Abstract
Hopanoids are triterpenoic, pentacyclic compounds that are structurally similar to sterols, which are required for normal cell function in eukaryotes. Hopanoids are thought to be an important component of bacterial cell membranes because they control membrane fluidity and diminish passive diffusion of ions, and a few taxons modulate their hopanoid content in response to environmental stimuli. However, to our knowledge, mutational studies to assess the importance of hopanoids in bacterial physiology have never been performed. Genome sequencing of the potato scab pathogen, Streptomyces scabies 87-22, revealed a hopanoid biosynthetic gene cluster (HBGC) that is predicted to synthesize hopene and aminotrihydroxybacteriohopane products. Hopene was produced by fully sporulated cultures of S. scabies on solid ISP4 (International Streptomyces Project 4) medium as well as by submerged mycelia grown in liquid minimal medium. The elongated hopanoid aminotrihydroxybacteriohopane was not detected under either growth condition. Transcription of the S. scabies HBGC was upregulated during aerial growth, which suggests a link between hopanoid production and morphological development. Functional analysis of the S. scabies Δhop615-1 and Δhop615-7 mutant strains, the first hopanoid mutants created in any bacterial taxon, revealed that hopanoids are not required for normal growth or for tolerance of ethanol, osmotic and oxidative stress, high temperature, or low pH. This suggests that hopanoids are not essential for normal streptomycete physiology.
Sterols are ubiquitous in eukaryotic cell membranes, where they function to control membrane fluidity and permeability (39). In contrast, most bacteria do not contain sterols; exceptions are members of the family Methylococcaceae (38). Bacteria, however, do produce triterpenoic, pentacyclic compounds known as hopanoids (19, 34). Like sterols, hopanoids act to control membrane fluidity and diminish passive diffusion of ions across the cell membrane (18, 19, 28). Therefore, hopanoids have been considered to be important components of bacterial membranes. This hypothesis is supported by the discovery that some bacteria modulate the hopanoid content of their membranes in response to environmental stimuli. For example, Frateuria aurantia increases its hopanoid content as temperature increases (17), and the acidophilic and thermophilic bacterium Alicyclobacillus (formally Bacillus) acidocaldarius increases its hopanoid content in response to temperature above 50°C and, to a lesser extent, low pH of <5.0 (30). However, the physiological importance of bacterial hopanoids has not been investigated in any species due to the lack of hopanoid mutant strains (19).
Hopanoids are produced by nearly half of all bacterial species analyzed but are entirely absent from archaea (33). Though long thought to be exclusively produced by aerobic organisms, hopanoids have recently been isolated from the strictly anaerobic Geobacter metallireducens and Geobacter sulfurreducens (9). Hopanoids are produced by multiple Streptomyces spp. (33), including the model streptomycete, Streptomyces coelicolor A3(2) (29).
Streptomycetes are saprophytic, high-G+C, gram-positive, soil-dwelling organisms that have a complicated developmental life cycle (11), the genetics of which have been of great interest (4, 5). After spore germination, vegetative growth proceeds to form a network of branched hyphal filaments called a vegetative mycelium. Upon nutrient deprivation and possibly other environmental signals, this vegetative mycelium undergoes a programmed cell death, which provides nutrients for the development of structures called aerial hyphae that then septate and form unigenomic exospores that disseminate into the environment to start the life cycle over again.
Hopanoids have been isolated from sporulated surface-grown cultures of S. coelicolor but could not be isolated from cells grown in liquid media (29). S. coelicolor produces two hopanoids, hopene and aminotrihydroxybacteriohopane, which are hypothesized to alleviate stress in aerial mycelia by decreasing water permeability of the cell membrane (29). However, the importance of hopanoids for normal cell function and tolerance of environmental stresses has not been tested in Streptomyces spp. or in any other bacteria.
The recent completion of a genome-sequencing project revealed the presence of a putative hopanoid biosynthetic gene cluster (HBGC) in the plant pathogen Streptomyces scabies 87-22 (http://www.sanger.ac.uk/Projects/S_scabies/). The objectives of this study were to demonstrate that the HBGC in S. scabies produces hopanoids and to analyze the importance of hopanoids in bacterial growth and tolerance to environmental stresses.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Escherichia coli strains were cultured as previously described (20, 36). Streptomyces strains were cultured at 28°C or room temperature using International Streptomyces Project 4 agar (ISP4) medium, mannitol-soya flour agar, R2, nutrient agar, tryptic soy broth (20), thaxtomin defined agar containing 0.7% cellobiose (15), or liquid minimal medium (MM), which contained NH4SO4 (2 g/liter), MgSO4·7H2O (0.6 g/liter), K2H2P04 (2.6 g/liter), CaCl2·2H2O(0.1 g/liter), glycerol (2.5% vol/vol), glucose (2.5 g/liter), yeast extract (0.05 g/liter), and trace element solution (20). All liquid cultures were shaken at ∼250 rpm. Media were supplemented with antibiotics at the following concentrations: apramycin, 100 μg/ml; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; and nalidixic acid, 25 μg/ml. All Streptomyces strains generated in this study were created by cross-genus conjugation from the nonmethylating E. coli stain ET12567/pUZ8002 as previously described (20).
Creation of mutant strains.
