Abstract
Campylobacter jejuni encodes all the enzymes necessary for a complete oxidative tricarboxylic acid (TCA) cycle. Because of its inability to utilize glucose, C. jejuni relies exclusively on amino acids as the source of reduced carbon, and they are incorporated into central carbon metabolism. The oxidation of succinate to fumarate is a key step in the oxidative TCA cycle. C. jejuni encodes enzymes annotated as a fumarate reductase (Cj0408 to Cj0410) and a succinate dehydrogenase (Cj0437 to Cj0439). Null alleles in the genes encoding each enzyme were constructed. Both enzymes contributed to the total fumarate reductase activity in vitro. The frdA::cat+ strain was completely deficient in succinate dehydrogenase activity in vitro and was unable to perform whole-cell succinate-dependent respiration. The sdhA::cat+ strain exhibited wild-type levels of succinate dehydrogenase activity both in vivo and in vitro. These data indicate that Frd is the only succinate dehydrogenase in C. jejuni and that the protein annotated as a succinate dehydrogenase has been misannotated. The frdA::cat+ strain was also unable to grow with the characteristic wild-type biphasic growth pattern and exhibited only the first growth phase, which is marked by the consumption of aspartate, serine, and associated organic acids. Substrates consumed in the second growth phase (glutamate, proline, and associated organic acids) were not catabolized by the the frdA::cat+ strain, indicating that the oxidation of succinate is a crucial step in metabolism of these substrates. Chicken colonization trials confirmed the in vivo importance of succinate oxidation, as the frdA::cat+ strain colonized chickens at significantly lower levels than the wild type, while the sdhA::cat+ strain colonized chickens at wild-type levels.
Campylobacter jejuni causes approximately two million cases of bacterial gastroenteritis in the United States annually (34). Humans are most often infected due to cross-contamination resulting from improper handling of poultry (27), which is the natural habitat of C. jejuni (28). The eradication of C. jejuni from poultry flocks is an important goal in reducing the number of campylobacteriosis cases.
C. jejuni can rely solely on catabolism of small organic acids and amino acids as a carbon and energy source, and the products of this catabolism are used for glycolysis and the tricarboxylic acid (TCA) cycle (15, 29). Fumarate and succinate are key intermediates in the TCA cycle, and the interconversion of these compounds is a vital process in organisms that use the TCA cycle for central carbon metabolism. C. jejuni encodes a complete oxidative TCA cycle, which converts TCA intermediates (carboxylic acids) to CO2, ATP, and reducing equivalents. One of the conversion steps, oxidation of succinate to fumarate, forms a reducing equivalent and is required for a complete cycle. Reduction of fumarate to succinate also occurs as part of the reductive TCA cycle, and this carbon fixation pathway has been proposed to be utilized by ɛ-proteobacteria found in deep-sea hydrothermal vents (3). C. jejuni encodes many of the reversible enzymes necessary for the reductive TCA cycle, including 2-oxoglutarate ferredoxin oxidoreductase (encoded by oorDABC) and pyruvate carboxylase (encoded by pycA and pycB) (29); however, C. jejuni does not encode an ATP citrate lyase, which is required for full cyclic reductive carboxylation (3). The fumarate-succinate interconversion is also involved in respiration (11), and fumarate has specifically been implicated as an electron acceptor that is an alternative to oxygen in other ɛ-proteobacteria (5, 17).
C. jejuni encodes an enzyme which is annotated as a fumarate reductase (Cj0408 to Cj0410) and an enzyme which is annotated as a succinate dehydrogenase (Cj0437 to Cj0439) (29). Both of these enzymes are part of a large family of proteins called the succinate:quinone oxidoreductases (SQRs). These compounds are membrane-bound enzymes that either catalyze the two-electron oxidation of succinate to the two-electron reduction of quinone/quinol or, in the reverse direction, couple the oxidation of quinol/quinone to the reduction of fumarate to succinate. The amino acid sequence, however, does not dictate the in vivo function (18), and in characterized organisms like Escherichia coli both enzymes are able to reduce fumarate and oxidize succinate, albeit with a preference for one substrate (6, 21).
