Abstract
The eukaryote-like DNA replication system of the model haloarchaeon Halobacterium NRC-1 is encoded within a circular chromosome and two large megaplasmids or minichromosomes, pNRC100 and pNRC200. We previously showed by genetic analysis that 2 (orc2 and orc10) of the 10 genes coding for Orc-Cdc6 replication initiator proteins were essential, while a third (orc7), located near a highly conserved autonomously replicating sequence, oriC1, was nonessential for cell viability. Here we used whole-genome marker frequency analysis (MFA) and found multiple peaks, indicative of multiple replication origins. The largest chromosomal peaks were located proximal to orc7 (oriC1) and orc10 (oriC2), and the largest peaks on the extrachromosomal elements were near orc9 (oriP1) in both pNRC100 and -200 and near orc4 (oriP2) in pNRC200. MFA of deletion strains containing different combinations of chromosomal orc genes showed that replication initiation at oriC1 requires orc7 but not orc6 and orc8. The initiation sites at oriC1 were determined by replication initiation point analysis and found to map divergently within and near an AT-rich element flanked by likely Orc binding sites. The oriC1 region, Orc binding sites, and orc7 gene orthologs were conserved in all sequenced haloarchaea. Serial deletion of orc genes resulted in the construction of a minimal strain containing not only orc2 and orc10 but also orc9. Our results suggest that replication in this model system is intriguing and more complex than previously thought. We discuss these results from the perspective of the replication strategy and evolution of haloarchaeal genomes.
Archaea are of considerable interest due to their unusual phylogenetic position and the similarity of their information transfer system to that of eukaryotes. In particular, studies of DNA replication in archaea have revealed characteristics of both bacterial and eukaryotic systems (1). While genome sequencing has shown that archaeal and bacterial genomes are composed of a single or few circular chromosomes, comparative genomic studies have found that most components of the archaeal DNA replication machinery, such as the origin recognition proteins, DNA polymerases, helicases, and primases, are similar to eukaryotic proteins. The hybrid nature of archaeal DNA replication systems raises important questions regarding the mechanism by which they select an origin(s) for initiation and coordinate orderly DNA replication and segregation into daughter cells.
Our understanding of DNA replication in archaea has thus far been based primarily on bioinformatic studies, with experimental analysis restricted to only a few tractable systems. An initial study of Pyrococcus species using GC (tetramer) skew analysis suggested that they use a single, unique origin of replication in their chromosomes. Subsequent [3H]uracil labeling analysis of Pyrococcus abyssi (21) showed that newly synthesized DNA mapped to the predicted replication origin region, which contained the only orc gene in the genome, a D family DNA polymerase gene, and a DNA sliding clamp loader subunit. In addition, two-dimensional gel analysis of replicating molecules confirmed the location of the DNA replication origin near the orc1 gene of P. abyssi, with predicted origin binding sequences and AT-rich DNA unwinding elements nearby (18). An investigation of DNA replication in Aeropyrum pernix used a combination of biochemical and two-dimensional gel electrophoresis and identified two potential sites of replication initiation, on opposite sides of the circular genome (14, 28). One of these sites (oriC1Ap) contained four origin recognition boxes and an AT-rich region and was shown to be bound by the ORC1 gene. The other site (oriC2Ap) contained repeat elements without an intervening AT-rich region and has been shown by two-dimensional gel electrophoresis to contain an active replication origin (28). An examination of replication in two Sulfolobus spp., Sulfolobus solfataricus and Sulfolobus acidocaldarius (16, 30), by use of a combination of bioinformatic and two-dimensional gel analysis and of marker frequency by use of DNA microarrays identified three well-separated replication origins per genome. Only two of the three origins were originally identified, due to their linkage to orc genes and conserved origin binding sequences, while the third was identified by marker frequency analysis (MFA). Using partially synchronized cells of S. acidocaldarius, the origins were shown to initiate DNA replication synchronously, indicating a highly coordinated and regulated process. Biochemical analysis has shown that either two or all three Orc proteins are able to bind to all Sulfolobus origins; however, binding at the third origin is considerably weaker (29). Replication origins were also recently identified in Methanothermobacter thermoautotrophicus (17).
Our laboratory has been investigating DNA replication in a halophilic archaeon capable of growth at saturating NaCl concentrations. The model system, Halobacterium sp. strain NRC-1, was one of the earliest archaeal genomes to be sequenced (23) and provided a DNA knockout method, utilizing the selectable and counterselectable ura3 gene, for genetic analysis (25). The NRC-1 genome was found to be organized into a 2-Mbp chromosome and two large and partially redundant extrachromosomal elements, pNRC100 and pNRC200. The genome sequence showed that the orc gene family was highly expanded, with four genes (orc6, -7, -8, and -10) distributed in the chromosome and six genes (orc1, -2, -3, -4, -5, and -9) in pNRC200, one of which (orc9) was also present in pNRC100. Three rep genes thought to be important for replication initiation were present in one (repJ in pNRC100) or both (repH and repI) of the extrachromosomal elements. Regions near two of these genes, orc7 and repH, were shown to harbor autonomous replicating ability and to contain inverted repeat sequences (IRs) and an AT-rich presumptive DNA unwinding region detectable by χ2 analysis (3, 22). Additionally, GC/oligomer skew analyses of Halobacterium sp. strain NRC-1 showed multiple inflection points in the chromosome, suggestive of multiple replication origins in this strain (15, 34).
Halobacterium sp. strain NRC-1 is the only archaeal system where gene mutation analysis has established which predicted DNA replication genes are essential to cells (2). As expected, two DNA polymerases (one B family and one D family polymerase), the MCM DNA helicase, DNA primase (Pri1/Pri2), the sliding clamp (PCNA), and flap endonuclease (Rad2) were all found to be essential. However, one B family DNA polymerase gene and 8 of the 10 orc and cdc6 genes, including the orc7 gene, were found to be nonessential by deletion analysis. Only the orc2 gene in pNRC200 and the orc10 gene in the chromosome were found to be essential, suggesting a critical role(s) for these genes in DNA replication.
In this study, we used a combination of MFA, employing whole-genome DNA microarrays, the ura3-based gene knockout method, and replication initiation point (RIP) analysis to further investigate DNA replication in Halobacterium sp. strain NRC-1. Our results indicate that initiation of DNA replication in NRC-1 is more complex than originally anticipated, with multiple origins likely present on the chromosome and the extrachromosomal elements.
