Abstract
The bacterial flagellar motor is a remarkable nanomachine that provides motility through flagellar rotation. Prior structural studies have revealed the stunning complexity of the purified rotor and C-ring assemblies from flagellar motors. In this study, we used high-throughput cryo-electron tomography and image analysis of intact Borrelia burgdorferi to produce a three-dimensional (3-D) model of the in situ flagellar motor without imposing rotational symmetry. Structural details of B. burgdorferi, including a layer of outer surface proteins, were clearly visible in the resulting 3-D reconstructions. By averaging the 3-D images of ∼1,280 flagellar motors, a ∼3.5-nm-resolution model of the stator and rotor structures was obtained. flgI transposon mutants lacked a torus-shaped structure attached to the flagellar rod, establishing the structural location of the spirochetal P ring. Treatment of intact organisms with the nonionic detergent NP-40 resulted in dissolution of the outermost portion of the motor structure and the C ring, providing insight into the in situ arrangement of the stator and rotor structures. Structural elements associated with the stator followed the curvature of the cytoplasmic membrane. The rotor and the C ring also exhibited angular flexion, resulting in a slight narrowing of both structures in the direction perpendicular to the cell axis. These results indicate an inherent flexibility in the rotor-stator interaction. The FliG switching and energizing component likely provides much of the flexibility needed to maintain the interaction between the curved stator and the relatively symmetrical rotor/C-ring assembly during flagellar rotation.
Flagellum-based motility plays a critical role in the biology and pathogenesis of many bacteria (3, 6, 17, 31). The well-conserved flagellum is commonly divided into three physical parts: the flagellar motor, the helically shaped flagellar filament, and the hook which provides a universal joint between the motor and the filament. In most bacteria, counterclockwise rotation of the flagella results in bundling of the helical flagella and propulsion of the cell through liquid or viscous environments. Clockwise rotation of the flagellar motor results in random turning of the cell with little translational motion (“tumbling”). Bacterial motility is thus a zigzag pattern of runs and tumbles, in which chemotactic signals favor running toward attractants and away from repellents (3).
Borrelia burgdorferi and other closely related spirochetes are the causative agents of Lyme disease, which is transmitted to humans via infected Ixodes ticks (40). Spirochetes have a distinctive morphology in that the flagella are enclosed within the outer membrane sheath and are thus called periplasmic flagella (6). The flagellar motors are located at both ends of the cell and are coordinated to rotate in opposite directions during translational motion and in the same direction (i.e., both clockwise or both counterclockwise) during the spirochete equivalent of tumbling, called “flexing” (6, 15). Spirochetes are also capable of reversing translational motion by coordinated reversal of the direction of motor rotation at both ends of the cell. Rotation of the flagella causes a serpentine movement of the entire cell body, allowing B. burgdorferi to efficiently bore its way through tissue and disseminate throughout the mammalian host, resulting in manifestations in the joints, nervous system, and heart (40).
The flagellar motor is an extraordinary nanomachine powered by the electrochemical potential of specific ions across the cytoplasmic membrane (3). Current knowledge of the flagellar motor structure and rotational mechanisms is based primarily on studies of Escherichia coli and Salmonella enterica and is summarized in several recent comprehensive reviews (3, 22, 31, 39, 42). The flagellar motor is constructed from at least 20 different kinds of proteins. The approximate location of these flagellar proteins has been determined by a variety of approaches and appears to be relatively consistent in a wide variety of bacteria. It can be divided into several morphological domains: the MS ring (FliF, the base for the flagellar motor); the C ring (FliG, FliM, and FliN, the switch complex regulating motor rotation); the export apparatus (multiple-protein complex located at the cytoplasmic side of the MS ring); the rod (connecting the MS ring and the hook); the L and P rings on the rod (thought to serve as bushings at the outer membrane and at the peptidoglycan layer, respectively); and the stator, which is the motor force generator embedded in the cytoplasmic membrane. Electron microscopy studies of the purified flagellar motor have provided a detailed view of the rotor/C-ring assembly (11, 44). However, there is no structural information on the stator and the export apparatus in these reconstructions, because these membrane-associated structures are not retained following detergent extraction during the extensive basal body purification process. The stator and the export apparatus were visualized by using freeze fracture preparations of cytoplasmic membranes. It appears that 10 to 16 stator units form circular arrays in the membrane (9, 20). Part of the export apparatus is located in the central space of the C ring (18). Recently a 7-nm-resolution structure of the intact flagellar motor in situ was revealed by averaging 20 structures obtained using cryo-electron tomography (cryo-ET) of Treponema primitia cells (32). Further analysis of the intact flagellar motor structure would lead to a better understanding of the motor protein distribution, the rotor-stator interaction, and the mechanism of bacterial motility.
Cryo-ET has emerged as a three-dimensional (3-D) imaging technique to bridge the information gap between X-ray crystallographic and optical microscopic methods (24, 30). This process involves rapidly freezing viable cells, collecting a series of electron micrographs at different angles, and computationally combining the resulting images into a 3-D density map. Cryo-ET allows investigation of the structure-function relationship of molecular complexes and supramolecular assemblies in their cellular environments without fixation, dehydration, embedding, or sectioning artifacts. Spirochetes are well suited for cryo-ET analysis because of their narrow cell diameter (typically 0.2 to 0.3 μm). Recently the cellular architecture of Treponema primitia, Treponema denticola, and B. burgdorferi, as well as the configuration of the B. burgdorferi periplasmic flagella, were revealed by cryo-ET (7, 16, 26, 33). In combination with advanced computational methods, cryo-ET is currently the most promising approach for determining the cellular architecture in situ at molecular resolution (30). We have developed novel strategies for capturing and averaging thousands of 3-D images of large macromolecular assemblies to obtain ∼2.0-nm-resolution structures (28, 29).
In this study, we present the molecular structures of infectious wild-type (WT) and mutant B. burgdorferi organisms and their flagellar motors in situ using high-throughput cryo-ET and 3-D image analysis. By averaging subvolumes of 1,280 flagellar motors from 322 cells, we obtained a ∼3.5-nm-resolution model of the intact flagellar motor, providing a detailed view of rotor-stator interactions. In addition, detergent treatment of intact cells provided a preliminary identification of the rotor and stator structures. Through the comparison of WT and mutant cells, we have also determined the location of the flgI gene product in the B. burgdorferi flagellar motor.