The S. scabies 87-22 Δhop strains were created using the λ RED-based PCR-targeting Redirect technology (8). A disruption cassette consisting of an oriT gene and the apramycin resistance gene, aac(3)IV, from pIJ773 was generated by PCR amplification. The forward primer contained 39 nucleotides (nt) (CCACGCTTCTGCCGTGCCCGCGCCGGGTGACGAGCGGTG) of S. scabies sequence including the putative start codon (bold) for SCAB13001 and 36 nt that were homologous to the region immediately upstream. The reverse primer contained 39 nt (CGCGCCGCAGTTCCTCGACCAGGCGCCGGGAGGAGACGC) that targeted the middle of SCAB12971 (Fig. 1). Primers used in deletion of the hopanoid biosynthetic genes were obtained from Integrated DNA Technologies, Coralville, IA. The resulting PCR product was gel purified and electroporated into E. coli BW25113 containing the λ RED gene-carrying plasmid pIJ790 and S. scabies 87-22 cosmid 1453. Transformants were screened for the presence of mutagenized cosmid by colony PCR. A KpnI and BglII double digest verified proper mutagenesis of cosmid 1453. The mutant cosmid was moved to wild-type S. scabies 87-22 via conjugation. Transconjugants were screened for apramycin resistance and kanamycin sensitivity, a phenotype associated with a double-crossover recombination event. Deletion of the HBGC was confirmed by Southern blot hybridization.
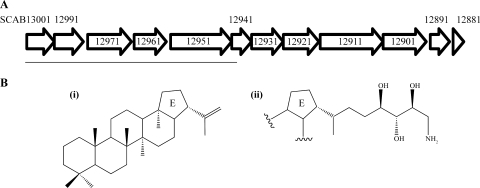
FIG. 1.
The S. scabies 87-22 HBGC. (A) Schematic representation of the HBGC locus in S. scabies (http://strepdb.streptomyces.org.uk/). The horizontal line under the gene arrows marks the region replaced by the apramycin resistance gene, aac(3)IV in the Δhop615-1 and Δhop615-7 mutant strains. (B) Putative products of the S. scabies 87-22 HBGC: (i), hopene; (ii), aminotrihydroxybacteriohopane. Chemical structures are adapted from reference 29.
RNA analysis.
RNA was isolated as previously described (16) from S. scabies wild-type cultures grown in liquid MM. Two micrograms of DNase-treated RNA was reverse transcribed using the Superscript III First Strand Synthesis system and 250 ng of random hexamer primers (Invitrogen). Control reactions in which enzyme was omitted from the reaction mixture were also performed. Twenty-seven cycles of PCR amplification were used to achieve suitable PCR products from cDNA. PCR products were cloned into pCR2.1TOPO (Invitrogen) and sequenced (Biotechnology Resource Center, Cornell University) using oligonucleotide primer M13r (Invitrogen).
Construction of the SCAB13001-xylE reporter plasmid and XylE activity assays.
Because of the difficulties of analyzing RNA from surface-grown cultures, an integrative xylE reporter plasmid was constructed to analyze the expression of SCAB13001. The tipAp promoter was removed from pIJ6902 (12) by digestion with AseI and BamHI, and a PCR-amplified, promoterless copy of the 2,3-catechol dehydrogenase gene, xylE, from pXE4 (14) was cloned in its place using the same restriction sites to create pRFSRL25. The oligonucleotide primers P1 (GGATCCACTAGTGACTGAGTGACCAGAAGGGAGCGGACATATGAACAAAGGTGTAATGC) and P2 (GATTAATTCAGGTCAGCACGGTCATGA) (Integrated DNA Technologies, Coralville, IA) were used to amplify the xylE gene from pXE4 (xylE sequences are in bold). P1 contained additional restriction enzyme sites for construction of a multicloning site as well as stop codons in all three reading frames to prevent translational fusions of cloned upstream sequences to the XylE protein. The pRFSRL25 plasmid integrates into streptomycete chromosomes at the ΦC31 attachment site. A ∼1.2-kb promoter probe fragment upstream and extending 81 bp into SCAB13001 was PCR amplified and cloned into the EcoRI and BamHI sites of pRFSRL25 to result in pRFSRL27. Both pRFSRL25 and pRFSRL27 were moved to wild-type S. scabies 87-22 via conjugation. Transconjugants were selected for apramycin resistance.
For XylE activity assays, cultures were grown on cellophane that was overlaid on top of ISP4 medium. Mycelia were harvested with a spatula and placed in a 2.2-ml microcentrifuge tube containing 0.5 ml of sample buffer (10% [vol/vol] acetone, 20 mM EDTA, 100 mM K2HPO4 buffer, pH 7.5). Samples were processed exactly as described previously (20). Absorbance at 375 nm was recorded at 1-min intervals for a total of 8 min to analyze the conversion of the colorless catechol substrate to the yellow 2-hydroxymuconic semialdehyde product. XylE activity was expressed as mU XylE activity, where 1 U is the amount of enzyme required to convert 1 μmol of substrate to product in 1 min. The molar extinction coefficient reported elsewhere (35) was used to calculate the amount of product formed. XylE activity was normalized by total protein. Protein was quantified using the Bradford method with bovine serum albumin as a standard.
Environmental stress tests.
Temperature, pH, and osmotic stress experiments were performed on ISP4 medium. Frozen spore stocks were used as the inoculum, and strains were allowed to grow for 7 days. Responses to temperature were observed after incubation at room temperature, 37°C, and 42°C. The response of strains to pH (7.3 to 5.0) was evaluated by adding increasing amounts of 1 M HCl. Osmotic shock was supplied in the form of KCl (0.1 to 0.7 M). Ethanol tolerance was assessed after 7 days of growth in the presence of ethanol concentrations ranging from 1 to 10% (vol/vol) in liquid MM. The oxidative stress disc diffusion assay was performed on mannitol-soya flour agar by seeding a plate with spores and dispensing either a water control or 0.3 to 3.0 μmol of H2O2 onto sterile filter paper discs placed on the agar. The diameters of the resulting zones of inhibited growth were measured after 2 days.