MATERIALS AND METHODS
Materials.
Restriction enzymes, calf intestinal phosphatase, T4 DNA polymerase, T4 polynucleotide kinase, T4 DNA ligase, Klenow fragment, Taq DNA polymerase, λ-exonuclease, and Vent (exo-) DNA polymerase were purchased from New England Biolabs, Beverly, MA. XL DNA polymerase was purchased from Applied Biosystems, Branchburg, NJ, and a fmol DNA cycle sequencing system was purchased from Promega, Madison, WI. Oligonucleotides were purchased from Sigma-Genosys, The Woodlands, TX. Gel extraction kits and plasmid purification kits were purchased from Macherey-Nagel, Easton, PA. Uracil dropout formula, nitrogen base, and benzoylated naphtholated DEAE-cellulose were purchased from Sigma-Aldrich, St. Louis, MO.
Strains and culturing.
Escherichia coli DH5α was grown in Luria-Bertani medium supplemented with 100 μg of ampicillin/ml at 37°C. Halobacterium sp. strain NRC-1 Δura3 and derivatives were cultured in CM+ medium containing 250 μg/ml of 5-fluoroorotic acid (5-FOA) at 42°C (4).
Gene knockouts.
To generate gene knockout-suicide plasmid vectors, regions surrounding the target gene were PCR amplified from wild-type Halobacterium sp. strain NRC-1 genomic DNA, and PCR products were digested with appropriate restriction enzymes and cloned into the multiple cloning site of plasmid pBB400, as previously described (2). Two independent suicide plasmid vector isolates for each gene were then transformed individually into Halobacterium sp. strain NRC-1 Δura3 derivatives via polyethylene glycol-EDTA methodology (4). Transformation cultures were then plated onto HURA+ solid medium and grown for 7 to 10 days at 42°C. DNAs from individual colonies were used as templates in PCRs to verify suicide plasmid integration into genomic DNA. Two independent isolates were then plated onto CM+ solid medium containing 250 μg/ml of 5-FOA and grown at 42°C for 7 days. Colonies were then picked and grown at 42°C for 7 days in liquid CM+ medium containing 250 μg/ml of 5-FOA. Genomic DNAs were extracted from these cultures and used as templates in PCRs to screen for knockout alleles, using primers flanking the target gene. Two independent isolates were selected for each knockout strain generated.
MFA.
Cultures (500 ml) of Halobacterium sp. strain NRC-1 and mutant strains were grown at 42°C in CM+ medium. Aliquots were removed at early log phase (optical density at 600 nm = 0.3) and stationary phase (optical density at 600 nm = 1.6 for NRC-1 and 0.8 for mutants), and DNAs were collected as previously described (9). All cultures were monitored by optical density measurement to ensure removal of aliquots at appropriate densities and growth phases. Genomic DNA was purified from Halobacterium sp. strain NRC-1 cultures at early log phase and stationary phase, followed by labeling with Cy3- and Cy5-dCTP, respectively, using random nonamers and the Klenow fragment of E. coli DNA polymerase I (27). Incorporation of the label was checked via gel electrophoresis and scanning on a Typhoon scanner. Labeled DNA was used for hybridization to an Agilent custom array containing 44,000 spots, synthesized in situ using ink-jet technology. Each array contained 16 oligomer (60-mer) probes for each open reading frame in the genome, designed utilizing OligoPicker (33). Washing and hybridization of the arrays were performed as recommended by Agilent (20). Both technical and biological replicates were performed, and after dynamic scanning (100 and 10 photomultiplier tube [PMT]) using an Agilent microarray scanner, the data were analyzed using Agilent Feature Extraction, where the signal from each channel was normalized using the LOWESS algorithm (5) to remove intensity-dependent effects within the calculated values. Our in-house software (6, 7) was used to determine which probes were statistically relevant for analysis. Low-quality spots (saturated spots and outliers) were excluded from analysis. The average ratio (exponential-phase value/stationary-phase value) for each strain was first calculated for each gene and then for replicate slides (6). The values were then normalized so that the lowest value was equal to 1. To determine the locations of replication origins, the averaged data for each gene from all replicate slides were represented graphically by plotting the average for a 100-gene window, with a 1-gene slide across the entire genome.
RIP mapping.
RIP mapping was performed as previously described (12), with minor modifications (26, 32). Briefly, replication intermediates were enriched by use of a benzoylated naphtholated DEAE-cellulose column and then treated with λ-exonuclease to digest the nicked DNA at 37°C for 24 h, whereas the nascent DNA was protected by its RNA primer and not digested. Vent (exo-) DNA polymerase was used to extend from a labeled primer to the DNA-RNA junctions of the nascent-strand templates in the replication intermediates. For primer extension, 500 ng of template DNA, 25 ng of radiolabeled primer (RIPF or RIPR, for the top or bottom strand, respectively), and 2 U of Vent (exo-) DNA polymerase were incubated in 25 μl of buffer provided by the manufacturer. After 30 cycles (1 min at 94°C, 1 min at 70°C, and 1.5 min at 72°C) of primer extension reaction, the samples were electrophoresed and analyzed in a 6% denaturing polyacrylamide gel containing 8 M urea and 1× Tris-borate-EDTA. Sequencing reactions were performed in parallel with a fmol DNA cycle sequencing system, using the same primers, and were analyzed side by side in the same gel.
Statistical and bioinformatic analysis.
For consensus building and searching, a frequency matrix with gap creation and extension penalties of 3.0 and 0.3, respectively, was constructed with the JEMBOSS program Prophecy, using the 31-bp IRs upstream of orc7 (3). Subsequently, the matrix was used to locate similar sequences in the genome, using Profit with a 75% threshold. To identify IRs distinct from the sequences reported by Berquist and DasSarma (3), dot plot analysis (Wisconsin Package) was performed with a window of 21 and a stringency of 14. Alignment of the conserved sequences was done using the CLUSTAL_W accessory application of BioEdit, and the consensus sequence was generated from the alignment by use of BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The consensus sequence logo was generated using the web-based program WebLogo3 (http://weblogo.berkeley.edu). χ2 and GC compositions were calculated in a sliding window by previously described methods (31), employing the SelfSim chi-square program. Pictorial representations of the genome were downloaded from the Halobacterium genome database (http://halo4.umbi.umd.edu). For genomic comparison of the oriC1 region, sequence and predicted gene data were downloaded from the NCBI genome project pages for Haloquadratum walsbyi, Natronomonas pharaonis, Haloarcula marismortui, Halorubrum lacusprofundi, and Halobacterium sp. strain R-1, the Archaeal Genome Browser (http://archaea.ucsc.edu) for Haloferax volcanii, and the Halobacterium genome database for Halobacterium sp. strain NRC-1.