MATERIALS AND METHODS
Cell preparation.
B. burgdorferi B31 5A18NP1 (19) was grown to late log phase in a humidified incubator at 37°C, 3% CO2, in Barbour-Stoenner-Kelly medium without gelatin (BSK II) prepared in-house. In most studies, organisms in late log phase (∼1 × 108/ml) were centrifuged in 1.5-ml tubes at 5,000 × g for 5 min and the resulting pellet rinsed gently with phosphate-buffed saline (PBS) prior to resuspension in 50 to 100 μl PBS (final concentration, ∼2 × 109 cells/ml). This simple wash procedure provided better contrast for cryo-ET by removing serum proteins that contribute to background “noise” while maintaining the structural integrity of the cell. For comparison, we also collected data from bacteria resuspended in BSK II medium. In detergent treatment studies, the pellet was gently suspended in 1% (vol/vol) nonionic detergent (Nonidet P-40 [NP-40], 4-nonylphenyl-polyethyleneglycol; Sigma Aldrich) in 100 μl PBS; following incubation at room temperature for 20 min, the organisms were centrifuged again at 5,000 × g for 5 min and the resulting pellet was suspended in 10 μl PBS. Proteinase K treatment of intact cells was carried out for 40 min as described previously (49).
Transposon mutants.
B. burgdorferi 5A18NP1 was subjected to transposon mutagenesis by a modification of the procedure described by Stewart et al. (41). The plasmid pMarGentKan was graciously provided by P. E. Stewart of the Rocky Mountain Laboratories, National Institutes of Health, Hamilton, MT. This version of pMarGent contains a kanamycin resistance cassette with a B. burgdorferi flaB promoter (flaB::aph1) in the “backbone” of the vector, outside the himar1-based transposable element (P. E. Stewart, unpublished data). pMarGentKan was further modified by inclusion of a 7-bp signature tag in the region between the ColE1 origin of replication and one of inverted repeats demarcating the transposable element (T. Lin., L. Gao, and S. J. Norris, unpublished data). Electroporation, recovery of transposon mutants, and precise determination of the insertion sites by plasmid rescue in E. coli were performed as described previously (4). The insertion sites of mutants T06TC173, T03TC042, and T09P01H08 were at nucleotides 813293, 813788, and 814017, respectively, in the B. burgdorferi B31 chromosome (NCBI reference sequence NC 001318.1). The morphology and motility of the mutants were examined by dark-field microscopy of B. burgdorferi cultured in BSK II medium and diluted 1:1 (vol/vol) with 2% methylcellulose in PBS to increase the medium viscosity. Organisms were observed using a CytoViva illumination system (Auburn, AL) associated with a Nikon E400 microscope.
Cryo-ET.
Aliquots of micron gold clusters were added to the resuspended viable B. burgdorferi samples as fiducial (reference) markers for image alignment (25). Four-μl samples were deposited on freshly glow-discharged holey carbon grids and rapidly frozen in liquid ethane maintained at −180°C, using a homemade gravity-driven plunger apparatus. Frozen specimens were imaged at liquid-nitrogen temperature using a Polara electron microscope (FEI Company) equipped with a 4K × 4K charge-coupled-device (16-megapixel) camera (TVIPS; GMBH, Gauting, Germany). The microscope was operated at 300 kV and magnification ×39,000, resulting in an effective pixel size of 4.5 Å after 2 × 2 binning. Using the FEI “batch tomography” software program, low-dose single-axis-tilt series were collected from each cell at −4- to −6-μm defocus with a cumulative dose of ∼100 e−/Å2 spread over 131 images. Tilt angles were in the range of −65° and +65°. Consecutively tilted images were aligned with respect to each other using fiducial markers and the IMOD software package (25). After further refinement using projection matching, 3-D cryo-tomograms were reconstructed using weighted back-projection implemented in the software package Protomo (46). In total, 322 cryo-tomograms were generated from 5A18NP1 cells, 281 from transposon mutants, 96 from 5A18NP1 cells treated with the nonionic detergent NP-40, and 68 from 5A18NP1 cells treated with proteinase K for 40 min.
3-D classification and averaging.
The subvolume processing of flagellar motors was carried out as follows. First, the positions and orientations of the flagellar motors in each tomogram were determined manually. For this purpose, three points were marked for each flagellum: the center of the basal body, the turning point of the hook, and a point on the flagellar filament. Based on the x, y, and z coordinates of these three points, the orientation of the flagellar motor was calculated and used as an initial estimate of the alignment parameters, which were subsequently refined iteratively.
After multiple cycles of alignment and averaging, a structure with 16-fold symmetry emerged. It should be noted that no symmetry was imposed during this process. To validate this result, which was obtained by the alignment to a single reference, several cycles of a reference-free alignment procedure were applied subsequently (47). One cycle of the procedure consisted of a classification step, in which the data set was separated into 20 classes. The class averages were then aligned with respect to each other and the alignment transformations of each class applied to the raw subvolumes of the respective members of the class, after which a new cycle was started. This procedure avoids the cross-correlation of subvolumes with a very low signal-to-noise ratio to references with a higher signal-to-noise ratio and thus increases the robustness of the alignment. If the motors occur in different orientations relative to the original tomograms, the missing wedge will also be filled with data due to the various orientations of the averaged subvolumes. The procedure also minimizes a potential reference bias, because the references are not subjectively selected but rather are dynamically created in the classification step.
Approximately 1,280 flagellar motors from 5A18NP1 cells, 1,100 from transposon mutants, and 454 from detergent-treated cells were used for the alignment, classification, and averaging. A difference map was calculated to quantify the location and shape of the protein FlgI by using the Protomo package.
Rotational symmetry of the flagellar motor.
The rotational symmetry of the motor was assessed by harmonic analysis (10). The subvolume average was resampled in cylindrical coordinates and then expanded into circular harmonics on planes perpendicular to the cylinder axis. The moduli of the coefficients are represented in Fig. S2 in the supplemental material as a function of the radius (perpendicular to the cylinder axis) and the z coordinate (along the cylinder axis) for n-fold symmetry, where n is in the range from 0 to 37. This approach allows discrimination of regions of the motor that possess strong or weak symmetries as well as the periodicity of symmetry.
Visualization of the flagellar motor.
