Abstract
Mycoplasmas that are known to exhibit gliding motility possess a differentiated tip structure. This polar organelle mediates cytadherence and gliding motor activity and contains a cytoskeleton-like component that provides structural support. Here, we describe gliding motility and a unique cytoskeleton in Mycoplasma insons, which lacks any obviously differentiated tip structure.
Bacterial cell polarity underlies a number of critical processes, including the appropriate clustering of chemotactic receptors (12), chromosome segregation (9), and placement of specialized polar structures (6). The bacterial cytoskeleton is an important component of the mechanism underlying the generation of polarity. The bacterial actin, MreB, is crucial for the morphology of elongated bacterial cells, directing peptidoglycan synthesis along the long axis of the cell (4). In most well-studied bacterial systems, cell shape is dictated by the interaction between the cytoskeleton and the cell wall biosynthetic machinery.
In contrast, bacteria of the class Mollicutes lack cell walls. Despite the absence of such a structure, many species maintain cell polarity and distinctive shapes. In Mycoplasma pneumoniae, one cell pole is organized as a terminal organelle. This tip structure, also called the attachment organelle, is readily distinguished from the more pleomorphic cell body by virtue of its slender, elongated appearance and regular dimensions. Adhesins are concentrated at the terminal organelle, which also contains a motor activity for gliding motility (1). Essential for the formation of the M. pneumoniae attachment organelle is a Triton X-100 (TX)-insoluble cytoskeletal structure, the electron-dense core (3), which contains novel proteins found only in M. pneumoniae and its close relatives (2). The distantly related species Mycoplasma mobile has a terminal organelle that also functions in gliding motility, with adhesin and cytoskeleton molecules that are completely unrelated to those of M. pneumoniae present at its cell-proximal end (1, 14). Interestingly, MreB is absent from both species (10, 11). The difference in composition between the terminal organelles of M. pneumoniae and M. mobile suggests that the two have evolved similar structures independently. Thus, mycoplasmas are innovators of cytoskeletons, and the variation among species provides a rich opportunity to learn a great deal about the generation of cell polarity.
Many mycoplasmas, including Mycoplasma insons, found in the respiratory tract and blood of healthy green iguanas, lack a distinct tip structure (13). M. insons shares with its two closest relatives, Mycoplasma cavipharyngis and Mycoplasma fastidiosum, a slender rod shape. A series of regularly spaced, electron-dense striations perpendicular to the long axis is visible in thin sections by transmission electron microscopy, and scanning electron microscopy reveals a somewhat twisted appearance along the long axis (13). To understand the underlying cellular organization, we investigated the ultrastructure and behavior of M. insons cells attached to glass.
The morphologies of whole cells and TX-insoluble material of M. insons were assessed by scanning electron microscopy. For whole-cell analysis, cells were grown for 24 h on glass coverslips in SP-4 broth at 30°C (7, 13, 15). Imaging was performed on a Zeiss Supra 35 FEG-VP scanning electron microscope (Carl Zeiss NTS GmbH, Oberkochen, Germany). M. insons cells appeared as nonbranching rods lacking distinguishable tip structures, as previously described (13) (Fig. 1A and B). After treatment of cells with A22, an MreB depolymerizing agent (5), cells remained rod shaped (not shown), suggesting an absence of MreB. The majority of cells lay flat on the coverslip; however, 8 to 10% of cells were attached at only one end, suggesting polar distribution of adhesins (Fig. 1C).
FIG. 1.
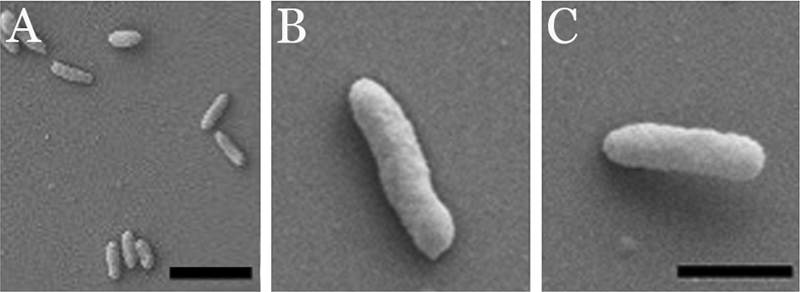
Scanning electron micrographs of whole cells of M. insons attached to a glass coverslip. (A) Low-magnification view. (B) Single cell lying flat on the coverslip. (C) Single cell attached by one pole to the coverslip. Bars: panel A, 2 μm; panel C, 0.5 μm; magnification of panel B is the same as for panel C.