RESULTS
MFA of the Halobacterium sp. strain NRC-1 chromosome.
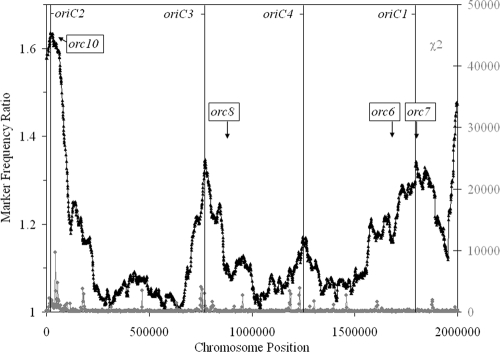
To determine the position(s) of replication initiation in Halobacterium sp. strain NRC-1, we used MFA, employing whole-genome DNA microarrays developed in this laboratory (20). By plotting the ratio of a marker's copy number in actively replicating cells to that in nonreplicating cells, characteristics such as the locations of origins, termini, and the directionality of replication may be discerned (16). For the Halobacterium sp. strain NRC-1 chromosome, we observed multiple peaks, indicating replication initiation from multiple origins (Fig. 1). Two peaks were located proximal to orc genes, with one corresponding to the previously mapped autonomously replicating sequence (ARS) (oriC1) near orc7 (2) and the other corresponding to a second origin (oriC2) mapped near orc10, the only chromosomal orc gene previously found to be essential for cell viability (2). Several additional peaks were also discernible, including two near vng1020 and vng1650 (named oriC3 and oriC4, respectively), as well as other smaller peaks. The symmetry of the peaks was consistent with bidirectional replication initiation from these sites.
FIG. 1.
Marker frequency and χ2 analysis of the chromosome of Halobacterium sp. strain NRC-1. The black points and line show the MFA results (the scale is shown on the y axis at the left), and the gray points and line show the χ2 values (the scale is shown on the y axis at the right) for the chromosome. The approximate locations of orc genes are indicated, and vertical lines show the approximate locations of oriC1, oriC2, oriC3, and oriC4 in relation to the chromosome position (the scale is shown on the x axis).
Chromosomal replication in Halobacterium orc deletion strains.
To study the genetic requirements for replication from oriC1, previously shown to harbor a highly conserved ARS element (2), we tested the replication properties of chromosomal orc deletion strains by MFA. As described above, early-log-phase and stationary-phase cultures were used for genomic DNA isolation, and DNA copy numbers were compared. The three nonessential orc genes were individually deleted, and the resulting strains (Δorc6, Δorc7, and Δorc8) were compared to wild-type NRC-1. The three deletant strains had similar marker frequency profiles to that of NRC-1; however, in the Δorc7 strain, the peak corresponding to oriC1 was greatly reduced (Fig. 2). These results indicated that the orc7 gene was required to initiate DNA replication at oriC1, while neither orc6 nor orc8 was required.
FIG. 2.
MFA of the chromosomes of orc deletion strains. Plots are labeled by the strain designation (Δorc6, Δorc7, Δorc8, Δorc68, and Δorc678), and the scale for MFA is shown at the left. Vertical lines show the approximate locations of oriC1, oriC2, oriC3, and oriC4 in relation to the chromosome position (positions are shown on the x axis).
To determine if one of the two genes, orc6 or orc8, was necessary along with orc7 for replication from oriC1, we constructed double-deletion (Δorc6 Δorc7, Δorc6 Δorc8, and Δorc7 Δorc8 strains [hereafter designated Δorc67, Δorc68, and Δorc78, respectively]) and triple-deletion (Δorc678, Δorc687, and Δorc786) strains lacking these genes (Table 1). Each of the triple-deletion strains contained only a single chromosomal orc gene, orc10, which was previously shown to be essential (2). The replication properties of two of the mutants, the Δorc68 double mutant and the Δorc678 triple mutant, were compared by MFA (Fig. 2). Both strains provided analogous results to those of the single mutants, with the absence of the peak at oriC1 in the triple mutant lacking orc7 but not in the double mutant containing orc7. These findings confirmed the requirement for orc7, but neither orc6 nor orc8, for replication initiation from oriC1 and also showed the lack of requirement for the three genes for replication initiation at the other major replication origins in the genome.
TABLE 1.
Orc gene knockout strains
| Strain or mutant | orc genes present | Reference |
|---|---|---|
| Halobacterium sp. strain NRC-1 | orc1, orc2, orc3, orc4, orc5, orc6, orc7, orc8, orc9, orc10 | 8 |
| Δura3 | orc2, orc3, orc4, orc5, orc6, orc7, orc8, orc9, orc10 | 25 |
| Δura3 Δorc1 Δorc6 | orc2, orc3, orc4, orc5, orc7,orc 8, orc9, orc10 | 2 |
| Δura3 Δorc1 Δorc7 | orc2, orc3, orc4, orc5, orc6, orc8, orc9, orc10 | 2 |
| Δura3 Δorc1 Δorc8 | orc2, orc3, orc4, orc5, orc6, orc7, orc9, orc10 | 2 |
| Δura3 Δorc1 Δorc6 Δorc7 | orc2, orc3, orc4, orc5, orc8, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc7 Δorc8 | orc2, orc3, orc4, orc5, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc8 | orc2, orc3, orc4, orc5, orc7, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc8 Δorc7 | orc2, orc3, orc4, orc5, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc7 Δorc8 | orc2, orc3, orc4, orc5, orc6, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc7 Δorc8 Δorc6 | orc2, orc3, orc4, orc5, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc7 Δorc8 Δorc4 | orc2, orc3, orc5, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc7 Δorc8 Δorc4 Δorc5 | orc2, orc3, orc9, orc10 | This work |
| Δura3 Δorc1 Δorc6 Δorc7 Δorc8 Δorc4 Δorc5 Δorc3 | orc2, orc9, orc10 | This work |
RIP at oriC1.
