on the coding sequence, being either synonymous or nonsynonymous. The ratio of the rate of nonsynonymous alterations (dN) to the rate of synonymous changes (dS) is often used as one indicator of selection pressure in a gene, with positive selection implied when dN/dS is >1 (27, 42, 53). Whereas the number of mutations within a single gene is generally small, the overall pattern seen for all mutations in all of the sequenced strains (dN/dS = 2.38) suggests positive selection for resistance to IR. For individual sequenced strains, the dN/dS ratio observed over all genes ranges from 1.24 (CB1014) to 4.67 (CB1012).
Although the sequencing technologies employed have the potential to miss much larger genomic rearrangements that invert or move substantial segments of the genome, we found no evidence for such changes. Genomic DNAs from CB1000, CB2000, and CB3000 were subjected to digestion with either NotI or XbaI, followed by PFGE (see Fig. S2 in the supplemental material). The banding patterns for the radioresistant strains are identical to that of the founder, except that the deletion of the e14 prophage can be seen in the NotI digest. A band of approximately 96 kbp for the founder is reduced in size by the expected 15 kbp corresponding to e14 for the evolved strains.
With the exception of the deletion of the e14 prophage, the mutational landscapes of all sequenced isolates from three different evolved populations are quite distinct. In particular, there are few recurrent mutations evident between the isolates derived from population IR-1-20 and the independently evolved CB2000 and CB3000 strains. These results suggest that there are multiple mechanisms that can contribute to radiation resistance and multiple evolutionary pathways leading to this phenotype. However, some clear patterns also emerge, as described below.
Genetic changes that contribute to IR resistance.
To identify genomic alterations that contribute to the radiation resistance phenotype, we focused on the purified strains derived from population IR-1-20. First, we examined Tables S4 and S5 in the supplemental material to identify genes or processes that represented the most common mutational targets in this evolved population. With this broad coverage, patterns become easier to discern and are likely to be meaningful. In the following discussion, note that in most cases (exceptions are noted later) we do not know if particular mutations inactivate, enhance, or otherwise affect the activity of the gene product. The strains reveal a diverse mutational landscape in which 382 of the detected mutations occur only once. Thus, mutations that are enriched—appearing in multiple sequenced strains from population IR-1-20—are relatively rare. These more common mutations may have been positively selected and thus significant. In some cases, we then isolated individual genetic alterations in otherwise wild-type backgrounds to better assess their individual contributions to IR resistance.
In Table 2, we list the genes where mutations appear in at least three of the seven sequenced isolates derived from IR-1-20 or where a cluster of mutations are evident in genes with closely related functions. In some cases, patterns are reinforced by mutations appearing in the independently evolved strains CB2000 and CB3000. Looking only at the isolates from IR-1-20, a number of prominent mutational targets are evident. Common mutations occur in recA (4/7 isolates, with an additional allele appearing in a fifth strain), ruvB (3/7 isolates, including two alleles), dnaT (4/7 isolates), gltS (4/7 isolates), bglH (3/7 isolates, including two different alleles), yjgL (3/7 isolates, including two alleles), and ylbE (3/7 isolates). Additional clusters of mutations are seen in the ftsW/ftsZ genes and the clpP/clpX genes. The RecA protein is a central player in double-strand-break repair (7, 29). RuvB contributes to the same process (6, 8). DnaT is part of the replication restart primosome, operating downstream of RecA and RuvB in double-strand-break repair (34, 35). The FtsW and FtsZ proteins are involved in cell division (23). The ClpP and ClpX proteins are components of a proteolytic system responsible for the turnover of at least 60 E. coli proteins, including chaperones, proteins involved in energy production, transcriptional regulators, and others (3, 18). GltS is a membrane-bound glutamate transporter (15, 48). BglH is an outer membrane porin for β-glucosides (2). YjgL and YlbE are predicted proteins about which little is known. All of the 19 different mutations detected within the genes featured in Table 2 are nonsynonymous.
TABLE 2.