For motility analyses, stocks of M. insons were prepared by growing cells in SP-4 broth to mid-exponential phase (phenol red indicator was orange; approximately 24 h) at 30°C. After incubation, cultures were centrifuged for 20 min at 17,400 × g, resuspended in 1 ml of fresh SP-4 broth, and frozen at −80°C in 50-μl aliquots. Subsequently, motility stocks were suspended in SP-4 broth plus 3% (wt/vol) gelatin, syringe-triturated seven times, passed through 0.45-μm syringe filters into chamber slides (Thermo Fisher Scientific, Rochester, NY), and incubated for 3 h at various temperatures before phase-contrast imaging with a Leica DM IRB inverted microscope (Leica Microsystems, Wetzlar, Germany). A heated chamber was used to maintain temperatures above ambient during analysis. Sequential frames were merged and analyzed as previously described (7).
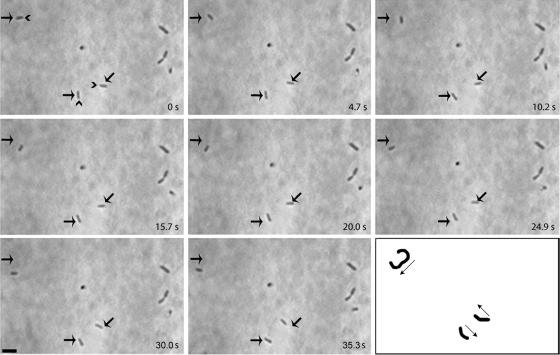
At 24°C, very few M. insons cells adhered to the glass surface. At 30°C, cells were capable of attaching and glided at an average speed of 25 ± 7 (mean ± standard deviation) nm s−1 with a range from 13 to 47 nm s−1 (n = 28). At 37°C, M. insons cells glided at an average speed of 30 ± 14 nm s−1 with a range from 10 to 88 nm s−1 (n = 45). These speeds were similar to the average speed of Mycoplasma pirum cells (28 nm s−1) (8), making these two the slowest gliding mycoplasmas characterized. Approximately 7% of cells glided at either temperature. This field and others were observed for a 2-h period. During this time, individual motile cells stopped and started moving and occasionally pivoted. However, no cell ever changed the polarity of its movement over this extended period of observation. Figure 2 shows eight sequential phase-contrast frames of M. insons cells incubated at 37°C taken approximately 5 s apart. Three cells within the field shown glided; their paths and directions are indicated in the bottom right panel. The cell indicated with the diagonal arrow shows an example of pivoting over the course of the first three frames. These observations constitute the first report of gliding motility in a mycoplasma lacking a differentiated tip structure. The unidirectionality exhibited by these cells, together with the observation of unipolar attachment to the substrate in a portion of cells similar to the portion of motile cells, suggested an underlying polarity.
FIG. 2.
Consecutive phase-contrast images of M. insons in a chamber slide at ∼5-s intervals at 37°C. Three representative cells are indicated by arrows. In the first frame (0 s), the carets point to the leading end of gliding cells. In the last frame, the paths of the gliding cells are represented, with arrows indicating the direction of gliding. Bar: 1 μm.