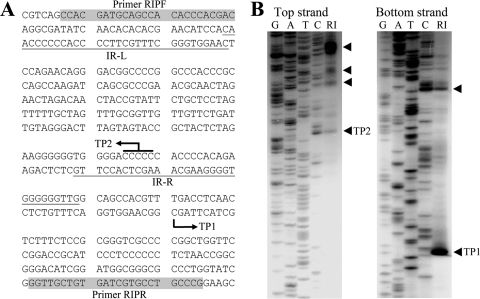
We previously showed that the 750 bp immediately upstream of orc7 (oriC1) contains a conserved inverted repeat sequence, an AT-rich element, and replication ability in the host organism (3). To further characterize the orc7-dependent replication origin at oriC1, we conducted RIP mapping. Primers located outside the AT-rich region, which is flanked by 31-bp IRs, were labeled at the 5′ ends with T4 polynucleotide kinase and [γ-32P]ATP and used for primer extension analysis to detect the DNA-RNA junctions of the nascent strand generated in DNA synthesis. The same primers were also used to produce Sanger sequencing ladders, which were electrophoresed in sequencing gels alongside the primer extension products (Fig. 3). The forward primer RIPF produced TP2, an extension product of approximately 250 nucleotides corresponding to a RIP at a C nucleotide within a sequence of five C's in the AT-rich region of oriC1. The reverse primer RIPR produced TP1, an extension product of 125 nucleotides that corresponded to a RIP at a C nucleotide 42 bp outside the AT-rich region. These findings confirmed that the orc7 ARS element is used for initiation of DNA replication in vivo and provided nearly exact locations where DNA replication starts within oriC1.
FIG. 3.
RIP mapping of Halobacterium sp. strain NRC-1. (A) DNA sequence upstream of the orc7 gene in Halobacterium strain NRC-1. The inverted repeats (IR-L and IR-R) flanking the AT-rich region are underlined. Primers (RIPF and RIPR) used for RIP mapping are shaded. Transition points (TPs) of leading and lagging strands are indicated by bent arrows. (B) RIP mapping of top and bottom strands with the RIPF and RIPR primers, respectively. Arrowheads pinpoint the initiation sites for DNA synthesis, and the transition points are indicated. Sequencing reactions (GATC) with RIPF and RIPR were run side by side in a 6% denaturing polyacrylamide gel. RI, replication intermediates.
Bioinformatic analysis of the oriC1 region.
We compared the orc7 gene regions of seven sequenced haloarchaea (Halobacterium sp. strain NRC-1, Halobacterium sp. strain R-1, Halorubrum lacusprofundi, Haloarcula marismortui, Haloquadratum walsbyi, Haloferax volcanii, and Natronomonas pharaonis). The upstream regions of the orc7 orthologs all contained IRs similar to the 31-bp IRs in NRC-1 (Fig. 4A and B). In addition, an 8-kb region surrounding the orc7 gene and oriC1 is largely syntenic. This conserved region contains gene homologs of COG3364 (vng2406), COG3365 (vng2408), COG1100 (gbp3), COG1474 (orc7), COG2259 (vng2413), COG0681 (sec11), and COG1311 (polD1), in order, in nearly all genomes (Fig. 5). One exception is N. pharaonis, where the sec11 and polD1 homologs are inverted and approximately 54 kb upstream of the vng2406 homolog. Additional differences include a duplication of the sec11 gene in H. marismortui, predicted extra genes between the gbp3 and orc7 homologs in both H. lacusprofundi and H. volcanii, and the absence of a short region containing vng2412 to vng2415 in all except the Halobacterium species.
FIG. 4.
Haloarchaeal IRs and regulatory regions of oriC1 and oriC2. (A) Alignment of 31-bp IRs located upstream of the orc gene homologs in seven sequenced haloarchaeal genomes. R and L indicate right and left sequences of IRs. (B) Logo representation of IRs found in oriC1 regions of seven haloarchaeal chromosomes. (C) Alignment of possible regulatory sequences found upstream of the orc gene homologs in seven sequenced haloarchaeal genomes. (D) Logo representation of regulatory sequences in panel C.
FIG. 5.
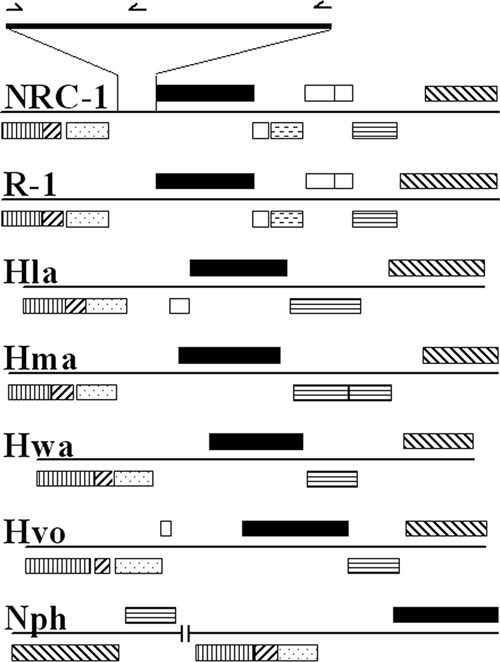
Map of oriC1 syntenic region in sequenced haloarchaea. The pictorial representations of an approximately 8-kb highly conserved region near oriC1 illustrate orc7 (black boxes), genes in COG3364 (boxes with vertical lines), COG3365 (boxes with left-to-right diagonal lines), COG1100 (boxes with dots), COG2259 (boxes with dashed lines), COG0681 (boxes with horizontal lines), and COG1311 (boxes with right-to-left diagonal lines), and genes not associated with COGs (white) of Halobacterium sp. strain NRC-1, Halobacterium sp. strain R-1, Halorubrum lacusprofundi (Hla), Haloarcula marismortui (Hma), Haloquadratum walsbyi (Hwa), Haloferax volcanii (Hvo), and Natronomonas pharaonis (Nph). The enlargement shows the locations of repeats and was drawn to scale based on the map of Halobacterium sp. strain NRC-1.