Mutations common in evolved strains
| Gene and functional category | Locus tag | Nucleotide position of mutation
|
Nucleotide in reference allele | Nucleotide in mutant allele | Change | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CB1000 population
|
CB2000 | CB3000 | |||||||||||
| CB1000 | CB1012 | CB1013 | CB1014 | CB1015 | CB1024 | CB1025 | |||||||
| Recombinational DNA repair | |||||||||||||
| ruvB | b1860 | 1943223 | T | C | D52G | ||||||||
| ruvB | b1860 | 1943323 | 1943323 | C | A | D19Y | |||||||
| recA | b2699 | 2820924 | C | A | A289S | ||||||||
| recA | b2699 | 2820962 | 2820962 | 2820962 | 2820962 | T | G | D276A | |||||
| recA | b2699 | 2820963 | C | T | D276N | ||||||||
| Replication restart primosome | |||||||||||||
| priA | b3935 | 4123174 | C | T | V553I | ||||||||
| priC | b0467 | 489549 | 489549 | A | G | L162P | |||||||
| dnaT | b4362 | 4599105 | 4599105 | 4599105 | 4599105 | G | A | R145C | |||||
| dnaB | b4052 | 4262560 | T | C | L74S | ||||||||
| dnaB | b4052 | 4262578 | C | A | P80H | ||||||||
| Cell division | |||||||||||||
| ftsW | b0089 | 98506 | A | G | E34G | ||||||||
| ftsW | b0089 | 99207 | 99207 | A | G | M268V | |||||||
| ftsZ | b0095 | 106214 | G | A | D303N | ||||||||
| Proteolysis | |||||||||||||
| clpP | b0437 | 456127 | 456127 | A | G | Y75C | |||||||
| clpP/clpXa | b0437/ b0438 | 456637 | G | A | - | ||||||||
| clpX | b0438 | 457803 | A | G | Y384C | ||||||||
| Glutamate transport | |||||||||||||
| gltS | b3653 | 3825922 | 3825922 | 3825922 | 3825922 | A | G | V255A | |||||
| Miscellaneous | |||||||||||||
| ylbEb | b4572 | 547836 | 547836 | 547836 | G | K85E | |||||||
| yjgL | b2453 | 4474024 | A | G | N188D | ||||||||
| yjgL | b2453 | 4475030 | 4475030 | A | G | D523G | |||||||
This mutation is intergenic.
This is a one-nucleotide insert that restores the full-length reading frame of an annotated pseudogene.
It is notable that for four of these genes, recA, ruvB, bglH, and yjgL, there are two different mutant alleles evident among the seven isolates. Given that only 1 to 2% of the loci in the sequenced strains were modified during their evolution, the probability of randomly generating different alleles in the same locus is vanishingly small. This observation reinforces the idea that changes in each of these loci provide the cell with a selective advantage.
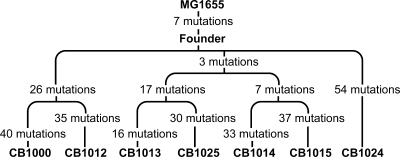
All of the strains derived from IR-1-20 are closely related. The presence of some shared genetic changes permits a phylogeny to be constructed based on genome sequences. This was done for the strains derived from population IR-1-20 and is shown in Fig. 6. In our initial analysis of these strains, we made the assumption that the closer two related IR-resistant strains are to each other, the more likely they are to share the same mechanisms of protection from IR. In a situation where the strains are very closely related, they are likely to carry identical genetic changes, and this overlap may identify those changes most relevant to IR resistance. Examination of CB1014 and CB1015, close relatives that share 10 identical base substitutions, illustrates this approach. Of these 10 changes, 6 result in synonymous changes that will not affect protein function and are unlikely to affect IR resistance. The four nonsynonymous mutations alter the amino acid compositions of the RecA, DnaT, GltS, and RimO proteins. The phylogeny presented in Fig. 6 indicates that among the sequenced strains, CB1013 and CB1025 most recently diverged from CB1014 and CB1015. CB1000 and CB1012 share 26 identical base substitutions—more than any other pair of isolates examined. CB1013 and CB1025 share 20 identical base substitutions. CB1013 and CB1025 also share three modifications with the CB1014-CB1015 pair, namely, the changes in RecA, DnaT, and GltS. The phylogeny reinforces the idea that different cells in the evolving population took different paths to radiation resistance.