Southern blot hybridization.
Genomic DNA was isolated from overnight tryptic soy broth-grown cultures using the MasterPure kit for gram-positive bacteria (Epicentre) according to the manufacturer's instructions. Five micrograms of genomic DNA was digested with PstI and electrophoresed on a 0.8% agarose gel. DNA was denatured with 0.5 M NaOH and 1.5 M NaCl and transferred to a nylon membrane (Whatman) by capillary transfer with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). DNA was UV cross-linked to the membrane by applying 120,000 μJ/cm2 for 4 min. The membrane was probed with a DNA fragment from the intergenic region between SCAB13001 and SCAB13011 that was labeled with dioxigenin-11-dUTP (Roche). Hybridization was performed overnight in a rotary oven at 68°C. Stripping conditions were as follows: 2× SSC at 68°C for 30 min and 0.1× SSC at 68°C for 30 min. Processing of the membrane from this point forward was performed according to the manufacturer's instructions (Roche).
Lipid extraction.
S. scabies 87-22 wild-type and Δhop615-1 mutant strains were grown until fully sporulated (7 days) on ISP4 medium. For hopanoid analysis from liquid culture, wild-type S. scabies 87-22 was grown in liquid MM for 5 days at room temperature. Cultures were freeze-dried, and total lipids were extracted from ∼0.8 g cells using chloroform-methanol (2:1, vol/vol) for 1 h on a rotary platform with intermittent vortexing. Chloroform-methanol extracts were evaporated to dryness, resuspended in 90% methanol, and subjected to high-performance liquid chromatography/multiple-response monitoring mass spectrometry (LC/MRM).
LC/MRM analysis.
LC/MRM analysis was performed by the Cornell University Biotechnology Resource Center. Hopanoids were separated using a Vydac C4 1- by 150-mm column attached to a Dionex Ultimate multidimensional LC and the effluent monitored with an Applied Biosystems 4000 Q Trap, with an atmospheric pressure chemical ionization ion source attached, operating in positive-ion MRM mode. For hopene (Q1 m/z 411.5 Da) and aminotrihydroxybacteriohopane (Q1 m/z 546.8 Da), two transition ions (Q3) were monitored, i.e., m/z 137.2 Da and m/z 163.4 Da, for detection of both molecules.
RESULTS
S. scabies 87-22 contains a putative HBGC.
Bioinformatic analysis of the S. scabies 87-22 genome (http://www.sanger.ac.uk/Projects/S_scabies/) revealed a cluster of 12 genes that were homologous to genes in the S. coelicolor A3(2) HBGC, which encodes production of hopene and aminotrihydroxybacteriohopane (Fig. 1) (1, 29). The genes of the S. scabies HBGC and their proposed functions are listed in Table 1. The presence of SCAB12911, which encodes a putative 1-deoxy-d-xylulose-5-phosphate synthase, is a hallmark for mevalonate-independent biosynthesis of hopanoids (32). It is interesting to note that an HBGC was also identified in the genomes of Streptomyces griseus (24) and Streptomyces avermitilis (13). The gene order and orientation of the HBGCs identified in the four streptomycetes are conserved, and the cluster is located ∼2 Mb from one end of the linear chromosome and is part of the core genome, consisting of genes with conserved synteny among streptomycetes (1, 13, 24). Based on amino acid homology with the S. coelicolor HBGC, hopene and aminotrihydroxybacteriohopane are the likely products of the S. scabies HBGC.
TABLE 1.
Proposed functions of the proteins encoded by the S. scabies HBGC
| S. scabies protein | Proposed function | S. coelicolor ortholog | Identity, similarity (%) |
|---|---|---|---|
| SCAB13001 | Squalene/phytoene synthase | SCO6759 | 81, 86 |
| SCAB12991 | Squalene/phytoene synthase | SCO6760 | 85, 91 |
| SCAB12971 | Squalene/phytoene dehydrogenase | SCO6762 | 82, 87 |
| SCAB12961 | Polyprenyl diphosphate synthase | SCO6763 | 85, 91 |
| SCAB12951 | Squalene-hopene cyclase | SCO6764 | 80, 86 |
| SCAB12941 | Lipoprotein | SCO6745 | 80, 85 |
| SCAB12931 | Hypothetical protein | SCO6766 | 90, 96 |
| SCAB12921 | 4-Hyroxy-3-methylbut-2-en-1-yl diphosphate synthase | SCO6767 | 88, 95 |