3-D tomograms were visualized and saved as images using IMOD (25). Surface rendering of 3-D maps was carried out using the software package UCSF Chimera (35). Watershed segmentation (45) was used to computationally separate the rotor, stator, C ring, and export apparatus from the intact flagellar motor. A model of the C ring (including the approximate locations of the three rotor proteins FliG, FliM, and FliN) was constructed based on the electron microscopy map.
RESULTS
Molecular details of intact B. burgdorferi.
In this study, we utilized a relatively high magnification (×39,000), a 4K × 4K charge-coupled-device camera, and 2 × 2 binning of pixels during the acquisition and processing of the micrographs. In addition, high-throughput methodologies were applied in both the acquisition and processing stages, permitting a relatively rapid collection of a large number of high-quality images. The comparatively small diameter of B. burgdorferi relative to that of other bacterial species provides reasonable contrast even at high tilted angles, which results in increased contributions of the high-angle micrographs to the tomographic reconstruction.
5A18NP1 is an infectious, transformable derivative of B. burgdorferi B31 (19); it is WT with regard to the flagellar structure and for simplicity is referred to as WT in this article. Suspensions of organisms were rapidly frozen on electron microscopy grids and were visualized by cryo-ET. B. burgdorferi has an unusual helical to flat plane wave morphology, as shown in dark-field imaging (Fig. 1A). A representative image from a B. burgdorferi cell obtained using cryo-ET (Fig. 1B) shows that the outer membrane sheath, the inner membrane, and a thick peptidoglycan layer are readily visible, as are the flagella that wrap around the ribosome-studded cell cylinder in the periplasmic space. The flagellar filaments extend from each flagellar motor and are clearly localized between the peptidoglycan layer and the outer membrane (Fig. 1B). The cells taper from an approximate diameter of 0.3 μm to 0.2 μm near the end of the cell. High-magnification views of the outer membrane indicate that the inner and outer leaflets of the lipid bilayer (4.0 nm in thickness) can be resolved in the image (Fig. 1C). In addition, an extra layer of density was observed on the external surface of the outer membrane. This layer was no longer visible following surface proteolysis of intact cells using proteinase K (see Fig. S1 in the supplemental material), indicating that it represents outer surface proteins of B. burgdorferi.
FIG. 1.
Molecular structures of intact B. burgdorferi. A dark-field image shows the typical helical to flat plane wave morphology (A). Scale bar, 1 μm. The tip of the cell outlined inside the small box at the top was imaged at high magnification by cryo-ET and is shown in panel B. The inner membrane (IM), the outer membrane (OM), a thick peptidoglycan layer (PG), and the periplasmic flagella are easily discernible in this representative slice of a cryo-ET reconstruction (B). The outer surface proteins (OSP) form an extra layer of density on the external surface of the outer membrane. The high-magnification views of the outer membrane indicate that the inner and outer leaflets of the lipid bilayer (4.0 nm in thickness) can be resolved in these images (C). In addition, an extra layer of density corresponding to OSP was observed on the outer surface of the lipid bilayer. At this magnification, inner and outer rings are also discernible in the flagellar motor structure underlying the flagellar hook (C).
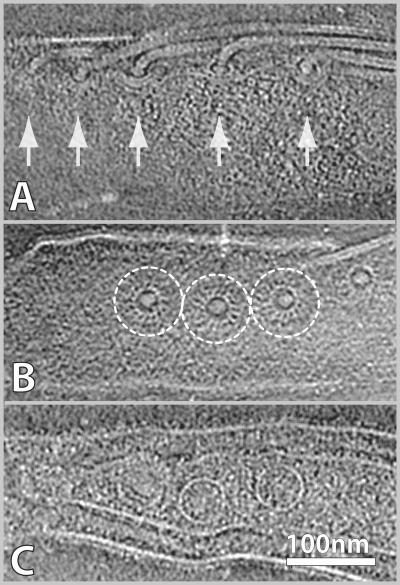
In this study, we focused on the flagellar motors, which are located at each end of the bacterium (Fig. 2). On average, about four flagellar motors were visible in the field of view (∼0.9 by 0.9 μm) of each cryo-ET 3-D reconstruction. Considerable detail is visible in cryo-ET views of flagellar motors (Fig. 2). Multiple flagellar motors are displayed in the side view (Fig. 2A); the characteristic bowl-shaped structure surrounding the rod is clearly visible in some sections. The turbine-like radial arrangement of the motor is apparent in the top view (Fig. 2B). In addition, the putative C rings are visible in the cytoplasmic space (Fig. 2C).
FIG. 2.
Cryo-ET sections of B. burgdorferi showing side and top views of multiple flagellar motors. (A) Central and tangential sections of five motors with hook and filament assemblies. The bowl-shaped rotor and P ring can be discerned in some views. (B) The turbine-like radial arrangement of three motors is visible in a top view. (C) Lower cryo-ET section of the same cell shown in panel A, showing putative C rings in the cytoplasm.
Molecular architecture of the intact flagellar motor.