The SQRs can be divided into three distinct classes based on function, all of which have similar subunit compositions and primary amino acid sequences. Class 1 SQRs couple the oxidation of succinate to the reduction of a high-redox-potential quinone like ubiquinone in vivo. Class 2 SQRs are the quinol:fumarate reductases, which couple the oxidation of menaquinol to the reduction of fumarate. And class 3 SQRs couple the oxidation of succinate to the reduction of a low-potential quinone, such as menaquinone, in vivo (11). Although each class has shared motifs, the in vivo function of an SQR enzyme cannot be resolved based on the primary sequence and must be determined experimentally. Fumarate reductase (Frd) activity has been reported to occur in the particulate fraction of C. jejuni cell lysates, and addition of formate to whole cells increased Frd activity (38), which implies that there is an active electron transport pathway. However, C. jejuni is unable to utilize fumarate as an alternative electron acceptor under anaerobic conditions (37, 41). C. jejuni can also use succinate as an electron donor to a respiratory quinone (12), which has been identified as either a menaquinone-6 or methylmenaquinone-6 (4). Yet succinate oxidation via menaquinone is an endergonic reaction; succinate has a redox midpoint potential (Em) of 30 mV, and menaquinone is more electronegative (Em = −80 mV). Although succinate oxidation coupled to menaquinone reduction would be an “uphill” reaction, class 3 SQRs can catalyze this reaction. Studies of gram-positive bacteria belonging to the genus Bacillus, as well as studies of sulfate-reducing bacteria, have shown that oxidation of succinate through menaquinone is driven by reverse transmembrane electron transport (18, 36, 45), and it is hypothesized that C. jejuni behaves similarly. The C. jejuni Frd enzyme contains three subunits, FrdC, FrdA, and FrdB, and the gene order in the operon is similar to that in Wolinella succinogenes (16, 19) and Helicobacter pylori (1, 9, 40). Based on Frd enzymes of other bacteria, FrdC (Cj0408) is the membrane anchor and diheme cytochrome b, FrdA (Cj0409) is a flavoprotein where the reduction of fumarate to succinate occurs, and FrdB (Cj0410) is an Fe-S protein (29). The succinate dehydrogenase of C. jejuni is also composed of three subunits, SdhABC encoded by Cj0437 to Cj0439 (29). SdhA is annotated as a succinate dehydrogenase flavoprotein subunit, SdhB is a putative succinate dehydrogenase Fe-S protein, and SdhC is a putative succinate dehydrogenase subunit C. According to ClustalW pairwise alignment, FrdA and SdhA of C. jejuni share 29% identity, FrdB and SdhB share 18% identity, and FrdC and SdhC share 13% identity.
A better understanding of the C. jejuni TCA cycle may help identify metabolic pathways that are crucial to C. jejuni's ability to thrive in poultry. The roles of the C. jejuni fumarate reductase and succinate dehydrogenase in the TCA cycle and respiration were investigated. Both enzymes contribute to the total fumarate reductase activity. We determined that the protein annotated as the fumarate reductase functions as the sole succinate dehydrogenase and that this enzyme is required for full colonization of chickens by C. jejuni. The sdh operon has been misannotated as the enzyme that it encodes exhibits no succinate dehydrogenase activity, as has recently been reported to be the case for the annotated succinate dehydrogenase of W. succinogenes (14).