FIG. 6.
Phylogeny of seven fully sequenced strains from the IR-1-20 population. Phylogeny was determined using the single nucleotide mutations that were shared among the different strains.
The parallel presence of mutations affecting RecA, DnaT, and GltS in four different purified strains has two possible explanations, as follows: (i) the modified proteins confer a selective advantage to the individuals in which they are expressed, providing protection against the lethal effects of IR (possibly acting together); or (ii) the modifications are neutral, and their appearance within the population is a manifestation of their clonal expansion from a mutational event that occurred early in the evolution of the population.
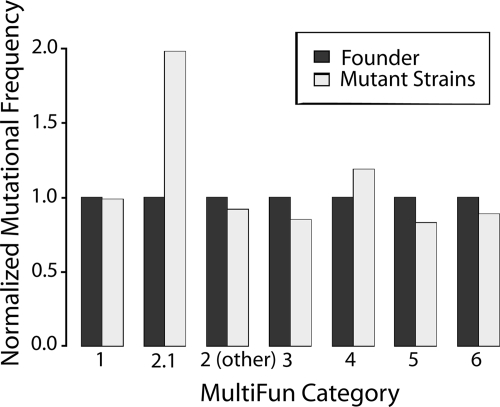
Table S7 in the supplemental material places all of the genes that carry nonsynonymous mutations in the three radioresistant strains into functional categories based on the “MultiFun” classification scheme developed by Serres and Riley (47). Examination of this table reveals that one-third to one-half of the mutations are in genes of unknown function. Although only a few of the mutations occur in genes encoding known DNA repair proteins, mutations in genes of this category are overrepresented in the genome by a factor of about 2 (Fig. 7).
FIG. 7.
Occurrence of mutations in particular gene classes (MultiFun) relative to the fraction of genes present in each genomic category. Although the total number of genes in each category shown is different (see Table S7 in the supplemental material), these numbers are effectively normalized to 1.0 in this analysis. The occurrence of mutations in particular gene classes was calculated as the total number of mutations in a particular category divided by the total number of scored mutations in all sequenced strains (if a particular mutation appeared in four different sequenced strains, then it was counted as four scored mutations). This ratio was divided by the fraction of the total genes present in that particular category. The results are shown by the light gray bars. If mutations are under- or overrepresented in a particular category, the calculation will yield values of <1.0 or >1.0, respectively. The MultiFun categories are as follows: 1, metabolism; 2, information transfer; 3, regulation; 4, transport; 5, cell processes; and 6, cell structure. The subgroup 2.1 includes genes involved in DNA replication, recombination, and repair.
Loss of e14 increases resistance to IR.