| SCAB12911 | 1-Deoxy-d-xylulose-5-phosphate synthase | SCO6768 | 87, 93 |
| SCAB12901 | Aminotransferase | SCO6769 | 89, 93 |
| SCAB12891 | Regulatory protein | SCO6770 | 83, 90 |
| SCAB12881 | Small hydrophobic protein | SCO6771 | 74, 92 |
S. scabies produces hopanoids during submerged growth.
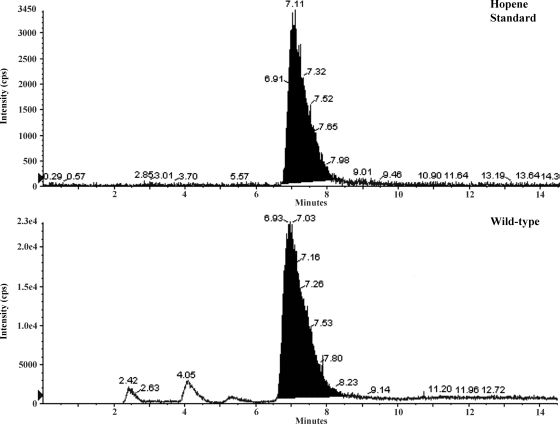
Transcriptional analysis of the squalene-hopene cyclase gene, SCAB12951, from S. scabies, suggested that hopanoids might be produced during submerged growth (Fig. 2). To assess this possibility, we extracted lipids from 5-day-old S. scabies wild-type cells grown in liquid MM. Lipids were subjected to LC/MRM. LC/MRM analysis detected transition ions for hopene (m/z 137.2 Da and m/z 163.4 Da) that had a retention time consistent with that of the standard (Fig. 3). However, we were unable to detect the other predicted hopanoid, aminotrihydroxybacteriohopane, in lipid extracts from S. scabies cells grown in liquid MM.
FIG. 2.
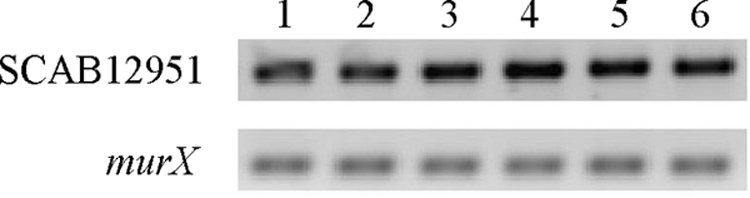
Reverse transcription-PCR analysis of the squalene-hopene cyclase gene (SCAB12951) from wild-type S. scabies 87-22 grown in liquid MM. The murX gene was used as a loading control, and reactions with the reverse transcriptase enzyme omitted did not yield any detectable PCR products (data not shown). cDNAs were from the following times: lane 1, day 2; lane 2, day 3; lane 3, day 4; lane 4, day 5; lane 5, day 6; lane 6, day 7.
FIG. 3.
Hopene is produced by S. scabies 87-22 in liquid MM. The panels display the extracted ion chromatogram from LC/MRM analysis. cps, counts per second.
Transcription of SCAB13001 is linked to morphological development.
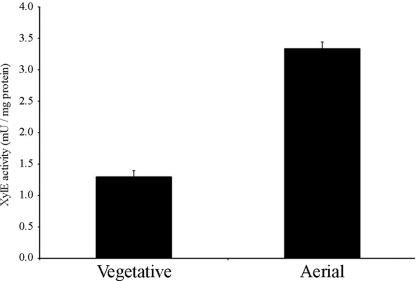
In order to assess transcription of SCAB13001 during surface growth conditions, we created the integrative reporter plasmid pRFSRL25, which contained a promoterless copy of the 2,3-catechol dehydrogenase gene, xylE. We transcriptionally fused a ∼1.2-kb DNA fragment upstream of SCAB13001 to the promoterless xylE gene in pRFSRL25. This reporter construct allowed us to analyze transcription of SCAB13001 spectrophotometrically. The SCAB13001 promoter was selected because of its position within the cluster (as the first gene in the cluster the promoter region was easily identifiable). XylE activity of the SCAB13001 reporter strain was examined under surface growth conditions on ISP4 medium, and a significant increase in enzyme activity was observed as the cultures progressed through a normal developmental cycle (Fig. 4), suggesting that transcription of SCAB13001, an important hopanoid biosynthetic gene, is upregulated during aerial growth.
FIG. 4.
Transcriptional analysis of the S. scabies 87-22 HBGC. XylE activity of the HBGC-xylE reporter strain was assessed during vegetative growth and aerial growth on ISP4 medium. XylE activity was not detected in S. scabies harboring the pRFSRL25 vector alone (data not shown). Results are the averages and standard deviations for three technical replicates from a single surface-grown culture. The values reported are statistically significantly different in a Student t test with a P value of <0.001. The experiment was performed twice with similar results.
Generation of a hopanoid null mutant.
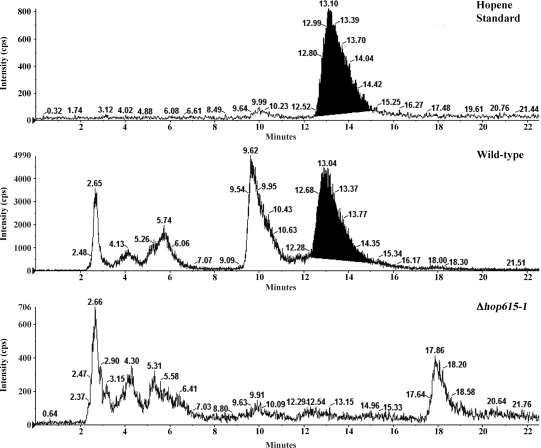
A hopanoid null (Δhop) mutant was created using the λ RED-based Redirect system for gene deletions in Streptomyces spp. (8). The hopanoid biosynthetic genes that were deleted are indicated in Fig. 1. Two mutant strains (Δhop615-1 and Δhop615-7 mutants) were isolated and verified by Southern blot hybridization (Fig. 5). Although the S. scabies 87-22 genome does not reveal an alternate means for hopanoid biosynthesis, we analyzed the hopanoid content of the Δhop615-1 mutant strain to confirm that the mutation had abolished hopanoid production. Lipids were isolated from fully sporulated ISP4-grown cultures and analyzed by LC/MRM. LC/MRM analysis revealed a peak in S. scabies lipid extracts that had a retention time and transition ions (m/z 137.2 Da and m/z 163.4 Da) that were consistent with the hopene standard, while the Δhop615-1 mutant strain did not (Fig. 6).