In order to determine the prevalence of the conserved 31-bp IRs in the oriC1 replication origin as a possible predictor of other origins in NRC-1, a search matrix was generated using the JEMBOSS program Prophecy and used for a gapped alignment search of the Halobacterium sp. strain NRC-1 genome, using Profit. The two top hits were similar to sequences immediately upstream of the orc7 and orc10 genes, with scores of 92% and 86%, respectively (Fig. 4C and D), in a region consistent with a role in gene regulation. Additional rounds of multiple sequence alignments, profile building, and searching did not identify other examples of the oriC1 repeated sequences with significant scores in the NRC-1 genome. However, similar arrangements were found immediately upstream of an orc7 homolog in the six other sequenced haloarchaea (Fig. 4C and D), indicating a conservation of the regulatory mechanism.
Statistical analysis of the chromosomal sequence.
In order to identify sequences that may be important for initiating replication at sites other than oriC1, we used χ2 and GC compositional analyses. For Halobacterium sp. strain NRC-1, deviant regions of the genome based on χ2 analysis have been shown to contain relatively high AT compositions, and in one case (near orc7 and oriC1) they were correlated with an inflection point in GC oligomer skew analysis (15). When the χ2 plot was superimposed on the MFA plot (Fig. 1), major χ2 peaks coincided with marker frequency peaks. These findings were consistent with a functional correlation between AT-richness, χ2, and DNA replication origins in Halobacterium sp. strain NRC-1. However, with the exception of the oriC1 region, no strict correlation could be observed between the MFA plot and the GC oligomer skew analysis (15).
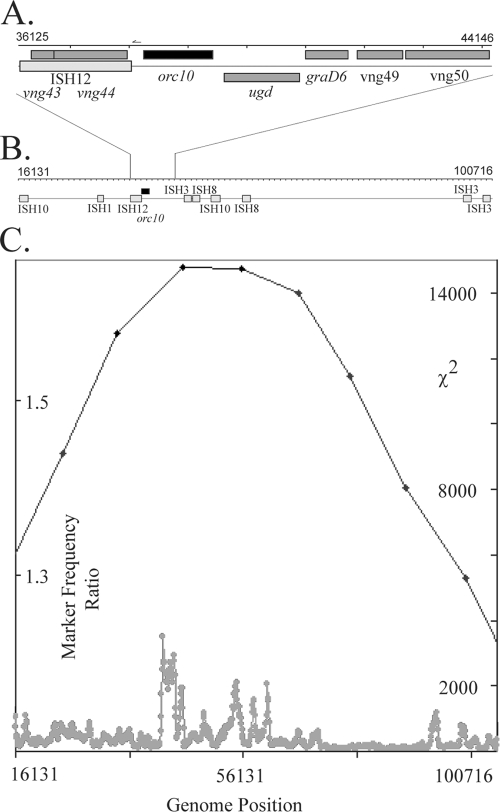
The region with both the highest peak of marker frequency and the highest χ2 value was near the essential orc10 gene (Fig. 1 and 6). Interestingly, a 40-kb segment around this region contained an abundance of ISH (insertion sequence from haloarchaea) elements, including two each of ISH8 and ISH10 and one each of ISH1, ISH3, and ISH12, the latter of which is located only 177 bp upstream of the orc10 coding region. The region of greatest χ2 deviation was found to be 3 to 4 kb downstream of orc10, in a highly AT-rich region carrying two nonconserved open reading frames (vng49 and vng50). This region and the oriC3 region contain small inflections in GC skew analysis (15).
FIG. 6.
Genome arrangement and χ2 analysis of oriC2. (A and B) Pictorial representations adapted from Halobacterium genome database illustrating orc10, insertion elements, hypothetical genes, and known genes. (A) Approximately 8-kb region near oriC2. The arrow depicts the relative position and direction of the consensus sequence found (not to scale). (B) Approximately 84-kb ISH-rich region surrounding oriC2. (C) MFA and χ2 analysis of chromosome region (drawn to scale). The black points and line show the marker frequency values (scale on y axis at left), and the gray points and line show the χ2 values (scale on y axis at right). The approximate location of the orc10 gene is indicated in relation to the chromosome position.
Replication of the pNRC100 minichromosome.
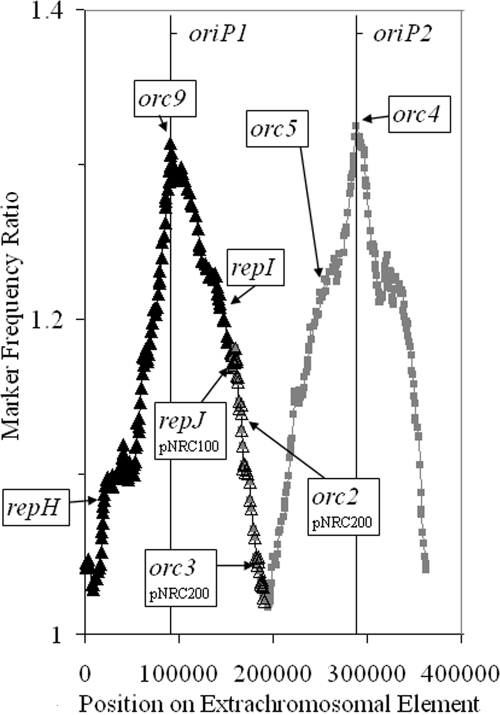
pNRC100 is the smaller (191 kb) of the two extrachromosomal elements and contains a single orc gene (orc9) previously shown to be nonessential through targeted gene knockout (2). Our previous study also identified an ARS in the region near the repH gene, which was shown to be required for replication of pNRC100 miniplasmid constructs by deletion and linker scanning mutagenesis (22). The finding of two other similar genes, repI and repJ, and an abundance of ISH elements suggested that this replicon evolved through the fusion of several smaller plasmids (22). The acquisition of several essential genes was underscored by the finding of a relatively GC-rich chromosomal region. In order to determine the location of the replication origin(s) on pNRC100, marker frequency data for Halobacterium sp. strain NRC-1 and orc gene deletion strains were plotted (Fig. 7 and data not shown). A single major peak was observed (oriP1) between orc9 and repI in every case, indicating a single replication origin. Moreover, no significant peak could be observed near repH, indicating that the nearby ARS previously identified experimentally was not used for initiating in vivo replication of pNRC100. Similar results were obtained for the Δorc6, Δorc7, Δorc8, Δorc68, and Δorc678 deletion strains (data not shown).