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Table 1 lists the strains of C. jejuni and E. coli and all of the plasmids and primers utilized in this study. Tryptic soy agar (Difco, Sparks, MD) supplemented with 10% defibrinated sheep blood (Gibson Laboratories, Inc., Lexington, KY) (TSA-B) was used for growth of C. jejuni. C. jejuni was routinely cultured at 37°C microaerobically in a tri-gas incubator (model 550D; Fisher Scientific) maintained with 5% CO2, 12% O2, and 83% N2. Liquid cultures were grown in Mueller-Hinton broth (MHB) (Difco, Sparks, MD) or modified Eagle's medium (MEM) lacking glucose, glutamine, phenol red, and sodium pyruvate (Sigma, St. Louis, MO). Liquid cultures were incubated at 37°C with shaking under a microaerobic atmosphere. Chloramphenicol (25 μg/ml) was added as indicated below. Genetic manipulations were performed with E. coli strain DH5α (laboratory stock). Luria-Bertani broth and agar were supplemented with ampicillin (100 μg/ml) or chloramphenicol (25 μg/ml) as noted below.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Description or sequence | Sourcea |
|---|---|---|
| C. jejuni strains | ||
| NCTC 11168 | Parent of all C. jejuni strains | NCTC |
| frdA::cat+ | cat insertion in frdA | This study |
| sdhA::cat+ | cat insertion in sdhA | This study |
| Cj0411::cat+ | cat insertion in Cj0411 | This study |
| E. coli strains | ||
| DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 φ80ΔlacZM15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Lab stock |
| One Shot Top10 | F−mcrA (mrr-hsdRMS-mcrBC) φ80ΔlacZM15 lacX74 recA1 ara139 (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG; chemically competent cells | Invitrogen |
| Plasmids | ||
| pCRTOPO2.1 | Cloning vector | Invitrogen |
| pJMA-001 | pGEM-T containing cat insert | Jay Andrus |
| pFrdA:TOPO | frdA cloned from NCTC 11168 into pCRTOPO2.1 | This study |
| pFrdA:cm | cat inserted into Eco47III site of pFrdA:TOPO | This study |
| pSdhA:TOPO | sdhA cloned from NCTC 11168 into pCRTOPO2.1 | This study |
| pSdhA:cm | cat inserted into SspI site of pSdhA:TOPO | This study |
| pCj0411:TOPO | Cj0411 cloned from NCTC 11168 into pCRTOPO2.1 | This study |
| pCj0411:cm | cat inserted into Eco47III site of pCj0411:TOPO | This study |
| Primers | ||
| FrdAF | 5′-GTTGTAGCAGGTATGATAGTGGGAG-3′ | IDT |
| FrdAR | 5′-AACTTCTGGCTCAATGCGTTC-3′ | IDT |
| SdhAF | 5′-TGATTGCTACAGGGGGATACAC-3′ | IDT |
| SdhAR | 5′-TCACTTCATTAGGATTTGGAATACG-3′ | IDT |
| Cj0411F | 5′-AAGCAAACTTGATACAGCCGAG-3′ | IDT |
| Cj0411R | 5′-GATTTACCACTTGAAAACTGCCC-3′ | IDT |
| RTGyrAF | 5′-TGGTTGTAACTATCACACATCGTGG-3′ | IDT |
| RTGyrAR | 5′-AATCATCATCATAAGTCGTAACGGC-3′ | IDT |
| RTFrdAF | 5′-GCAGCCATCGCTCTTGAAACAG-3′ | IDT |
| RTFrdAR | 5′-CGACAGCCCTCAGTAAGCAAAATAC-3′ | IDT |
| RTCj0411F | 5′-CAGGGCTTGATGATGTTGTAGTTC-3′ | IDT |
| RTCj0411R | 5′-GACAATGCACCAAAAAGTCTGC-3′ | IDT |
NCTC, National Collection of Type Cultures; IDT, Integrated DNA Technologies, Coralville, IA.
Succinate dehydrogenase assay.
Succinate dehydrogenase activity was measured by measuring 2,6-dichlorophenol-indophenol (DCPIP)-dependent reduction by succinate, as described by Schirawski and Unden (35). Briefly, a closed quartz cuvette containing 50 mM NaPO4 buffer (pH 7.2), 0.25 mM DCPIP, 0.4 mM phenazine methosulfate, and 100 μl of cell extract was made anoxic by sparging it with N2 gas. The reaction was started by addition of 20 mM sodium succinate (pH 7.4), and the DCPIP-dependent reduction kinetics were recorded with a spectrophotometer at 600 nm. The rate of reduction was expressed in μmol DCPIP reduced min−1 mg protein−1 using a molar extinction coefficient for DCPIP of 2.1 × 104 cm−1.
Fumarate reductase assay.