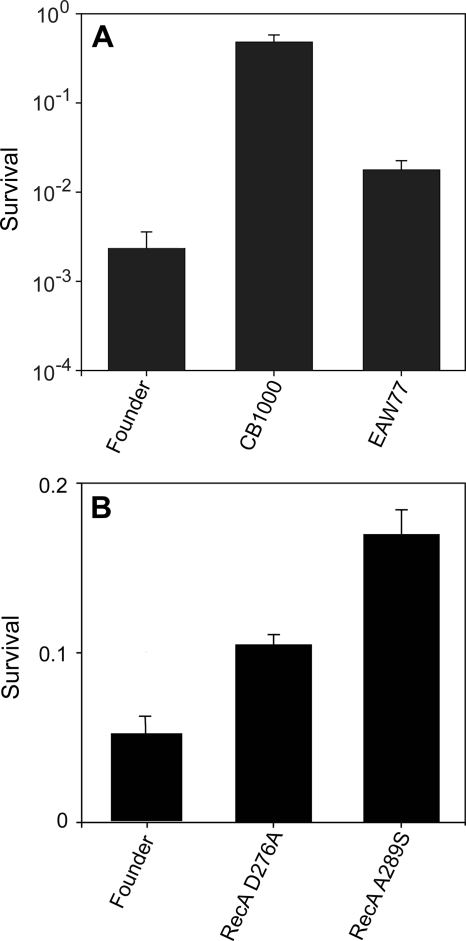
We set out to determine directly if some of the common mutations contributed substantially to the acquired phenotype. We isolated the IS1 insertion in the fimE gene in an otherwise wild-type background, but this genomic alteration had no evident effect on IR resistance (data not shown). A much different result was obtained when we took a closer look at the deletion of the e14 prophage. This prophage is known to undergo deletion in response to an induction of the SOS response (21), indicating that the SOS response had been initiated at some point (probably repeatedly) in the protocols leading to our four isolates. Deleting e14 from the founder increased the IR resistance of the resulting strain approximately eightfold at 3,000 Gy (Fig. 8A), suggesting that loss of this cryptic phage contributes to IR resistance. The e14 prophage expresses a toxic inhibitor of several E. coli metabolic processes, called Lit (25, 26, 38). Its absence, or the absence of some other toxic factor produced by the prophage, may explain the increase in radiation resistance in the e14 deletion strains.
FIG. 8.
Effects of selected mutations on survival of E. coli. Cell cultures were irradiated to 3,000 (A) or 2,000 (B) Gy and plated to measure survival as described in Materials and Methods. In panel A, strain EAW77 contains the e14 deletion in an otherwise wild-type background. Results in panel A were obtained with a 60Co irradiator (19 Gy/min); results in panel B were obtained with a 137Cs irradiator (7.8 Gy/min).
A role for modified RecA in IR resistance.
As part of a broader survey to determine if other mutations (mostly mutations present in CB1000) were present in the population (Table 3), 14 additional strains purified from population IR-1-20 were surveyed for the presence of the D276A alteration in RecA. This recA mutation was observed in 11 of these strains. Furthermore, additional RecA alleles were detected: CB1024 expresses RecA A289S, and CB2000, which was isolated from an independently derived population, encodes RecA D276N. The appearance of a recA allele in so many IR-resistant isolates (modified RecA proteins appear in 17 of 23 IR-resistant strains examined), coupled with the appearance of multiple alleles including a different change at the same nucleotide in CB2000, strongly argues that these modifications to RecA contribute to IR resistance. To test this possibility, the RecA D276A and A289S alleles were moved into the founder. In both cases, the change resulted in a small but significant increase in IR resistance in the resulting strain relative to that of the founder (Fig. 8B). The A289S allele was consistently the more effective of the two in its enhancement of radiation resistance. Once again, the recA mutations are evident contributors to radiation in the strains in which they occur, but they do not explain the entire phenotype. We note that the positive effects of these recA alleles are dependent on the presence of an intact recX gene in its normal location immediately downstream of recA. No effects of the recA alleles on IR resistance were observed in some early trials using a recA gene replacement procedure that left a genomic scar inactivating recX. To illustrate the importance of RecA to the overall repair process, we deleted recA from CB1000 and demonstrated that the resulting strain was as sensitive to IR as a recA derivative of MG1655 (data not shown). Along with the e14 results, the recA mutations clearly telegraph a situation in which multiple contributing mechanisms are present. This in turn is consistent with the multiple steps in the increase in radiation resistance in the populations monitored and shown in Fig. 1.
TABLE 3.