FIG. 7.
MFA of the megaplasmids of Halobacterium sp. strain NRC-1. Filled and empty black triangles and lines show the marker frequency values for pNRC100. Filled black triangles and filled gray squares show the marker frequency values for pNRC200. Filled black triangles represent the marker frequency values for the region shared by pNRC100 and pNRC200. Empty black triangles represent the marker frequency values for the unique region of pNRC100. Gray squares represent the marker frequency values for the unique region of pNRC200. Vertical lines show the approximate locations of oriP1 and oriP2 in relation to the position on pNRC200.
Replication of the pNRC200 minichromosome.
pNRC200 is the larger (365 kbp) of the two extrachromosomal elements, containing six orc genes, orc1 to orc5 and orc9, also present in pNRC100. pNRC200 contains an additional 220-kb unique region with many important genes, including the essential orc2 gene, the only arginyl-tRNA synthetase gene (argS), and others, that define this element as a minichromosome (8). Analysis of the marker frequency data for Halobacterium sp. strain NRC-1 indicated that in addition to the origin in the common region with pNRC100 (oriP1), pNRC200 contains a second origin in its unique region (oriP2), near the nonessential orc4 and polB2 genes (Fig. 7). No difference was observed in the Δorc6, Δorc7, Δorc8, Δorc68, and Δorc678 deletion strains (data not shown).
Construction of a minimal orc strain.
Previously, single deletions of orc genes had shown that all but two, orc2 and orc10, were nonessential (2). In order to determine the minimum number and identity of orc genes necessary for DNA replication and cell viability in Halobacterium sp. strain NRC-1, we proceeded to serially delete the orc genes. Starting with the Δorc678 mutant, which also contained a natural deletion of orc1, we successively deleted orc4, orc5, and finally orc3 to obtain a septuple-deletion strain (Δorc1 Δorc3 Δorc4 Δorc5 Δorc6 Δorc7 Δorc8) containing only three orc genes, orc2, orc9, and orc10 (Table 1). Several rounds of attempted deletion of orc9, which had previously been deleted singly, were unsuccessful. Up to 40 excisants were screened in each attempt, which provides a probability of >99.999% of identifying a knockout (2) and strongly suggests the requirement for this gene in the absence of other deleted genes.
DISCUSSION
Genetic studies of DNA replication together with MFA have provided a complex picture of a fundamental genetic process in a model haloarchaeon. The initial finding of an ARS near the orc7 gene in Halobacterium sp. strain NRC-1 and the conservation of an orthologous gene in all haloarchaea and other archaeal organisms (Aeropyrum, Pyrococcus, and Sulfolobus spp.) where replication origins have been identified experimentally indicated that this region serves as a major chromosomal origin of replication (3, 14, 16, 21). However, our subsequent study showed that the orc7 gene was dispensable for cell viability in NRC-1, while the chromosomal orc10 gene and the pNRC200 orc2 gene were essential (2). With the results of MFA and RIP analysis from our current investigation, the ARS region near orc7, now named oriC1, has been confirmed to serve as an orc7-dependent and major, albeit nonessential, origin of replication in the chromosome of the model archaeon Halobacterium sp. strain NRC-1.
The current results establish the use of multiple replication origins in the NRC-1 chromosome and show that oriC1 is not the sole replication origin. Multiple replication origins are also used for replication of the larger extrachromosomal element (pNRC200), with only a single origin likely used for the smaller one (pNRC100). MFA plots showed that the copy numbers of markers around the chromosome display four or more major symmetric peaks, suggestive of at least four bidirectional replication origins; however, the exact number of origins remains unresolved due to the presence of additional, smaller peaks. Significantly, the second largest peak corresponds to the orc7 gene and oriC1, previously shown to harbor an orc7-dependent ARS activity (3). Our deletion analysis coupled with MFA results also clearly shows that the loss of orc7, but not orc6 or orc8, leads to the reduced use or abandonment of oriC1 as a replication origin and establishes the dispensability of this origin for cell viability. This result was reinforced by the use of double- and triple-deletion mutants. Our comparative genomic results demonstrated that the oriC1 region is highly conserved among the haloarchaea, despite its dispensability for Halobacterium sp. strain NRC-1.
We also established the exact locations of bidirectional RIPs in the oriC1 region by RIP mapping. Primers in the regions flanking the ARS AT-rich/IR region upstream of orc7 clearly showed two divergently oriented initiation points within the ARS. The initiation point moving downstream from the region with respect to the orientation of orc7 gene transcription occurs within the AT-rich likely DNA unwinding element, while the initiation point moving upstream occurs between the AT-rich region and the start of the orc7 gene. These results suggest that the leading strands of replicated DNA are nonsymmetric with respect to the origin, perhaps reflecting an asymmetry in DNA-protein complexes and/or inherent melting properties of the unwinding regions (12, 19).
The strongest peak apparent by MFA for Halobacterium sp. strain NRC-1 and all of the orc gene mutants corresponds to the essential orc10 gene region. As a result, we concluded that the orc10 gene region contains a second major chromosomal origin. However, oriC2, unlike the oriC1 region, is not highly conserved in other haloarchaea. The oriC2 region is distinguished by being the most AT-rich sequence and having a highly deviant composition based on χ2 analysis, as well as having the highest abundance of ISH elements in the NRC-1 chromosome. No less than seven recognized ISH elements are present in a 40-kb region, and this region was previously proposed to be “plasmid-like” (23). It is possible that the oriC2 region is derived from a plasmid that integrated into the chromosome, bringing along its own replication origin that became functional and essential in the integrant. If so, oriC2 probably evolved to become an essential component of the chromosomal replication system only in Halobacterium species, resulting in the dispensability of the oriC1 region. oriC2 is functional in a Δorc678 background, indicating that the replication machinery involved in initiation at this origin is distinct from that used for oriC1.