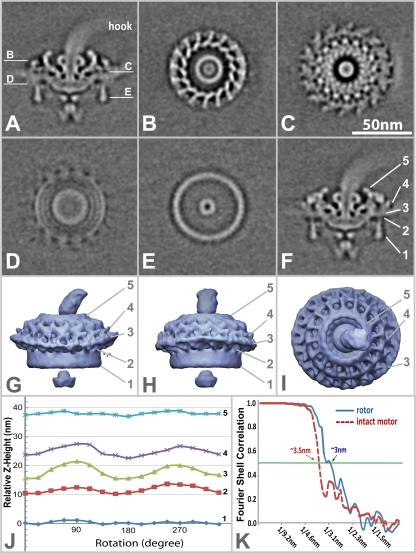
A 3-D reconstruction (Fig. 3) of the intact flagellar motor was obtained after 90 cycles of alignment and averaging of 3-D density maps of ∼1,280 flagellar motors. No symmetry was imposed during the processing and the construction of the model. Two central sections of the flagellar motor observed at different radial angles show the locations of the putative hook, rod, rotor, stator, C ring, and export apparatus structures (Fig. 3A and 3F; see also Movie S1 in the supplemental material). The hook structure is visible at the top, attached to the central rod. Its indistinct appearance in the reconstruction most likely reflects variability of the hook position relative to the motor. The rod has a lower density in the middle, which is consistent with the presence of a central channel, as seen in purified rotor-C-ring complexes (44). The distal portion of the rod is curved slightly in the direction of the hook (Fig. 3A). A hollow torus-shaped structure is present near the rod-hook junction (Fig. 3A, B, and F). The 16-fold symmetry of the outer portion of the upper region of the motor is clearly demarcated in Fig. 3B, C, and I. The radial extrusions of the motor are angled like the blades of a turbine. Most structural elements extend outward from the center in a clockwise direction when viewed from the top (Fig. 3B and C). The 16-fold symmetry is no longer dominant at the level of the putative MS ring (Fig. 3D), yielding to the appearance of concentric rings. The most central ring in this view is the edge of the MS ring, and the outermost ring is in the vicinity of the stator-C-ring interface. The two concentric rings in between the MS-ring and C-ring structures are thought to represent aspects of the interaction between the MS ring, the C ring, and the stator and may represent FliG domains; corresponding regions of density connecting the MS ring and the C ring are also visible at level D of the central sections in Fig. 3A and F. While the outermost ring in Fig. 3D retains some degree of 16-fold symmetry (most likely due to the stator), weak periodicity of the middle two rings is suggestive of higher orders of symmetry (∼32-fold). Figure 3E shows the uniform appearance of the C ring. The centrally located export apparatus is visible on the cytoplasmic side of the MS ring (Fig. 3A, E, and F). The B. burgdorferi C ring appears very similar to the C rings of E. coli and Salmonella but with a larger diameter (∼57 nm compared to 45 nm). The accuracy of this asymmetric, averaged model of the flagellar motor is reinforced by the similarity to individual motors in cryo-ET tomograms (Fig. 2).
FIG. 3.
Molecular architecture of the intact flagellar motor. Center sections of an unsymmetrized 3-D average of the flagellar motor are shown in panels A (0° relative to the direction of the filament) and F (10°). Panels B, C, E, and F are horizontal cross sections through a flagellar motor, and the locations of the sections are indicated in panel A. Section B is located near the top of the motor, and the central rod, the P ring, and the uppermost portion of the rotor are visible. Section C transects the middle of the rotor body and the stator, just above the cytoplasmic membrane. Section D is at the level of the cytoplasmic side of the MS ring and the top of the C ring. Two ring-like features are visible between the MS ring and the C ring in this section (C). Density consistent with the bottom of the C ring is visible in section E. A portion of the export apparatus is visible in the center of the C ring. The surface rendering of the 3-D average is displayed in three different orientations as shown in panels G, H, and I. Panel G is the side view of motor, with the flagellar hook pointing to the right (0° view). Panel H is the view along the cell cylinder from the tip of the cell (90° view). The top view of the flagellar motor is shown in panel I. Panel J shows the positions of five key structural features (indicated in panel H) traced along a 360° rotation, relative to the bottom of the C ring at 0°. Panel K represents the Fourier shell correlation calculated for the intact asymmetric motor (red curve) and the central portion (putative rotor) of the motor (blue curve), indicating 3.5-nm and 3.0-nm resolutions, respectively. For additional sections and enlarged views of the flagellar motor structure, see Fig. S4 and PowerPoint file S1 in the supplemental material.
Surface views of the B. burgdorferi flagellar motor are represented in Fig. 3G, H, and I. An unexpected finding of this study was that the most external portion of the motor exhibits curvature corresponding to that of the cytoplasmic membrane. The vertical locations of points 1 through 5 in Fig. 3F, G, and H were plotted relative to the bottom of the C ring at a constant radius relative to the motor axis. The top of the motor (point 5) and the bottom of the C ring (point 1) remain relatively constant, indicating very little vertical change around the motor. However, there is an obvious vertical modulation of three structural features: the putative stator at the level of the membrane (point 3) and the structural features on both sides of the membrane (points 2 and 4). Additional views of this curvature are provided in the supplemental material (see PowerPoint file S1 in the supplemental material). Thus, the 16 putative stator units are symmetrically distributed around the motor but are vertically displaced by up to 6 nm relative to one another. This finding provides evidence that the portion of the motor representing the proposed stator follows the curvature of the inner membrane. This is consistent with the symmetry analysis (see Fig. S2 in the supplemental material), which shows a weak 16-fold symmetry in the stator region. The stator is most affected by the curvature modulation, while the most pronounced symmetry is located near the collar portion of the rotor, which is least affected by the vertical modulation.
Flagellar motor from detergent-treated cells.
Following treatment with the nonionic detergent NP-40, B. burgdorferi cells maintained their overall shape but became nonmotile and appeared relatively transparent (“ghostlike”) by dark-field microscopy. Cryo-ET images show that the cells have no outer membranes, and the internal macromolecular complexes, such as ribosomes, are more visible (see Fig. S3A in the supplemental material). Although the periplasmic flagella are no longer closely associated with the cell cylinder, the flagellar motors are still attached to the cell envelope. By averaging of 454 basal bodies from 96 tomograms, we obtained the 3-D structure of the motor after detergent treatment (Fig. 4). The difference between the motor structure before and after detergent treatment is striking (Fig. 4; see also Fig. S3 and S4 in the supplemental material). The remaining structure is 45 nm in diameter and closely resembles the central portion of the intact flagellar motor. We propose that this structure represents the NP-40-insoluble core of the B. burgdorferi rotor. It is much larger than the well-characterized basal body purified from Salmonella but is similar to the central region of the Treponema primitia flagellar motor (32). The putative rotor has a bowl-like structure and is reinforced by internal “truss-like” elements that are visible as thin arms of density in central sections (Fig. 4C and E). The flagellar rod sits in the middle of the rotor and interacts with the MS ring at the bottom of the bowl. There is an overall striking similarity but also noticeable differences in the putative rotor structures before and after the detergent treatment, as observed by comparing Fig. 4E and the corresponding structure colored green in Fig. 4H. In particular, there is a groove in the detergent-treated structure that is filled in the intact motor; in addition, a thin isthmus of density extends from the C ring to the same region of the rotor (Fig. 4H and I). We postulate that together these regions of density represent the N-terminal portion of FliG polypeptides, interconnecting the rotor and the C ring. The torus-shaped structure associated with the distal rod is virtually absent following NP-40 treatment (compare Fig. 4A and C), indicating that it is labile in the presence of detergent. A portion of the putative export apparatus is still visible below the MS ring following detergent treatment (Fig. 4E and F).