Survey of mutations present in independent evolved isolatesa
| Gene | Presence of allele
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CB1000 | CB1012 | CB1013 | CB1014 | CB1015 | CB1016 | CB1017 | CB1018 | CB1019 | CB1020 | CB1021 | CB1022 | CB1023 | CB1024 | CB1025 | CB1026 | CB1027 | CB1028 | CB1029 | CB1030 | CB1031 | |
| recA | − | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − |
| recB | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| fimE | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + |
| e14 prophage | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ygaH | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| clpP | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| ybdL | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
All strains listed were single colony isolates derived from the same population that gave rise to CB1000. All single nucleotide polymorphisms surveyed here were confirmed by direct Sanger sequencing methods. The e14 deletions and fimE insertions were confirmed by PCR analysis (see Fig. S3a and S3b in the supplemental material). The alleles tested were identical to those found in CB1000 (see Tables S4 and S5 in the supplemental material), except that the recA allele tested was that encoding the RecA D276A mutant protein. The symbols + and − indicate the presence and absence of the tested allele, respectively.
The RecA D276A and D276N proteins have both been purified. Both exhibit wild-type levels of DNA strand exchange and ATP hydrolytic activities in vitro (V. Petrova and M. Cox, unpublished data). There is no indication that these proteins have been inactivated or impeded in their normal recombinogenic functions in any way. Both D276 and A289 are located on the protein surface that defines the major groove of a RecA filament structure. We speculate that the mutations affect an as yet unidentified protein-protein interaction, possibly with the LexA protein.
Is there a role for the proteins that mediate replication restart in IR resistance?
Strains CB1013, CB1014, CB1015, and CB1025 all express DnaT R145C. DnaT is a component of the E. coli restart primosome, which includes PriA, PriB, PriC, DnaT, and DnaB. This complex of proteins is needed to reestablish replication forks after stalled forks are repaired by recombinational DNA repair (10, 24, 36). Mutations in other components of the restart primosome were observed. Among the IR-resistant strains whose genomes were sequenced, CB1000 and CB1012 carry an allele of priC, CB1024 and CB2000 alleles of dnaB, and CB3000 an allele of priA. In other words, every IR-resistant strain for which we have a complete genome sequence carries a mutation in one of the loci that encode the E. coli restart primosome, and four of the five proteins contributing to this complex are affected.
When the most common of the restart primosome alleles—the dnaT allele—was transferred to the founder, the resulting strain was substantially more sensitive to IR than the founder (data not shown). Given that four of the sequenced genomes carry this mutation, the genetic context must determine the effect of this dnaT allele on the radioresistance phenotype.
DISCUSSION
We generated four extremely radiation-resistant populations of Escherichia coli, all derived from the same MG1655 founder strain. The adaptations are generally specific for IR resistance. Characterization of these populations, including the complete genomic sequencing of nine strains independently isolated from them, provides information about the mechanisms underlying the acquired phenotype. As shown in Fig. 4, an enhanced capacity to promote recombinational DNA repair after extreme genomic degradation plays a substantial role in the IR resistance seen in these strains. Passive protection from damage inflicted by IR is not in evidence. Major mutational targets include the recA and ruvB genes and the genes encoding components of the replication restart primosome. All of these are involved in recombinational DNA repair. Multiple mutations are found in the clpP and clpX genes, encoding components of an important proteolytic system, and the ftsW and ftsZ genes, encoding proteins involved in cell division. Additional processes may contribute to radiation resistance, including those involving the gltS, bglH, yjgL and ylbE genes, the deletion of the e14 prophage, and perhaps other genes where contributing mutations appeared too late in the selection to become common in the population.
Evidence has been obtained that the e14 deletion and at least two of the recA mutant alleles contribute directly to radiation resistance. Whereas this begins to account for the acquired phenotype, these genomic alterations do not account for all of the improvement in IR resistance. These results argue strongly that multiple processes contribute to the acquired phenotype. Given the diversity evident in population IR-1-20 and the unique genomic alterations observed in isolates derived from other evolved populations, it seems likely that different cells in these populations may pursue often distinct pathways to IR resistance. This view is reinforced by the phylogeny in Fig. 6.