While the location of the oriC1 origin of replication was defined by MFA, ARS, and RIP mapping at the nucleotide level, the location of oriC2 was mapped to a relatively broad 40-kb region, based solely on MFA. To determine whether this region contained any structural similarities to oriC1, we performed extensive bioinformatic analysis. We found that a 2.4-kb region downstream of orc10 was highly AT-rich (>50%, compared to the 67% GC average for the chromosome), but it did not contain flanking IRs similar in arrangement to those in oriC1. Upstream of orc10 (3.6 kb), imperfect direct repeats (12 of 19 bp) were identified, with Profit scores of 71 and 75%, flanking a 276-bp relatively (>50%) AT-rich region. However, whether either of these regions contains the oriC2 replication origin is not known. We also identified the most closely related sequences similar to single copies of the IRs immediately upstream of the orc7 and orc10 start codons (4 and 57 bp, respectively), in regions likely to be involved in regulation of these genes. These findings suggest that orc7 and orc10 may either be autoregulated or regulated by another common protein binding at these sites. The size of the oriC1 IRs is somewhat larger, especially in the 5′ region, than the orc7 and orc10 putative regulatory sequences, suggesting that an additional replication factor may bind at the oriC1 IRs. Conservation of these features at the orc7/oriC1 origin in all sequenced haloarchaea suggests the occurrence of common mechanisms for replication initiation within the oriC1 region.
Several other regions of the chromosome appear to harbor replication origins not predicted in previous Z-curve (34) and GC skew (15) analyses, based on MFA. Like the oriC2 region, we lack further mapping information for those regions. Based on our genetic and MFA studies, we do know that these likely origins are independent of orc6, orc7, and orc8, but which genes are necessary for initiation at these sites is currently unknown. The requirement for orc10 as well as orc2 and orc9 for initiation at these sites is a possibility. Replication origins in eukaryotes have been shown to be of the following two general types: (i) specifically recognized origins with localized initiation regions and (ii) randomly recognized origins whose initiation may be dispersed across a large region (13). It is possible that oriC1 is similar to the former type, while oriC2 and the other origins may be reminiscent of the latter type. This is further highlighted by the observations that the positions of oriC3 and oriC4 seem to differ slightly in the different orc mutant strains and that oriC2 and oriC3 contain several inflection points in GC skew analysis (Fig. 2) (10). Additional studies are necessary to establish the genes and the specificity of origin sequences that are involved in replication of haloarchaeal genomes.
Halobacterium sp. strain NRC-1, like most haloarchaea, harbors multiple extrachromosomal replicating elements in its genome, proposed to constitute minichromosomes (11). Interestingly, the pNRC100 and pNRC200 elements share an extensive region of identity with one another (23). The common region displayed a peak in MFA, corresponding to one replication origin, oriP1. The peak is located near orc9 and not far from repI, a gene related to repH, which is required for replication of the pNRC minireplicon constructs, suggesting the possibility of different modes of replication in these regions (22, 23). The possibility of closely spaced origins of diverse types is consistent with the previously proposed hypothesis of the evolution of pNRC100 and pNRC200 from the fusion of multiple smaller plasmids.
Previously, only a single ARS region, which contained repH, could be cloned from pNRC100 (22), but this region did not contain a significant peak in MFA consistent with a function as a replication origin. However, the replication ability of the repH gene region has been shown amply by the development and utilization of a family of vectors. The successful replication ability of these vectors may be the result of a lack of inhibition of replication from incompatibility with the resident pNRC plasmids, which do not use the same replication system for their propagation in Halobacterium sp. strain NRC-1 (10). The mechanism that keeps the repH ARS region inactive for replication initiation on pNRC100 and pNRC200 is not known but may involve a cis-acting factor not present on the pNRC miniplasmid constructs.
pNRC200 displays a large region of identity to pNRC100, as well as a large distinct unique region, and it showed more complex replication characteristics by MFA. The unique region contained a second MFA peak, centered near orc4, consistent with a second replication origin (oriP2). Interestingly, the polB2 gene, which we found to be nonessential in previous work, is also found near oriP2 (2). The arrangement is similar to that in P. abyssi, where the DNA polymerase gene polD was previously found near its replication origin (21).
Our deletion analysis showed that most of the orc genes are nonessential, either in strains with single deletions or after sequential deletions. Only two orc genes were essential in single deletions, namely, orc2 (on pNRC200) and orc10 (on the chromosome). We were able to delete the other three chromosomal orc genes, orc6, orc7, and orc8, and subsequently three pNRC200 genes, orc3, orc4, and orc5. The orc1 gene had been lost earlier by a natural deletion in the Δura3 strains used for construction of gene knockouts, so the resulting strain had 7 of 10 orc genes deleted. However, the orc9 gene, found on both pNRC100 and pNRC200, was not deleted in several attempts in a strain with deletion of all of the other genes except for orc2 and orc10, suggesting that it may be required when certain other orc genes are absent. Therefore, the minimal orc gene strain we were able to construct contained orc2, orc9, and orc10. It is possible that these genes are essential for replication of the chromosome and/or possibly pNRC100/200. Since the haloarchaeal orc genes are homologs of both the eukaryotic origin recognition and helicase loader proteins, one possibility is that orc2 and orc10 (and possibly orc9) function in these two different processes. Studies of A. pernix and S. solfataricus have shown that not all Orc proteins bind at the origin during initiation of replication (30). The two Orc proteins in S. solfataricus and one in A. pernix that bind during initiation are members of a separate archaeal Orc1/Cdc6 family from the family of Orc proteins that do not, supporting the idea of individual Orc proteins serving the function of origin recognition or helicase loading (2). Since the replication of the pNRC megaplasmids was unaffected by any of the chromosomal orc genes, it is possible that each one uses its own locally encoded Orc protein(s) for replication, which is supported by the essentiality of orc2, orc9, and orc10. Therefore, the orc10 gene may be essential for replication of the chromosome, while orc2 and orc9 may be required for replication of pNRC200 and pNRC100. However, additional experimentation will be required to test this hypothesis.