FIG. 4.
Comparison of the B. burgdorferi flagellar motor before (A and B) and after (C and D) treatment with the nonionic detergent NP-40. All panels represent asymmetric reconstructions (without imposed rotational symmetry). Panels A and C are central section views at 0° relative to the flagellar filament, whereas panels B and D are horizontal cross-section views at the same level. 3-D isosurface views of the detergent-treated flagellar motor are shown in panels E (central section), F (side view), and G (top view). A “groove” at the edge of the MS ring is indicated. For comparison, similar views of the intact flagellar motor are shown in panels H, I, and J. The segmented structure was color coded as follows: rotor, green; C ring and stator, blue; export apparatus, gray. The density that fills in the groove in the intact motor (H) may represent the N-terminal domain of FliG interacting with the bottom of the rotor (MS ring). Bar, 50 nm.
The most peripherally located elements (Fig. 4H, I, and J) of the motor lost after NP-40 treatment are thought to represent the stator, which is composed of the membrane-associated proteins MotA and MotB and is embedded in the curved cytoplasmic membrane (Fig. 4H and I). The C ring (Fig. 4H and I) also was not visible following detergent treatment.
flgI mutants and their flagellar motor structures.
Transposon mutants of the infectious, transformable B. burgdorferi strain 5A18NP1 were generated as part of a project to identify virulence determinants (T. Lin, L. Gao, and S. J. Norris, unpublished data). The insertion sites of three mutants used in this study are indicated in the map of the Borrelia burgdorferi genetic locus encoding the P-ring protein FlgI (Fig. 5A). Two of these mutations are located within the flgI open reading frame (Fig. 5A), whereas a third mutation is located downstream of flgI in an open reading frame recently identified as flgJ (C. Li and N. W. Charon, unpublished data). The morphology and motility of the mutants as examined by dark-field microscopy were indistinguishable from those of WT cells. The three transposon mutants were visualized and reconstructed using cryo-ET and compared with the parental strain 5A18NP1.
FIG. 5.
Localization of the P-ring protein FlgI in the B. burgdorferi flagellar motor, as determined by cryo-ET-derived models from transposon mutants. Panel A is a map of the Borrelia burgdorferi genetic locus encoding the P-ring protein FlgI and other flagellar proteins. Gold-colored genes are flagellum related, and hypothetical genes are shown in blue. The alternative sigma factor gene rpoS is shown in red. The insertion sites of three transposon mutants used in this study are shown. The dashed arrow indicates the predicted transcript of a multicistronic operon with a promoter upstream of flhO. Sections through 3-D asymmetric averages of flagellar motors from WT cells (B) or three mutants, T09P01H08 (C), T03TC042 (D), and T06TC173 (E), are shown. The corresponding surface renderings are from WT (F) or flgI mutant T09P01H08 (G and H) cells. The difference map for the WT and the flgI mutant reveals the P ring (colored beige), shown in the inset and in the context of the motor (H).
3-D structures of the intact flagellar motors from the three transposon mutants were determined (Fig. 5). The overall structures of the flagellar motors from all of the mutants are similar to that of WT cells (Fig. 5B, C, D, and E). However, motors in the two mutants with insertion sites in the flgI open reading frame (Fig. 5C and D) lacked a hollow, torus-shaped structure around the rod. In contrast, the hollow torus-shaped structure is clearly visible in the flgJ mutant (Fig. 5E), and its overall structure was indistinguishable from that of WT cells (Fig. 5B). The surface rendering of the flgI mutant T09P01H08 is presented in Fig. 5G. The difference map of the WT and the flgI mutant reveals the structural shape (inset figure in Fig. 5) and the location of the FlgI proteins (colored beige of Fig. 5H), corresponding to the P ring of this organism.
The two flgI mutants examined were motile in both BSK II medium and the same medium with 1% methylcellulose, a reagent that increases medium viscosity. As with WT cells, the mutant cells consistently exhibited rapid cellular rotation in the BSK II medium and translational motion in the viscous medium. However, one of the flgI mutants (T09P01H08) had obvious defects in the flagellar assembly as revealed by cryo-ET. Interestingly, this mutant (T09P01H08) is noninfectious in C3H/HeN mice (T. Lin, L. Gao, and S. J. Norris, unpublished data), indicating that flgI (or due to polar effects, the downstream gene flgJ) is required for full infectivity. Further analysis with complemented mutants is needed to understand these findings.
DISCUSSION
We have developed a method for visualizing hundreds of cellular structures at molecular resolution, using a combination of automated cryo-ET data collection and high-throughput image processing. The ability to rapidly produce and process hundreds of tilt series has improved both the throughput and the quality of the resulting 3-D reconstructions. To effectively align and process the data, we have combined two methodologies: fiducial alignment implemented in IMOD (25) and fiducial free alignment in Protomo (46). Tomographic 3-D reconstructions of 767 intact bacteria were generated in this study, and more than 2,800 flagellar motors were utilized in structural analyses.
Tomograms of intact bacteria rapidly frozen in vitreous ice are free of the chemical fixation artifacts and thus provide a representative view of the organism's native architecture. Our studies reveal many of the same structural features described in recent cryo-ET studies of B. burgdorferi and other spirochetes (7, 16, 26, 33), including the inner and outer membranes and the peptidoglycan layer. As in the detailed analysis by Kudryashev et al. (26), we observed the 4-nm lipid bilayer of the outer membrane and an additional electron-dense layer, which they described as a slime layer. In this study, we found that this additional layer was removed by treatment with proteinase K (see Fig. S1 in the supplemental material). We therefore hypothesize that this layer represents the outer surface proteins of B. burgdorferi, which in organisms cultured in vitro at 37°C consist primarily of the outer surface proteins A (OspA), -B, -C, and -D (13). The largest of these lipoproteins, OspB, is approximately 100 Å in length (2), which corresponds well with the observed thickness of the surface layer. Periplasmic flagellar filaments and motors are also discernible in the cryo-tomograms. Our data confirm that flagellar filaments reside in the periplasmic space between the peptidoglycan layer and the outer membrane. This result is consistent with a prior report (26) but differs from the recent tomographic results of Izard et al. (16), who reported that the T. denticola flagella were located between the peptidoglycan layer and the inner membrane. A central region of the flagella has low density in most views, consistent with the presence of a central channel involved in transport of subunits to the nascent end during flagellar assembly (48). As described previously by Kudryashev et al. (26), the hook assembly and the proximal portion of the flagellar filament were found to cause a bulging of the B. burgdorferi outer membrane, as well as an indentation of the inner membrane in the direction of flagellar extension.