Benzyl viologen-linked reductase assays were carried out with sonicated cell extracts using a 1-ml assay mixture as described previously (37). Reagents were added with a syringe through the stopper, while N2 gas was flushed through the cuvette. The reaction mixture in 1-ml quartz cuvettes with stoppers (reagents were kept anaerobic during the course of the assay) contained 75 mM sodium phosphate buffer (pH 6.8), 0.2 mM benzyl viologen, and 1 to 5 μg of cell extract. Freshly made 20 mM sodium dithionite was then injected into each cuvette until the absorbance at 585 nm reached 0.8 to 0.9, which represented half-reduced benzyl viologen. An anaerobic solution of sodium fumarate (final concentration, 5 mM) was added, and the benzyl viologen (extinction coefficient, 8.65 cm−1 mM−1) oxidation kinetics were recorded with a spectrophotometer at 585 nm. Fumarate reductase activity was expressed in nmol of benzyl viologen oxidized min−1 mg−1.
O2 uptake assay.
O2 uptake experiments were performed using a Clarke-type electrode and a YSI model 5300 oxygen monitor (YSI, Yellow Springs, OH) as described previously (44). Briefly, washed whole cells were resuspended in phosphate-buffered saline (PBS), added to the constantly stirred chamber, and allowed to equilibrate until no change in O2 consumption was seen for several minutes. Lactate or succinate (final concentration, 5 mM) was added to the chamber through a capillary tube via a Hamilton syringe, a chart recorder documented the rate of oxygen consumption, and the slope of the line showed the rate of oxygen uptake. The rate was expressed in nmol of O2 consumed min−1 mg−1.
Cloning and construction of C. jejuni mutants.
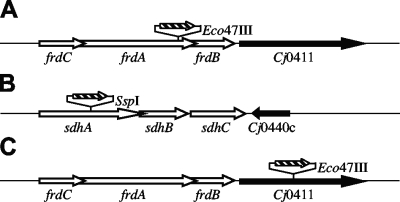
Oligonucleotide primers for cloning genes of interest were designed using sequenced strain NCTC 11168 (29) and are listed in Table 1. PCR amplification was performed with Taq DNA polymerase (Promega, Madison, WI) using chromosomal DNA isolated from C. jejuni NCTC 11168 as the template. The PCR products (1,154 bp of frdA, 904 bp of sdhA, and 2,327 bp of Cj0411) were cloned into pCR2.1-TOPO vectors (Invitrogen, Carlsbad, CA) and confirmed by restriction analysis. The coding region of a gene of interest was disrupted by insertion of a chloramphenicol resistance gene (cat), which was originally isolated from Campylobacter coli (Fig. 1) (43). The restriction endonuclease recognition sequences utilized were Eco47III for frdA, SspI for sdhA, and Eco47III for Cj0411 (Table 1). Electrocompetent C. jejuni cells were transformed with each construct to obtain the corresponding C. jejuni mutant as described previously (44). Transformed cells were spotted onto cold nonselective TSA-B plates, and the plates were incubated microaerobically for 24 h, after which the cells were transferred to TSA-B plates containing chloramphenicol. Resistant colonies that appeared after 2 to 5 days were passed on selective TSA-B plates, and correct insertion of the cassette was confirmed by isolation of chromosomal DNA from the candidate strains and by PCR amplification of the gene. Agarose gel electrophoresis of the PCR product was used to monitor the increase in the size of the gene of interest with the antibiotic cassette insertion (data not shown).
FIG. 1.
(A) Gene organization in the fumarate reductase operon (frdCAB) of C. jejuni. (B) Gene organization in the succinate dehydrogenase operon (sdhABC) of C. jejuni. (C) Directly downstream of the frdCAB operons is Cj0411, which encodes a putative GTPase. The insertion sites of the chloramphenicol cassette (striped arrows) used for mutagenesis are indicated. The directions of the arrows indicate the transcriptional orientations of the genes. Unrelated genes directly downstream of the genes of interest are indicated by black arrows.
Quantitative RT-PCR.
PCR primers used in this study are listed in Table 1 and were designed to amplify gene fragments that were the following sizes: 115 nucleotides for gyrA, and 136 nucleotides for Cj0411. Total RNA was isolated from the C. jejuni parent strain and the frdA::cat+ strain using a MasterPure Complete RNA purification kit (Epicentre Biotechnologies, Madison, WI). Quantitative reverse transcriptase PCR (RT-PCR) was performed by using a Quantitect SYBR green RT-PCR kit (Qiagen, Valencia, CA). The PCR mixture (20 μl) contained 40 ng RNA, 10 μl 2× QuantiTect SYBR green RT-PCR master mixture, and 0.2 μl QuantiTect RT mixture. The RT cycle step consisted of 50°C for 30 min, and this was followed by an initial PCR activation step of 95°C for 15 min. The mixtures were then amplified using 30 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s in an automated thermal cycler (iCycler; Bio-Rad, Hercules, CA). The iCycler software was used to determine the threshold cycle at which each transcript could be detected. Threshold cycles were then compared to a standard curve, which was generated independently for each gene, to determine the number of RNA molecules at the start. The total RNA in each sample was normalized using the internal control gene gyrA (Cj1027c).