Recent work on replication origins in a distantly related haloarchaeon, H. volcanii, showed that only a fraction of the 14 orc and cdc6 genes were near replication origins, and the essentiality of these genes for cell viability was not established. Using ARS and RIP mapping analysis, five origins were identified, with two on the chromosome and one each on three of the four smaller plasmids. Furthermore, of the two chromosomal origins detected, one was found to be less efficient than the other, similar to oriC1 and oriC2 (24). The relatively different peak heights for the Halobacterium origins of replication suggest that they may be used similarly, with different efficiencies.
The multiple replication origins in the genome and the variation in their efficiencies, as well as the presence of multiple copies of the orc genes, could suggest that different origins and perhaps Orc proteins are preferentially used under different environmental stresses or growth conditions. We have previously shown via biochemical, genetic, and transcriptomic data that this is true for some of the proteins involved in promoter selection (7), and it is tantalizing to speculate a similar regulatory mechanism for the replication machinery in NRC-1. Consistent with this hypothesis, our preliminary microarray analysis suggested that the orc genes are differentially regulated under different growth conditions (6). It should also be noted that the variation in height of the marker frequency peaks found for the wild-type and deletant strains could indicate asynchronous replication or heterogeneity in the population.
Studies of DNA replication in diverse archaea have shed considerable light on this fundamental genetic process in the third domain of life. While the genome organization and structure are similar to those of bacteria, the DNA replication machinery carrying out the initiation process is more closely related to that of eukaryotes. The findings reported in the present work and previous studies (16, 24, 30) suggest that multiple replication origins are commonly used in the archaeal branch of life and provide another significant similarity between higher organisms and archaea. Whether and how the actions of multiple replication origins can be regulated and coordinated remain problems of significant interest.
Acknowledgments
We thank Steven Salzberg for providing software and Nancy Fossett for a careful reading of the manuscript.
This work was supported by NSF grant MCB-0296017 and NASA grant NNX08AT70G to S.D. and by the National Basic Research Program of China (2004CB719603) to H.X.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Berquist, B., and S. DasSarma. 2008. DNA replication in archaea, p. 96-120. In P. Blum (ed.), Archaea: new models for prokaryotic biology. Horizon Press, Norwich, CT.
- 2.Berquist, B. R., P. DasSarma, and S. DasSarma. 2007. Essential and non-essential DNA replication genes in the model halophilic archaeon, Halobacterium sp. NRC-1. BMC Genet. 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berquist, B. R., and S. DasSarma. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185:5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berquist, B. R., J. A. Müller, and S. DasSarma. 2006. Genetic systems for halophilic archaea, p. 649-680. Extremophiles 35:649-680. [Google Scholar]
- 5.Cleveland, W. S. 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74:829-839. [Google Scholar]
- 6.Coker, J. A., P. DasSarma, J. Kumar, J. A. Muller, and S. DasSarma. 2007. Transcriptional profiling of the model archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Syst. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coker, J. A., and S. DasSarma. 2007. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic archaeon: coordinate action of TbpD and TfbA. BMC Genet. 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DasSarma, S. 2004. Genome sequence of an extremely halophilic archaeon, p. 383-399. In C. M. Fraser, T. D. Read, and K. E. Nelson (ed.), Microbial genomes. Humana Press, Inc., Totowa, NJ.
- 9.DasSarma, S. 1995. Halophiles, p. 179-184. In S. DasSarma and E. M. Fleischmann (ed.), Archaea, a laboratory manual—halophiles. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 10.DasSarma, S. 1995. Natural plasmids and plasmid vectors of halophiles, p. 243-250. In S. DasSarma and E. M. Fleischmann (ed.), Archaea, a laboratory manual—halophiles. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 11.DasSarma, S., M. Capes, and P. DasSarma. 2008. Haloarchaeal megaplasmids, p. 3-32. In E. Schwartz (ed.), Microbial megaplasmids, vol. 11. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 12.Gerbi, S. A., and A. K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271-280. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grainge, I., M. Gaudier, B. S. Schuwirth, S. L. Westcott, J. Sandall, N. Atanassova, and D. B. Wigley. 2006. Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J. Mol. Biol. 363:355-369. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, S. P., W. V. Ng, S. L. Salzberg, L. Hood, and S. DasSarma. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundgren, M., A. Andersson, L. Chen, P. Nilsson, and R. Bernander. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. USA 101:7046-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majernik, A. I., and J. P. Chong. 2008. A conserved mechanism for replication origin recognition and binding in archaea. Biochem. J. 409:511-518. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga, F., P. Forterre, Y. Ishino, and H. Myllykallio. 2001. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA 98:11152-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga, F., C. Norais, P. Forterre, and H. Myllykallio. 2003. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller, J. A., and S. DasSarma. 2005. Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J. Bacteriol. 187:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myllykallio, H., P. Lopez, P. Lopez-Garcia, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:2212-2215. [DOI] [PubMed] [Google Scholar]
- 22.Ng, W. L., and S. DasSarma. 1993. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. J. Bacteriol. 175:4584-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norais, C., M. Hawkins, A. L. Hartman, J. A. Eisen, H. Myllykallio, and T. Allers. 2007. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 26.Pei, H., J. Liu, J. Li, A. Guo, J. Zhou, and H. Xiang. 2007. Mechanism for the TtDnaA-Tt-oriC cooperative interaction at high temperature and duplex opening at an unusual AT-rich region in Thermoanaerobacter tengcongensis. Nucleic Acids Res. 35:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollack, J. R., C. M. Perou, A. A. Alizadeh, M. B. Eisen, A. Pergamenschikov, C. F. Williams, S. S. Jeffrey, D. Botstein, and P. O. Brown. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, N. P., and S. D. Bell. 2007. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl. Acad. Sci. USA 104:5806-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson, N. P., K. A. Blood, S. A. McCallum, P. A. Edwards, and S. D. Bell. 2007. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 26:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, N. P., I. Dionne, M. Lundgren, V. L. Marsh, R. Bernander, and S. D. Bell. 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116:25-38. [DOI] [PubMed] [Google Scholar]
- 31.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, C., M. Zhou, Y. Li, and H. Xiang. 2006. Molecular characterization of the minimal replicon and the unidirectional theta replication of pSCM201 in extremely halophilic archaea. J. Bacteriol. 188:8136-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., and B. Seed. 2003. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics 19:796-802. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, R., and C. T. Zhang. 2005. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea 1:335-346. [DOI] [PMC free article] [PubMed] [Google Scholar]