Flagellar motor.
This study was focused primarily on the 3-D structure of the intact B. burgdorferi flagellar motor, resulting in a model at ∼3.5-nm resolution. We decided to concentrate our efforts on an asymmetric model, i.e., one developed without applying rotational symmetry or a preexisting model. As shown in the analysis of purified S. enterica flagellar rotors (44), rotational symmetry assumptions may degrade the resolution and structural details of subassemblies due to variability in the symmetric pattern in different parts of the motor. In addition, we were surprised to find that there were structural asymmetries in stator, rotor, and C-ring assemblies, as discussed in detail below. The model described herein has many similarities to the T. primitia structure at 7-nm resolution obtained with the application of 16-fold symmetry, as reported by Murphy et al. (32), indicating that the overall flagellar motor structure is well conserved within spirochetes and perhaps in other bacteria. Features of the B. burgdorferi flagellar motor are described below from the most central to the most distal aspects.
The flagellar motor is connected to the filament through the hook, which must maintain flexibility through an almost 90-degree turn across the peptidoglycan layer. The rod is curved slightly in the direction of the hook (Fig. 3A), most likely reflecting the tension placed upon the hook-filament assembly due to its constrained location within the periplasmic space. The rod, as in the flagellar filament, has a lower density in the middle, consistent with the presence of a central channel essential for translocation of flagellar proteins.
In an effort to discern the boundary between the rotor and the stator, we utilized treatment of intact B. burgdorferi with the nonionic detergent NP-40. Treatment with detergents has been utilized in early steps of bacterial flagellar rotor purifications and removes the stator structure (1, 11). NP-40 treatment of Borrelia resulted in ghost cells in which the outer membrane and portions of the cytoplasmic membrane were removed (see Fig. S3 in the supplemental material). The outermost ring of the motor and the C ring were also solubilized, leaving behind a bowl-shaped structure surrounding the central rod still attached to the flagellar hook and filament (Fig. 4; see also Fig. S3 and S4 in the supplemental material). We propose that the remaining bowl-shaped structure represents the rotor whereas the solubilized outer ring is the stator. The upper portion of the C ring (most likely comprised of FliG homologs) is in close proximity with both the putative stator and rotor structures, in keeping with structures of purified rotor/C-ring assemblies as well as biophysical and genetic findings for other organisms (5, 44). The remainder of the discussion will refer to the proposed rotor, stator, and C-ring structures shown in Fig. 4.
Rotor structure.
The flagellar rotor is a complex, bowl-shaped structure 45 nm in diameter that surrounds the central rod. The overall structure closely parallels the flagellar rotor of T. primitia described by Murphy et al. (32). The bottom of the bowl is attached to the rod and corresponds to the MS ring identified in purified flagellar rotor/C-ring assemblies from other bacteria. As such, it is likely composed primarily of the MS ring protein FliF (BB0291). No discernible symmetry was visible in this portion of the B. burgdorferi rotor in our current 3-D reconstruction. However, the analysis of the Salmonella rotor by Thomas et al. (44) indicated a rotational symmetry of 24- to 26-fold within the MS ring, suggesting the possible arrangement of the FliF subunits. The sides of the rotor (collar) exhibited strong 16-fold symmetry (see Fig. S2 in the supplemental material) and appeared to consist of 16 identical subassemblies joined together at the top of the rotor and to the MS ring at the bottom. Additional structures extend diagonally from the central part of the MS ring to the middle of the collar (Fig. 4A and F) and may represent trusses that reinforce and stabilize the rotor structure. The proteins that comprise the collar and truss structures have not as yet been identified but may include proteins specific for spirochetes (e.g., FlbB, encoded by BB0286 in the major flagellar operon). It is not known whether a similar bowl-like rotor structure exists in bacteria other than spirochetes. Cryo-ET or other approaches to studying the flagellar motor in situ in E. coli, Salmonella, and other organisms have not been reported as yet, due in part to a ∼0.5-μm specimen thickness limit in cryo-ET.
The overall structure of the rotor in NP-40-treated cells closely resembles the corresponding region of the intact flagellar motor (Fig. 4). However, a groove is present in the detergent-treated structure (Fig. 4E and F) that is filled in with the electron-dense material in the intact rotor (Fig. 4H and I). We postulate that this density represents the N-terminal region of FliG that forms a connection with the MS ring. In other organisms, it has been suggested that the FliG N terminus is linked to the MS ring of the rotor (11, 43). In addition, the motif EHPQ of FliG domain I and a hydrophobic patch in domain II interact with FliM, thus connecting the rotor to the C ring (5). Another portion of the FliG C-terminal domain II interacts with MotA, thereby conveying the thrust stroke to the rotor/C-ring assembly. B. burgdorferi has two FliG orthologs, called FliG1 and FliG2 (12). fliG2 is located in the major flagellar operon, and its predicted product more closely resembles the functional FliG in other bacteria. For example, FliG2 contains the EHPQ motif and key residues of the hydrophobic patch, whereas these sequences are not well conserved in FliG1. Thus, it is likely that FliG2 is primarily involved in flagellar rotation. All spirochetal genomes sequenced to date (Treponema pallidum, T. denticola, Leptospira interrogans, and several Borrelia species) contain at least two fliG orthologs (12, 13, 34, 36, 38); Leptospira genomes contain three. The multiple FliG homologs therefore may be involved in a motility mechanism common to spirochetes. The flagellar motors rotate in opposite directions at each end of the cell during translational motion (6); it is possible that the multiple FliG homologs influence the “default” direction of flagellar rotation. A fliG1 transposon mutant was isolated previously that remained motile but was noninfectious in mice (4); complementation to verify the role of fliG1 has not been reported to date. It is thus possible that FliG1 may fulfill an as yet unrecognized role in motility or another aspect of infectivity.