FIG. 5.
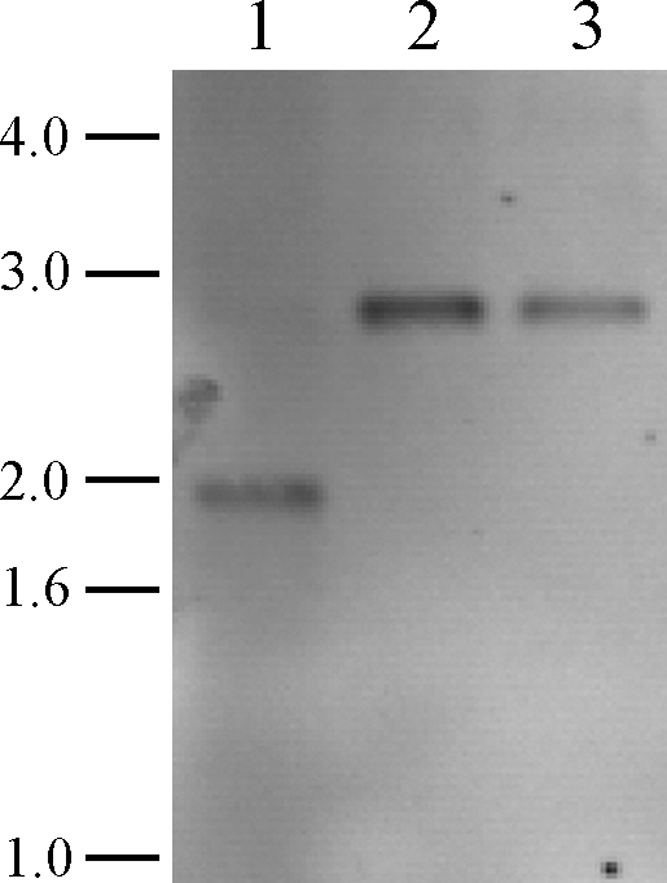
Southern analysis of S. scabies 87-22 hopanoid mutants. Chromosomal DNAs from the wild-type and Δhop615-1 and Δhop615-7 mutant strains were digested with PstI and after transfer to nylon membrane were probed with a PCR-generated dioxigenin-11-dUTP-labeled fragment upstream of SCAB13001. Hybridization of the probe to wild-type and Δhop615-1 and Δhop615-7 DNAs resulted in visualization of the anticipated 1,888-bp (wild-type) and 2,716-bp (mutant) bands. Sizes of the DNA markers are indicated to the left in kb. Lanes: 1, wild-type S. scabies 87-22; 2, Δhop615-1 mutant; 3, Δhop615-7 mutant.
FIG. 6.
Deletion of the S. scabies 87-22 HBGC abolishes hopene production. The panels show the extracted ion chromatograms from LC/MRM analysis. cps, counts per second.
Hopanoids are not required for normal morphological differentiation or tolerance of environmental stresses.
Since there is a link between hopanoids and aerial growth of S. scabies and hopanoids are produced during the transition from vegetative to aerial growth in S. coelicolor (29), we analyzed the importance of hopanoids for aerial hypha formation by S. scabies. The production of aerial mycelia by the Δhop615-1 and Δhop615-7 mutant strains on ISP4 medium was indistinguishable from that by the wild-type strain (Fig. 7), suggesting that hopanoids are not required for aerial hypha growth. To ensure that this result was medium independent, we analyzed growth on a variety of solid media, including mannitol-soya flour medium, glucose MM, thaxtomin defined agar, and R2, all of which yielded indistinguishable aerial hypha growth for the wild-type and Δhop mutant strains. In addition, the enumeration of spores formed by the wild-type and the Δhop615-1 and Δhop615-7 mutant strains indicated that all three strains produced similar numbers of spores (108 spores per plate for ISP4), suggesting that hopanoids are not required for spore formation or viability in S. scabies.
FIG. 7.
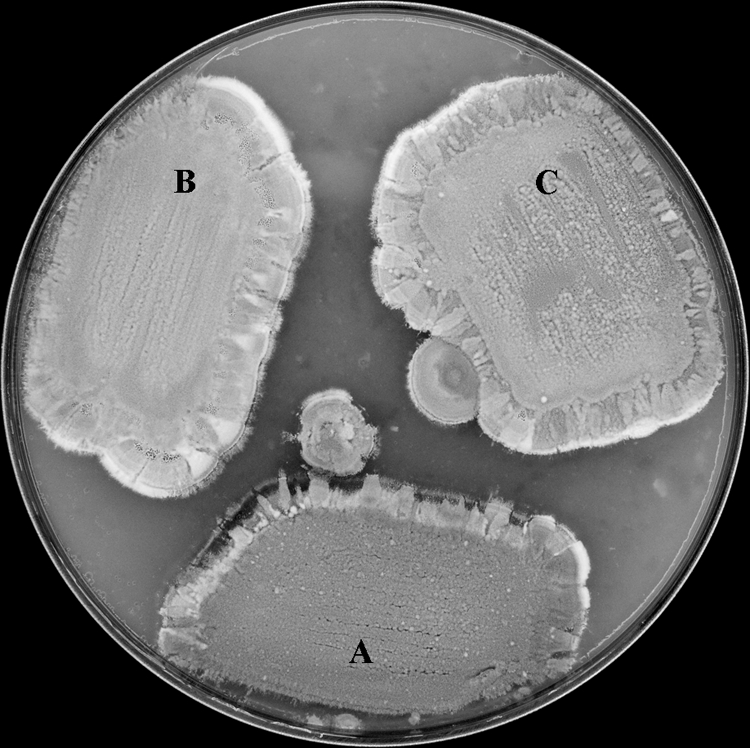
Hopanoids are not required for normal development of S. scabies 87-22. (A) Wild type; (B) Δhop615-1 mutant; (C) Δhop615-7 mutant. Strains were grown on ISP4 medium at 28°C for 7 days.