Sonication.
Cells in PBS (resuspended to an optical density of 5.0) were disrupted by sonication. Sonication was performed on ice with a W-370 horn cup sonicator (Heat Systems-Ultrasonics, Inc., Farmingdale, NY) using four 45-s pulses at 60% power and an output control setting of 7.0. Cell debris was removed by centrifugation at 1,000 × g for 10 min at 4°C, and the supernatant was removed. The resulting preparation is referred to below as the cell extract.
Protein assay.
Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as a standard.
Chicken colonization.
Campylobacter-free 1-day-old broiler chicks were supplied by the Lake Wheeler Poultry Facility operated by the North Carolina State University Poultry Department. Birds were housed in isolation rooms at the Dearstyne Avian Health Center (Department of Poultry Science, North Carolina State University) in isolation brooder batteries (10 chicks per battery). The chicks were fed Purina Mills Start & Grow SunFresh Recipe (Purina Mills LLC, St. Louis, MO) and given water ad libidum. One-week-old chicks were inoculated by oral gavage with 0.1 ml of C. jejuni in PBS harvested from TSA-B plates which had been incubated at 37°C microaerobically. Control chicks were inoculated with 0.1 ml sterile PBS (pH 7.4). Two weeks postinoculation the chickens were humanely sacrificed by CO2 asphyxiation, and approximately 1 g of cecal contents was collected, serially diluted (in PBS), and plated on selective TSA-B containing 40 μg/ml cefoperazone, 40 μg/ml vancomycin, 10 μg/ml trimethoprim, and 100 μg/ml cycloheximide. All samples were incubated microaerobically at 37°C. After 2 days of incubation, colonies were counted, and the number of CFU/g of cecal contents was calculated. Data were analyzed by using a one-tailed Mann-Whitney test and a 95% confidence interval.
RESULTS
Fumarate reductase mutants do not enter the second C. jejuni growth phase.
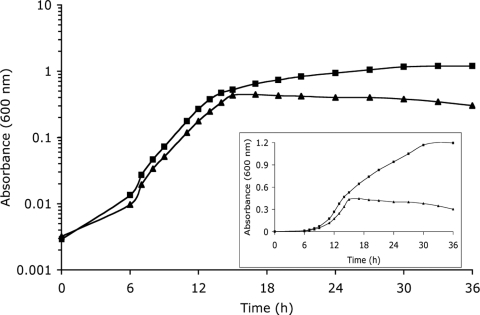
Typical growth curves for the wild type and the frdA::cat+ strain are shown in Fig. 2. The wild type exhibits a characteristic C. jejuni biphasic growth pattern when it is grown in MHB under microaerobic conditions. The frdA::cat+ strain behaves like the wild type during the first growth phase, and the generation time of this strain is 2.2 ± 0.7 h, compared to a growth rate of 2.0 ± 0.4 h for the wild type. When wild-type C. jejuni reaches the mid-log phase (absorbance at 600 nm of approximately 0.3 in MHB), there is a shift to a second growth phase and a longer generation time, 10.3 ± 1.2 h. The frdA::cat+ strain never enters the second growth phase and stops growing after the first growth phase. The inset in Fig. 2 shows the growth curve with a linear scale to better visualize the marked difference in growth characteristics between the wild type and the frdA::cat+ strain. The sdhA::cat+ strain grows like the wild type, with two distinct growth phases with generation times of 2.2 ± 0.05 h and 12 ± 1 h.
FIG. 2.
Microaerobic growth of C. jejuni NCTC 11168 (▪) and the frdA::cat+ strain (▴). Cultures grown in MHB were incubated at 37°C. (Inset) Growth curves with a linear scale to better visualize the marked difference in growth characteristics. The growth curves shown are examples of five independent growth curves.