We utilized a series of transposon mutants to confirm that the P ring of B. burgdorferi is composed of the protein FlgI (Fig. 5). Transposon mutants were selected in which flgI (BB0772) was disrupted either 67% or 90% into the open reading frame; a third strain carrying a mutation in an overlapping, downstream open reading frame of unknown function (flgJ) was used as a control for possible polar effects. Disruption of flgI resulted in a loss of the torus-shaped ring associated with the rod (Fig. 5C and D), whereas the downstream mutant had a wild-type flagellar motor structure (Fig. 5E). The flgI mutants are fully motile, indicating that the P ring is not required for flagellar rotation. However, some of these mutants have an increased proportion of motors with defects in hook or flagellar filament assembly and are noninfectious in C3H/HeN mice (T. Lin, L. Gao, and S. J. Norris, unpublished data). Further analysis of the mechanisms behind these phenotypes is ongoing, including gene complementation studies. The fact that the P ring was also dissociated by NP-40 treatment (Fig. 4) indicates that it is a detergent-labile structure in B. burgdorferi. As discussed previously in the context of the T. primitia flagellar motor (32), the location of the P ring within the rotor appears to preclude its interaction with the peptidoglycan layer in spirochetes. Rather, the collar of the rotor may act as a bushing.
Stator structure.
This study provides an unprecedented view of the stator portion of the motor. By not imposing rotational symmetry, asymmetric aspects of the flagellar motor were revealed, including curvature of the stator in concordance with the cylindrical shape of the cell (see Movie S1 in the supplemental material). The rotor structure (as derived from NP-40-treated preparations) was used as a guide in the segmentation and estimation of the boundary between the closely apposed stator and rotor structures in the intact motor (Fig. 4). The stator has 16 radially arranged units, similar to the 16-fold symmetry of the rotor. The 16 stator units not only were present in the averaged structure but were also observed consistently in individual reconstructions of flagellar motors (e.g., see Fig. 2B). Recent studies with E. coli, Salmonella, and Vibrio organisms indicate that the stator is a dynamic structure in which the functional units (MotA4MotB2, or PomA4PomB2 in sodium gradient-driven motors) are freely interchanged between flagellar motor and stator units “circulating” in the cytoplasmic membrane (14, 27). Our results, as well as those obtained with T. primitia (32), indicate that these spirochetal flagellar motors have a consistent number of stator units. It is possible that the stator in these organisms is relatively stable or that interchange occurs without disrupting the overall arrangement of stator units. The precise locations of MotA and MotB in the stator and FliG in the stator/C-ring interface of the B. burgdorferi motor are not known and will be the subject of future studies. However, it is reasonable to assume that the top part of the putative stator structure may be the C-terminal periplasmic domain of MotB (8), which contains the peptidoglycan binding domain (37). The bottom portion of the stator-associated density is expected to be primarily MotA (BB0281), a transmembrane protein embedded in the cytoplasmic membrane. Biochemical and structural studies imply that the motor torque is generated by conformational changes in the stator upon the protonation of a critical aspartate residue in MotB (corresponding to D24 in B. burgdorferi BB0280) (23, 50). This motion is transmitted to the rotor by means of interactions between the rotor and FliG in the C-ring assembly.
C-ring structure.
B. burgdorferi exhibits a large C ring ∼57 nm in diameter, compared to the 45-nm C ring of E. coli and Salmonella. This enlarged C ring correlates well with the larger rotor size observed in B. burgdorferi. Interestingly, the borrelial C ring has features remarkably similar to those of the Salmonella structure (44): a cylindrical structure with a slightly bulky bottom and a Y-shaped extension at the top (see the cross-sectional view in Fig. 3A). It is thus likely that the C ring is composed of multiple copies of the proteins FliG1/2, FliM, and FliN arranged in a manner similar to that in Salmonella. The bottom of the C ring (Fig. 3F) contains FliN and is slightly bigger than the middle portion, which is composed of FliM. A model of the C ring was proposed based on the cryo-ET map (Fig. 6C and D).
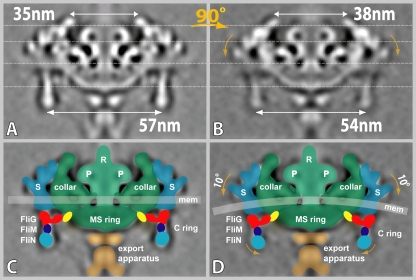
FIG. 6.
Asymmetric arrangement of stators and the role of FliG. Two central sections of flagellar motor are displayed, one oriented perpendicular (A) and the other parallel (B) to the cell axis. The vertical displacement of the outermost density caused by the membrane curvature results in a 10° “tilt” of the stator (D), a slight narrowing of the C-ring diameter from 57 nm to 54 nm, and a little widening of the collar region from 35 nm to 38 nm. A cartoon model of the flagellar motor was overlaid on the density map in panels C and D. The C ring is composed of FliG (red), FliM (dark blue), and FliN (cyan). The yellow region is the proposed location of the N terminus of FliG. The major components (the rod, the stator [S], the P ring [P], the MS ring, and the export apparatus) are labeled.
Stator curvature and FliG “transaxle.”
The outer aspects of the stator assembly (3 and 4 in Fig. 3G and H) were found to be ∼6 nm lower at the lateral sides of the motor than at the sides along the cell axis, corresponding precisely with the membrane curvature (see PowerPoint file S1 in the supplemental material). This vertical “displacement” also resulted in a 10° “tilt” of the stator structures and (surprisingly) a slight narrowing of the C ring in the lateral dimension (54 nm) compared to that in the cell axis dimension (57 nm) (Fig. 6: see also PowerPoint file S1 in the supplemental material). The observed curvature in the stator (Fig. 3) was an unexpected finding but makes sense given the strong association of MotA and MotB with the cytoplasmic membrane. MotA has four predicted transmembrane regions (21), and MotB has a single transmembrane segment; these hydrophobic helices are conserved in the B. burgdorferi proteins. Thus, the MotA and MotB proteins are firmly embedded in the membrane, and their vertical locations would therefore follow those of the membrane surface. Because of the small cell diameter at the tip of the B. burgdorferi cell (∼0.2 μm), there is considerable curvature in the cytoplasmic membrane perpendicular to the cell axis. The rotor, however, exhibits no rotational curvature either in our analyses or in those of prior studies of purified rotor/C-ring assemblies. We therefore postulate that flexibility in the FliG-MS-ring interconnection (as well as the C ring) is required to maintain this connection during rotation. In an automotive analogy, the FliG/C-ring assembly operates as a transaxle, acting not only as the transmission (representing the switching and force-transmitting functions) but also as the ball joints of the flagellar motor. This flexibility was reflected to some extent as density changes in the region of expected FliG-MS-ring interconnection at 0° and 90° of rotation relative to the direction of the flagellum (Fig. 6; see also PowerPoint file S1 in the supplemental material). Forces caused by the vertical displacement of the stator also result in the observed narrowing of the C ring in the direction perpendicular to the cell axis during its rotation. Thus, both FliG and the C ring appear to accommodate the tension placed on the rotor/C-ring assembly as it rotates. A similar, but reduced, effect would be expected to occur in bacteria with a larger diameter. However, a higher-resolution structure of this region is needed to better understand the vital role of FliG in stator-rotor interactions, the relative movement between these structures during rotation, the mechanism of switching, and maintenance of the stator/rotor/C-ring linkage.