Since hopanoids are hypothesized to reduce the passive diffusion of ions across the plasma membrane, analogous to sterols in eukaryotic cells (25, 28), we analyzed whether hopanoids are important for tolerance of osmotic stress in S. scabies. We grew the S. scabies wild-type and Δhop615-1 and Δhop615-7 mutant strains on ISP4 medium supplemented with KCl (0.0 to 0.7 M), and aerial growth of all three strains was identical at all concentrations of KCl (data not shown). Sporulation of both wild-type and mutant strains decreased at KCl concentrations of above 0.3 M, with sparse growth of aerial hyphae in the presence of 0.7 M KCl. These data suggest that hopanoids are not important for tolerance of osmotic stress in S. scabies.
Since some bacteria are known to modulate their hopanoid content during growth at high temperature or low pH (17, 30), we analyzed whether hopanoids in S. scabies are important for growth during these conditions. We assessed growth of the S. scabies wild-type and Δhop615-1 and Δhop615-7 mutant strains on ISP4 medium at either 28°C, 37°C, or 42°C. Growth of wild-type S. scabies and the two Δhop mutants was indistinguishable at 28°C or 37°C (data not shown), and all three strains failed to grow at 42°C, even when the strains were allowed a 1-day preincubation at 28°C. To analyze the importance of hopanoids in S. scabies for growth under low-pH conditions, we incubated the wild-type and Δhop615-1 and Δhop615-7 mutant strains on ISP4 medium adjusted to different pH levels (pH 5.0 to 7.3) (S. scabies does not grow at a pH of <5.0 [21]) and found that the three S. scabies strains grew at the same rate at each pH level tested. Sporulation of aerial hyphae was less abundant as the pH decreased; however, this trend was observed for both wild-type S. scabies and the mutants. Taken together, these results indicate that the S. scabies hopanoids are not essential for growth under high-temperature conditions and are not required for tolerance of acidic conditions.
There is conflicting evidence regarding modulation of membrane hopanoid content in the presence of ethanol in bacteria (3, 10, 23, 37). In order to assess the possibility that hopanoids are important for ethanol tolerance in S. scabies, we grew wild-type S. scabies or the Δhop615-1 and Δhop615-7 mutant strains in liquid MM supplemented with ethanol (1 to 10%, vol/vol). Growth of the wild-type and Δhop615-1 and Δhop615-7 mutant strains was identical at ethanol concentrations of ≤4% after 7 days, whereas all S. scabies strains failed to grow at ethanol concentrations of >4%.
Since transcription of a SCAB13001 ortholog, SCO6759, was induced by oxidative stress (26) and a polycyclic terpenoid compound from Bacillus subtilis was involved in oxidative stress protection (2), we hypothesized that S. scabies hopanoids may be involved in oxidative stress tolerance. To assess this possibility, we performed a disc diffusion assay with 0.3 μmol, 1.5 μmol, and 3.0 μmol of hydrogen peroxide. All hydrogen peroxide concentrations produced a circular zone of inhibited growth and did so in a dose-dependent manner (data not shown). There was not a difference in the diameter of the zone of inhibition between the wild-type and Δhop615-1 and Δhop615-7 mutant strains, suggesting that S. scabies hopanoids are not required for tolerance to oxidative stress.
DISCUSSION
Sterols are ubiquitous in eukaryotes and are required for normal cell function (6, 31). However, with few exceptions, these molecules are absent in bacteria. The similarity in structure between sterols and hopanoids and the presence of hopanoids in the membranes of many bacterial species support a long-standing hypothesis that hopanoids function for bacteria as sterols do for eukaryotes and predict that hopanoids are required for normal growth. However, this hypothesis has not been explicitly tested until now. Here we demonstrate that hopanoids, though conserved in the filamentous genus Streptomyces, are not required for growth in liquid or solid media or for development of aerial hyphae or sporulation. Furthermore, we were unable to demonstrate a role of hopanoids in tolerance of environmental stresses, including ethanol, osmotic and oxidative stress, high temperature, or low pH.
The presence of an HBGC in the potato scab pathogen S. scabies is not surprising, given that hopanoids are produced by other Streptomyces spp. (29, 33), and the genome sequences of S. griseus (24) and S. avermitilis (13) contain putative HBGCs. Previous work with S. coelicolor demonstrated a link between hopanoids and development (29). A link between hopanoids and development also exists in S. scabies, as the SCAB13001-xylE reporter strain showed higher XylE activity during aerial growth than during vegetative growth. The precise relationship between hopanoids and development is unclear but is possibly related to structural changes in the cell membrane induced by the formation of aerial hyphae.
Our LC/MRM analysis showed that S. scabies produces hopene during submerged growth in liquid MM and during surface growth on ISP4 medium. These data suggest that hopanoid production in S. scabies might not be lifestyle dependent (i.e., surface growth versus submerged growth), which is seemingly the case for S. coelicolor (29). Direct comparison of our work with work previously conducted with S. coelicolor is difficult because different growth media were used. Curiously, we were unable to detect the other predicted hopanoid, aminotrihydroxybacteriohopane, during growth in liquid MM or on ISP4; however, its detection was complicated by lack of a suitable standard. It is possible that aminotrihydroxybacteriohopane may not be produced by S. scabies under these growth conditions or, alternatively, that the method of analysis used is not suitable for its detection. SCAB12901, which encodes a putative aminotransferase that is presumably involved in addition of the amino group present in aminotrihydroxybacteriohopane, is transcribed during growth in liquid MM (data not shown).