The results provide a strong case that an enhancement of recombinational DNA repair makes a major contribution to the acquired phenotype. Mutations in genes involved in this process occur at a high rate and are often prominent in the evolved population. As shown in Fig. 5, the evolved strains exhibited a much-improved capacity to reconstitute their genomes. The recA mutations make a clear positive contribution to the IR resistance phenotype, and three different recA mutant alleles were found within the nine strains that were sequenced in their entirety. Mutations were also found in another component of the recombinational DNA repair system, ruvB. It is possible, of course, that some of the other common mutations make an indirect contribution to recombinational DNA repair by favorably altering the cellular environment in which it is carried out.
The dnaT allele provides an example of apparent mutational epistasis. The presence of the dnaT allele does not adversely affect the overall IR resistance of the radioresistant strains in which it occurs. We must assume that the dnaT mutation cannot have appeared in the genome before acquisition of one or more compensating mutations permitting the dnaT strains to survive irradiation. In other words, the effect of the dnaT allele appears to be conditional, depending on the genetic background. Assuming that dnaT contributes to radioresistance, this result suggests that at least some of the steps in building an IR-resistant strain must occur in a temporal sequence. If a cell deviates from that sequence and creates the dnaT mutation before its compensating mutation, irradiation will select against that cell.
The other common mutations may signal additional mechanisms of IR resistance but may also contribute indirectly to the same repair process. The ftsW and ftsZ mutations may alter the regulation or timing of cell division, perhaps providing needed time for repair. The clpP and clpX mutations might slow the turnover of proteins needed for energy production, cell division, detoxification, and/or protein folding, enhancing the environment for repair. The gltS and bglH alterations may also change the cellular environment in which the repair occurs. Each of these sets of mutations will require more detailed analysis.
The newly robust capacity of these strains for genome reconstitution relies on both the RecF and RecBCD pathways for recombinational DNA repair. Inactivation of either the recB or recF gene in CB1000 eliminated much of the radiation resistance of that strain, although the sensitivity of the recB knockout strain was substantially greater (data not shown). Inasmuch as Deinococcus lacks homologues of the recB and recC genes, it is unlikely that the double-strand-break repair pathways used for genome reconstitution in the evolved E. coli strains faithfully replicate the pathways documented for Deinococcus (54).
The extreme adaptations to IR exposure exhibited by these strains are specific to IR. Whereas some of the strains exhibit improved resistance to other DNA damaging agents, those improvements are generally more modest and do not occur in every strain derived from the evolved populations. The adaptations also do not appear to reinforce prominent hypotheses put forward to explain radiation resistance in Deinococcus radiodurans. The evolved E. coli strains do not have unusually condensed nucleoids, a property of Deinococcus nucleoids highlighted by Minsky and colleagues (29, 40). There is also no indication of the presence of elevated levels of Mn ion or reduced levels of protein oxidation, as seen in Deinococcus (11). In both cases, the proposed mechanisms may still be applicable to IR resistance in Deinococcus. However, the results of the current study indicate that neither mechanism should be considered a universal requirement for extreme IR resistance. One major message of the present research is that IR resistance is a complicated phenotype with potentially many components. A focus on just one contributing mechanism is unlikely to yield a complete understanding of the phenomenon.
Supplementary Material
Acknowledgments
We thank Andrew Whitehead for providing many helpful comments on an early draft of the manuscript, Trang D. Nguyen (LSU) for assistance in isolating IR-resistant strains, and Hiromi Brown (LSU) for determining DNA content. We thank Cassandra Jenson for carrying out some of the early direct sequencing/CGS validation efforts. We also thank Victor Ruotti, James Thomson, and Michael Sussman for arranging and/or providing access to and assistance with Illumina instrumentation.