To what extent can the flagellar motor structural information obtained with B. burgdorferi and other spirochetes be applied to other bacteria, including the well-studied flagellar systems of E. coli and Salmonella? Given the high degree of conservation among the proteins involved, it is likely that many of the same structural features are present in other systems. Other aspects, such as the bowl shape of the rotor, may be specific for spirochetes and not found in other bacteria. Broadening the cryo-ET analysis of the intact flagellar motor in other organisms may help to answer this question and to thereby delineate both the similarities and differences in this fascinating machine within the bacterial kingdom.
Supplementary Material
Acknowledgments
We thank David Blair, Angel Paredes, Hong Zhou, and James K. Stoops for their help and suggestions in this project.
This work was supported by the UT Houston Medical School Dean's Fund (to J.L.) and NIH grant no. AI059048 to (to S.J.N.).
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aizawa, S. I., G. E. Dean, C. J. Jones, R. M. Macnab, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M., J. Bunikis, B. D. Lade, J. J. Dunn, A. G. Barbour, and C. L. Lawson. 2005. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J. Biol. Chem. 280:17363-17370. [DOI] [PubMed] [Google Scholar]
- 3.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 4.Botkin, D. J., A. N. Abbott, P. E. Stewart, P. A. Rosa, H. Kawabata, H. Watanabe, and S. J. Norris. 2006. Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 74:6690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, P. N., M. Terrazas, K. Paul, and D. F. Blair. 2007. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47-73. [DOI] [PubMed] [Google Scholar]
- 7.Charon, N. W., S. F. Goldstein, M. Marko, C. Hsieh, L. L. Gebhardt, M. A. Motaleb, C. W. Wolgemuth, R. J. Limberger, and N. Rowe. 2009. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J. Bacteriol. 191:600-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 9.Coulton, J. W., and R. G. Murray. 1978. Cell envelope associations of Aquaspirillum serpens flagella. J. Bacteriol. 136:1037-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowther, R. A., and L. A. Amos. 1971. Harmonic analysis of electron microscope images with rotational symmetry. J. Mol. Biol. 60:123-130. [DOI] [PubMed] [Google Scholar]
- 11.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 12.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, C. Fujii, S. Garland, B. Hatch, K. Horst, K. Roberts, M. Sandusky, J. Weidman, H. O. Smith, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 14.Fukuoka, H., T. Wada, S. Kojima, A. Ishijima, and M. Homma. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825-835. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, S. F., N. W. Charon, and J. A. Kreiling. 1994. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc. Natl. Acad. Sci. USA 91:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izard, J., C. E. Hsieh, R. J. Limberger, C. A. Mannella, and M. Marko. 2008. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J. Struct. Biol. 163:10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 18.Katayama, E., T. Shiraishi, K. Oosawa, N. Baba, and S. Aizawa. 1996. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J. Mol. Biol. 255:458-475. [DOI] [PubMed] [Google Scholar]
- 19.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 21.Kim, E. A., M. Price-Carter, W. C. Carlquist, and D. F. Blair. 2008. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry 47:11332-11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93-134. [DOI] [PubMed] [Google Scholar]
- 23.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 24.Koster, A. J., R. Grimm, D. Typke, R. Hegerl, A. Stoschek, J. Walz, and W. Baumeister. 1997. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120:276-308. [DOI] [PubMed] [Google Scholar]
- 25.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 26.Kudryashev, M., M. Cyrklaff, W. Baumeister, M. M. Simon, R. Wallich, and F. Frischknecht. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol. Microbiol. 71:1415-1434. [DOI] [PubMed] [Google Scholar]
- 27.Leake, M. C., J. H. Chandler, G. H. Wadhams, F. Bai, R. M. Berry, and J. P. Armitage. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355-358. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., D. W. Taylor, E. B. Krementsova, K. M. Trybus, and K. A. Taylor. 2006. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature 442:208-211. [DOI] [PubMed] [Google Scholar]
- 30.Lucic, V., F. Forster, and W. Baumeister. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833-865. [DOI] [PubMed] [Google Scholar]
- 31.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, G. E., J. R. Leadbetter, and G. J. Jensen. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062-1064. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, G. E., E. G. Matson, J. R. Leadbetter, H. C. Berg, and G. J. Jensen. 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol. Microbiol. 67:1184-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-De-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. De Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 36.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 37.Roujeinikova, A. 2008. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. USA 105:10348-10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshadri, R., G. S. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E. Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, and I. T. Paulsen. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA 101:5646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sowa, Y., and R. M. Berry. 2008. Bacterial flagellar motor. Q. Rev. Biophys. 41:103-132. [DOI] [PubMed] [Google Scholar]
- 40.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, P. E., and P. A. Rosa. 2008. Transposon mutagenesis of the Lyme disease agent Borrelia burgdorferi. Methods Mol. Biol. 431:85-95. [DOI] [PubMed] [Google Scholar]
- 42.Terashima, H., S. Kojima, and M. Homma. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39-85. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, D., D. G. Morgan, and D. J. DeRosier. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183:6404-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, D. R., N. R. Francis, C. Xu, and D. J. DeRosier. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volkmann, N. 2002. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J. Struct. Biol. 138:123-129. [DOI] [PubMed] [Google Scholar]
- 46.Winkler, H., and K. A. Taylor. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106:240-254. [DOI] [PubMed] [Google Scholar]
- 47.Winkler, H., P. Zhu, J. Liu, F. Ye, K. H. Roux, and K. A. Taylor. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165:64-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.