The nonessential role of hopanoids in S. scabies is inconsistent with reduced viability of hopanoid-producing bacteria treated with squalene-cyclase inhibitors; however, Streptomyces spp. were not analyzed in that study (7). Hopanoids may play a more important role in normal growth of the bacteria tested in that study than in S. scabies, or squalene-cyclase inhibitors may have uncharacterized toxic effects on some bacteria. Our data demonstrate that the Δhop615-1 and Δhop615-7 mutant strains are not compromised in their tolerance to ethanol, which is in agreement with the most recent reports regarding modulation of membrane hopanoid content in response to ethanol in Zymomonas mobilis (10, 23). The Δhop615-1 and Δhop615-7 mutant strains are also not compromised in their tolerance to osmotic stress, high temperature, or low pH under the conditions tested. Other studies suggest that some bacteria may modulate their hopanoid content in response to these conditions (17, 30). It has been postulated the condensing effect of hopanoids on cell membranes is required to minimize diffusion of water out of the cytoplasm while hyphae grow away from vegetative mycelium into the air (29). Our data do not support this hypothesis, as the Δhop615-1 and Δhop615-7 mutant strains form aerial hyphae indistinguishable from those of the wild-type strain. The severity of osmotic stress experienced during aerial growth remains to be determined, and the strategies that streptomycetes employ to cope with this stress still need to be elucidated. As previously suggested, uptake of compatible solutes could alleviate this stress (29).
Curiously, deletion of the HBGC did not result in an increased sensitivity to oxidative stress. This was surprising, because in S. coelicolor a hopanoid biosynthetic gene (SCO6759) was upregulated in response to oxidative stress (26), and a polycyclic terpenoid from B. subtilis was involved in oxidative stress protection (2). Increased transcription of SCO6759 in response to oxidative stress is correlative and does not indicate that hopanoids are involved in oxidative stress protection in S. coelicolor. However, the polycyclic terpenoid involved in oxidative stress protection in B. subtilis differs significantly from that of hopene produced by S. scabies; the fifth ring in the B. subtilis metabolite is open, and there is a methylphenyl that terminates the side chain (2). The fact that hopene from S. scabies is not involved in oxidative stress protection could potentially be explained by these structural differences.
The presence of hopanoids in many, but not all, bacteria (33) is consistent with an important but nonessential function. However, the presence of an HBGC in phylogenetically diverse streptomycetes suggests that hopanoids have an important role in streptomycete physiology. We were unable to demonstrate an essential role of hopanoids in normal growth or tolerance to ethanol or other environmental stresses. In light of these data, the long-standing hypothesis that hopanoids are important components of bacterial membranes must be reevaluated. It is important to note that our analysis does not discount the possibility that hopanoids are important for tolerance to cold temperatures or are important for tolerance to desiccation or survival of Streptomyces spp. in a soil environment, which will be the subject of future investigation. Besides their role in enhancing membrane integrity in eukaryotes, sterols are components of lipid microdomains or lipid rafts, which are important for membrane protein localization and function (27). Lipid microdomains occur in bacteria and are involved in recruitment of membrane proteins and in cell division (22). It is possible that hopanoids play a role in the formation of lipid microdomains in bacteria as sterols do in eukaryotes.
Acknowledgments
We thank Gregg S. Pettis for kindly providing pXE4 plasmid DNA and Sheng Zhang and Robert Sherwood at the Cornell Biotechnology Resource Center for performing LC/MRM analysis. We also thank Dawn R. D. Bignell and Danny Vereecke for helpful suggestions concerning the manuscript.
This project was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service, grant no. 2008-35319-19202. Financial support was also provided to R.F.S. through the USDA Multidisciplinary Graduate Education Traineeship Program.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A.-M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra†, C. W. Chen§, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Bosak, T., R. M. Losick, and A. Pearson. 2008. A polycyclic terpenoid that alleviates oxidative stress. Proc. Natl. Acad. Sci. USA 105:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bringer, S., T. Hartner, K. Poralla, and H. Sahm. 1985. Influence of ethanol on the hopanoid content and fatty acid pattern in batch and continuous cultures of Zymomonas mobilis. Arch. Microbiol. 140:312-316. [Google Scholar]
- 4.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. 2001. Reglation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Eisenkolb, M., C. Zenzmaier, E. Leitner, and R. Schneiter. 2002. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol. Biol. Cell 13:4414-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flesch, G., and M. Rhomer. 1987. Growth inhibition of hopanoid synthesizing bacteria by squalene cyclase inhibitors. Arch. Microbiol. 147:100-104. [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartner, T., K. L. Straub, and E. Kannenberg. 2005. Occurrence of hopanoid lipids in anaerobic Geobacter species. FEMS Microbiol. Lett. 243:59-64. [DOI] [PubMed] [Google Scholar]
- 10.Hermans, M. A. F., B. Neuss, and H. Sahm. 1991. Content and composition of hopanoids in Zymomonas mobilis under various growth conditions. J. Bacteriol. 173:5592-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodgson, D. A. 1992. Differentiation in actinomycetes, p. 407-440. In S. Mohan, C. Down, and J. A. Cole (ed.), Prokaryotic structure and function: a new perspective. Cambridge University Press, Cambridge, United Kingdom.