This work was supported by grant GM067085 from the National Institutes of Health to M.M.C. and by U.S. Department of Energy grants DEFG0201ER63151 and CSP2009.796601 to J.R.B.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, C., B. Rak, and R. Benz. 1999. The gene bglH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of beta-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol. Microbiol. 31:499-510. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. A., and R. T. Sauer. 2006. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, V. J. Collado, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bridges, B. A. 2005. Error-prone DNA repair and translesion synthesis: focus on the replication fork. DNA Repair (Amsterdam) 4:618-619, 634. [DOI] [PubMed] [Google Scholar]
- 6.Cox, M. M. 1999. Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acids Res. Mol. Biol. 63:310-366. [DOI] [PubMed] [Google Scholar]
- 7.Cox, M. M. 2007. The bacterial RecA protein: structure, function, and regulation, p. 53-94. In R. Rothstein and A. Aguilera (ed.), Topics in current genetics. Springer-Verlag, Heidelberg, Germany.
- 8.Cox, M. M. 2002. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 510:107-120. [DOI] [PubMed] [Google Scholar]
- 9.Cox, M. M., and J. R. Battista. 2005. Deinococcus radiodurans--the consummate survivor. Nat. Rev. Microbiol. 3:882-892. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 11.Daly, M. J., E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, R. D. Leapman, B. Lai, B. Ravel, S. M. Li, K. M. Kemner, and J. K. Fredrickson. 2007. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly, M. J., E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, A. Venkateswaran, M. Hess, M. V. Omelchenko, H. M. Kostandarithes, K. S. Makarova, L. P. Wackett, J. K. Fredrickson, and D. Ghosal. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 13.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, R., and A. J. Sinskey. 1973. Radiation-resistant mutants of Salmonella typhimurium LT2: development and characterization. J. Bacteriol. 113:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty, T. J., J. A. Thanassi, and M. J. Pucci. 1993. The Escherichia coli mutant requiring d-glutamic acid is the result of mutations in two distinct genetic loci. J. Bacteriol. 175:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elena, S. F., and R. E. Lenski. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457-469. [DOI] [PubMed] [Google Scholar]
- 17.Erdman, I. E., F. S. Thatcher, and K. F. Macqueen. 1961. Studies on the irradiation of microorganisms in relation to food preservation. II. Irradiation resistant mutants. Can. J. Microbiol. 7:207-215. [DOI] [PubMed] [Google Scholar]
- 18.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 19.Gerrish, P. J., and R. E. Lenski. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 103:127-144. [PubMed] [Google Scholar]
- 20.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 21.Greener, A., and C. W. Hill. 1980. Identification of a novel genetic element in Escherichia coli K-12. J. Bacteriol. 144:312-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gresham, D., M. M. Desai, C. M. Tucker, H. T. Jenq, D. A. Pai, A. Ward, C. G. DeSevo, D. Botstein, and M. J. Dunham. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genetics 4:e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haeusser, D. P., and P. A. Levin. 2008. The great divide: coordinating cell cycle events during bacteria growth and division. Curr. Opin. Microbiol. 11:94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller, R. C., and K. J. Marians. 2006. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell. Biol. 7:932-943. [DOI] [PubMed] [Google Scholar]
- 25.Hiom, K., S. M. Thomas, and S. G. Sedgwick. 1991. Different mechanisms for SOS induced alleviation of DNA restriction in Escherichia-coli. Biochimie 73:399-405. [DOI] [PubMed] [Google Scholar]
- 26.Kao, C., and L. Snyder. 1988. The Lit gene product which blocks bacteriophage T4 late gene expression is a membrane protein encoded by a cryptic DNA element, e14. J. Bacteriol. 170:2056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, M. 1977. Preponderance of synonymous changes as evidence for neutral theory of molecular evolution. Nature 267:275-276. [DOI] [PubMed] [Google Scholar]