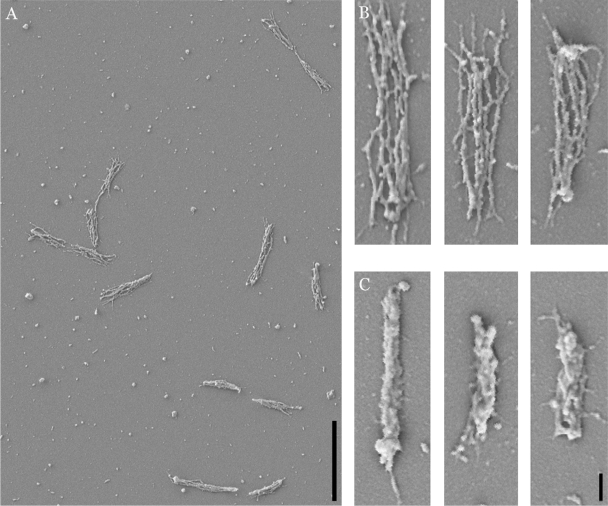
Other mycoplasmas that exhibit polarity contain cytoskeletal structures associated with the differentiated pole (2, 14). To test whether M. insons cells contained cytoskeletons, coverslips with attached cells were incubated in TX in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl for 30 min at 37°C and then fixed and further processed as described above for whole cells. Treatment with 3.2% TX revealed loose bundles of 5 to 8 roughly parallel filaments ∼30 nm in width (Fig. 3A and B). Extraction with only 1.2% TX revealed the presence of filaments that were partially obscured by remnants of the cell membrane (Fig. 3C), reminiscent of the twisted appearance previously described (13). Filament bundles from 3.2%-TX-extracted cells were an average length of 1.18 ± 0.30 μm with a range of 0.65 to 1.85 μm (n = 140), whereas whole cells had an average length of 0.78 ± 0.28 μm with a range of 0.30 to 2.09 μm (n = 180). The slightly larger average in filament bundle length might indicate that the cytoskeleton is under longitudinal compression in intact cells. No clear evidence of polarity of these filaments was observed in these analyses, although further study is necessary to understand whether they have a role in the polarization of the adherence machinery. Structures corresponding to the electron-dense bands observed by transmission electron microscopy (13) were not observed.
FIG. 3.
Scanning electron micrographs of M. insons attached to glass coverslips and extracted with TX. (A) Low-magnification view of cells extracted with 3.2% TX. (B) High-magnification view of three separate cells extracted with 3.2% TX. (C) High-magnification view of three separate cells extracted with 1.2% TX. Bars: panel A, 2 μm; panel C, 200 nm; magnification of panel B is the same as for panel C.
This report constitutes the first description of motility in a mycoplasma lacking an obviously differentiated tip structure and with an undifferentiated cytoskeleton extending throughout the entire cell. The internal organization of M. insons is entirely distinct from that of any previously described mycoplasma, representing a novel paradigm for mycoplasma cell polarity. These results suggest that the evolution of polarity in mycoplasmas has taken multiple independent pathways.
Acknowledgments
This work was supported by National Institutes of Health grant AI073994.
We thank D. Brown and M. May (University of Florida) for many helpful discussions during the preparation of this research and Y. Brun (Indiana University) for his generous gift of A22.
Footnotes
Published ahead of print on 12 June 2009.
REFERENCES
- 1.Balish, M. F. 2006. Subcellular structures of mycoplasmas. Front. Biosci. 11:2017-2027. [DOI] [PubMed] [Google Scholar]
- 2.Balish, M. F., and D. C. Krause. 2006. Mycoplasmas: a distinct cytoskeleton for a wall-less bacteria. J. Mol. Microbiol. Biotechnol. 11:244-255. [DOI] [PubMed] [Google Scholar]
- 3.Biberfeld, G., and P. Biberfeld. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Blaauwen, T., M. A. de Pedro, M. Nguyen-Distèche, and J. A. Ayala. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321-344. [DOI] [PubMed] [Google Scholar]
- 5.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 6.Goley, E. D., A. A. Iniesta, and L. Shapiro. 2007. Cell cycle regulation in Caulobacter: location, location, location. J. Cell Sci. 120:3501-3507. [DOI] [PubMed] [Google Scholar]
- 7.Hatchel, J. M., R. S. Balish, M. L. Duley, and M. F. Balish. 2006. Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology 152:2181-2189. [DOI] [PubMed] [Google Scholar]
- 8.Hatchel, J. M., and M. F. Balish. 2008. Attachment organelle ultrastructure correlates with phylogeny, not gliding motility properties, in Mycoplasma pneumoniae relatives. Microbiology 154:286-295. [DOI] [PubMed] [Google Scholar]
- 9.Hazan, R., and S. Ben-Yehuda. 2006. Resolving chromosome segregation in bacteria. J. Mol. Microbiol. Biotechnol. 11:126-139. [DOI] [PubMed] [Google Scholar]
- 10.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lybarger, S. R., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.May, M., G. J. Ortiz, L. D. Wendland, D. S. Rotstein, R. F. Relich, M. F. Balish, and D. R. Brown. 2007. Mycoplasma insons sp. nov., a twisted mycoplasma from green iguanas (Iguana iguana). FEMS Microbiol. Lett. 274:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakane, D., and M. Miyata. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 104:19518-19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tully, J. G., D. L. Rose, R. F. Whitcomb, and R. P. Wenzel. 1979. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J. Infect. Dis. 139:478-482. [DOI] [PubMed] [Google Scholar]