- 12.Huang, J., J. Shi, V. Molle, B. Sohlberg, D. Weaver, M. J. Bibb, N. Karoonuthaisiri, C.-J. Lih, C. M. Kao, M. J. Buttner, and S. N. Cohen. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276-1287. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinrose, H. Kikuchi, T. Shiba, Y. Sakai, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 14.Ingram, C., M. Brawner, P. Youngman, and J. Westpheling. 1989. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 171:6617-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, E. G., M. V. Joshi, D. M. Gibson, and R. Loria. 2007. Cello-oligosaccharides released from host plants induce pathogenicity in scab-causing Streptomyces species. Physiol. Mol. Plant Pathol. 71:18-25. [Google Scholar]
- 16.Joshi, M. V., D. R. D. Bignell, E. G. Johnson, J. P. Sparks, D. M. Gibson, and R. Loria. 2007. The AraC/XylS regulator TxtR modulates thaxtomin biosynthesis and virulence in Streptomyces scabies. Mol. Microbiol. 66:633-642. [DOI] [PubMed] [Google Scholar]
- 17.Joyeux, C., S. Fouchard, P. Llopiz, and S. Neunlist. 2004. Influence of the temperature and the growth phase on the hopanoids and fatty acids content of Frateuria aurantia (DSMZ 6220). FEMS Microbiol. Ecol. 47:371-379. [DOI] [PubMed] [Google Scholar]
- 18.Kannenberg, E., K. Poralla, and A. Blume. 1980. A hopanoid from the thermo-acidophilic Bacillus acidocaldarius condenses membranes. Naturwissenschaften 67:458-459. [Google Scholar]
- 19.Kannenberg, E. L., and K. Poralla. 1999. Hopanoid biosynthesis and function in bacteria. Naturwissenschaften. 86:168-176. [Google Scholar]
- 20.Kieser, T., M. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 21.Lambert, D. H., and R. Loria. 1989. Streptomyces scabies sp. nov., nom. rev. Int. J. Syst. Bacteriol. 39:387-392. [Google Scholar]
- 22.Matsumoto, K., J. Kusaka, A. Nishibori, and H. Hara. 2006. Lipid domains in bacterial membranes. Mol. Microbiol. 61:1110-1117. [DOI] [PubMed] [Google Scholar]
- 23.Moreau, R. A., M. J. Powell, W. F. Fett, and B. D. Whitaker. 1997. The effect of ethanol and oxygen on the growth of Zymomonas mobilis and the levels of hopanoids and other membrane lipids. Curr. Microbiol. 35:124-128. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ourisson, G., and M. Rohmer. 1992. Hopanoids. 2. Biohopanoids: a novel class of bacterial lipids. Acc. Chem. Res. 25:403-408. [Google Scholar]
- 26.Paget, M. S. B., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 27.Panwar, S. L., R. Parsija, and R. Prasad. 2008. Membrane homoeostasis and multidrug resistance in yeast. Biosci. Rep. 28:217-228. [DOI] [PubMed] [Google Scholar]
- 28.Poralla, K., E. Kannenberg, and A. Blume. 1980. A glycolipid containing hopane isolated from the acidophilic, thermophilic Bacillus acidocaldarius, has a cholesterol-like function in membranes. FEBS Lett. 113:107-110. [DOI] [PubMed] [Google Scholar]
- 29.Poralla, K., G. Muth, and T. Hartner. 2000. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189:93-95. [DOI] [PubMed] [Google Scholar]
- 30.Poralla, K., T. Hartner, and E. Kannenberg. 1984. Effect of temperature and pH on the hopanoid content of Bacillus acidocaldarius. FEMS Microbiol. Lett. 23:253-256. [Google Scholar]
- 31.Rodriguez, R. J., F. R. Taylor, and L. W. Parks. 1982. A requirement for ergosterol to permit growth of yeast sterol auxotrophs on cholestanol. Biochem. Biophys. Res. Commun. 106:435-441. [DOI] [PubMed] [Google Scholar]
- 32.Rohmer, M. 1999. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16:565-574. [DOI] [PubMed] [Google Scholar]
- 33.Rohmer, M., P. Bouvier-Nave, and G. Ourisson. 1984. Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 130:1137-1150. [Google Scholar]
- 34.Sahm, H., M. Rohmer, S. Bringer-Meyer, G. A. Sprenger, and R. Welle. 1993. Biochemistry and physiology of hopanoids in bacteria. Adv. Microb. Physiol. 35:247-273. [DOI] [PubMed] [Google Scholar]
- 35.Sala-Trepat, J. M., and W. C. Evans. 1971. The meta cleavage of catechol by Azotobacter species 4-oxalocrotonate pathway. Eur. J. Biochem. 20:400-413. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Schmidt, A., S. Bringer-Meyer, K. Poralla, and H. Sahm. 1986. Effect of alcohols and temperature on the hopanoid content of Zymomonas mobilis. Appl. Microbiol. Biotechnol. 25:32-36. [Google Scholar]
- 38.Schouten, S., J. P. Bowman, W. I. C. Rijpstra, and J. S. Sinninghe Damste. 2000. Sterols in a psychrophilic methanotroph, Methylosphaera hansonii. FEMS Microbiol. Lett. 186:193-195. [DOI] [PubMed] [Google Scholar]
- 39.Volkman, J. K. 2003. Sterols in microorganisms. Appl. Microbiol. Biotechnol. 60:495-506. [DOI] [PubMed] [Google Scholar]