- 28.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 29.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lusetti, S. L., E. A. Wood, C. D. Fleming, M. J. Modica, J. Korth, L. Abbott, D. W. Dwyer, A. I. Roca, R. B. Inman, and M. M. Cox. 2003. C-terminal deletions of the Escherichia coli RecA protein—characterization of in vivo and in vitro effects. J. Biol. Chem. 278:16372-16380. [DOI] [PubMed] [Google Scholar]
- 32.Manjunatha, U. H., H. Boshoff, C. S. Dowd, L. Zhang, T. J. Albert, J. E. Norton, L. Daniels, T. Dickl, S. S. Pang, and C. E. Barry. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in A cell population. J. Bacteriol. 179:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marians, K. J. 2000. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 25:185-189. [DOI] [PubMed] [Google Scholar]
- 35.Marians, K. J. 2000. Replication and recombination intersect. Curr. Opin. Genet. Dev. 10:151-156. [DOI] [PubMed] [Google Scholar]
- 36.Marians, K. J. 2008. Understanding how the replisome works. Nat. Struct. Mol. Biol. 15:125-127. [DOI] [PubMed] [Google Scholar]
- 37.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehta, P., S. Casjens, and S. Krishnaswamy. 2004. Analysis of the lambdoid prophage element e14 in the E. coli K-12 genome. BMC Microbiol. 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. H., A. Suthar, J. Tai, A. Yeung, C. Truong, and J. L. Stewart. 1999. Direct selection for mutators in Escherichia coli. J. Bacteriol. 181:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minsky, A. 2003. Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50:367-376. [DOI] [PubMed] [Google Scholar]
- 41.Moseley, B. E., and A. Mattingly. 1971. Repair of irradiation transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J. Bacteriol. 105:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen, R., and Z. H. Yang. 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 20:1231-1239. [DOI] [PubMed] [Google Scholar]
- 43.Nyman, K., K. Nakamura, H. Ohtsubo, and E. Ohtsubo. 1981. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature 289:609-612. [DOI] [PubMed] [Google Scholar]
- 44.Parisi, A., and A. D. Antoine. 1974. Increased radiation resistance of vegetative Bacillus pumilus. Appl. Microbiol. 28:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rainey, F. A., K. Ray, M. Ferreira, B. Z. Gatz, M. F. Nobre, D. Bagaley, B. A. Rash, M. J. Park, A. M. Earl, N. C. Shank, A. M. Small, M. C. Henk, J. R. Battista, P. Kampfer, and M. S. da Costa. 2005. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 71:5225-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sargent, R. G., and C. K. Mathews. 1987. Imbalanced deoxyribonucleoside triphosphate pools and spontaneous mutation rates determined during dCMP deaminase-defective bacteriophage T4 infections. J. Biol. Chem. 262:5546-5553. [PubMed] [Google Scholar]
- 47.Serres, M. H., and M. Riley. 2000. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5:205-222. [DOI] [PubMed] [Google Scholar]
- 48.Szvetnik, A., J. Gal, and M. Kalman. 2007. Membrane topology of the GltS Na+/glutamate permease of Escherichia coli. FEMS Microbiol. Lett. 275:71-79. [DOI] [PubMed] [Google Scholar]
- 49.Tassotto, M. L., and C. K. Mathews. 2002. Assessing the metabolic function of the MutT 8-oxodeoxyguanosine triphosphatase in Escherichia coli by nucleotide pool analysis. J. Biol. Chem. 277:15807-15812. [DOI] [PubMed] [Google Scholar]
- 50.Witkin, E. M. 1946. Inherited differences in sensitivity to radiation in Escherichia coli. Proc. Natl. Acad. Sci. USA 32:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodgate, R. 1999. A plethora of lesion-replicating DNA polymerases. Genes Dev. 13:2191-2195. [DOI] [PubMed] [Google Scholar]
- 52.Xie, C.-X., A. Xu, L.-J. Wu, J.-M. Yao, J.-B. Yang, and Z.-L. Yu. 2004. Comparison of base substitutions in response to nitrogen ion implantation and 60Co-gamma ray irradiation in Escherichia coli. Genet. Mol. Biol. 27:284-290. [Google Scholar]
- 53.Yang, Z. H., and J. P. Bielawski. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zahradka, K., D. Slade, A. Bailone, S. Sommer, D. Averbeck, M. Petranovic, A. B. Lindner, and M. Radman. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569-573. [DOI] [PubMed] [Google Scholar]
- 55.Zerbino, D. R., and E. Birney. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmerman, J. M., and J. R. Battista. 2005. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.