The frdA::cat+ strain does not utilize substrates that precede succinate oxidation in the TCA cycle.
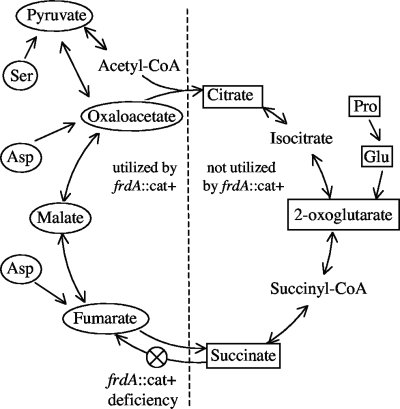
Although fumarate reductase can be used as an alternative electron acceptor in place of oxygen by other Campylobacter species and the ɛ-proteobacterium W. succinogenes (5, 17), C. jejuni is unable to grow anaerobically with fumarate (37, 38). Fumarate reductase is also an essential enzyme in the reverse (reductive) TCA cycle. TCA cycle intermediates were added to MHB to see if the wild-type growth phenotype could be restored to the frdA::cat+ strain. Addition of several metabolic intermediates increased the length in the first growth phase (and thus the terminal optical density) of the frdA::cat+ strain; however, no substrate produced the second growth phase typically seen with wild-type C. jejuni (data not shown). Table 2 shows the terminal optical density at 600 nm (OD600) of the frdA::cat+ strain after 26 h of microaerobic growth after addition of TCA substrates. The TCA intermediates fumarate, malate, oxaloacetate, and pyruvate (which can be converted to oxaloacetate via oxaloacetate decarboxylase) did extend the first growth phase and thus increased the terminal optical density of the frdA::cat+ strain. However, the terminal optical density of the frdA::cat+ strain was not affected by addition of acetate, citrate, 2-oxoglutarate, and succinate, which fall in the first half of the TCA cycle (Fig. 3). When grown in media containing amino acids, C. jejuni utilizes aspartate, glutamate, proline, and serine until they are depleted (10, 20). Both aspartate (a precursor of fumarate via AspA) and serine (via serine dehydratase, which deaminates serine to pyruvate) increased the growth of the frdA::cat+ strain (Table 2). Glutamate (a precursor of 2-oxoglutarate) and proline (proline dehydrogenase oxidizes proline to glutamate) had no effect on the frdA::cat+ strain. The substrates were also tested using the defined medium MEM, which has been shown to support growth when it is supplemented with amino acids (23). The results were essentially the same; none of the non-growth-promoting substrates were utilized by the frdA::cat+ strain, while all of the substrates (except serine) that enhanced growth in MHB supported growth (Table 2). These results are summarized in Fig. 3. TCA intermediates that are utilized by the frdA::cat+ strain group together immediately following the fumarate-succinate interconversion. The intermediates that had no effect on the frdA::cat+ strain group together immediately prior to the fumarate and succinate enzymatic reactions (Fig. 3).
TABLE 2.
Final absorbance at 600 nm of frdA::cat+ strain cultures grown in MHB or MEM (with or without substrate) for 26 h
| Substrate (28 mM) | Absorbance at 600 nm in MHB | Absorbance at 600 nm in MEM | Growth stimulusa |
|---|---|---|---|
| None | 0.24 ± 0.01 | 0.01 ± 0.00 | |
| Citrate | 0.30 ± 0.02 | 0.01 ± 0.00 | N |
| α-Ketoglutarate | 0.28 ± 0.01 | 0.01 ± 0.00 | N |
| Succinate | 0.29 ± 0.01 | 0.01 ± 0.00 | N |
| Glutamate | 0.24 ± 0.01 | 0.01 ± 0.00 | N |
| Proline | 0.24 ± 0.01 | 0.01 ± 0.00 | N |
| Acetate | 0.27 ± 0.01 | 0.01 ± 0.00 | N |
| Pyruvate | 0.56 ± 0.02 | 0.26 ± 0.02 | Y |
| Oxaloacetate | 0.52 ± 0.01 | 0.31 ± 0.01 | Y |
| Malate | 0.69 ± 0.04 | 0.49 ± 0.02 | Y |
| Fumarate | 0.67 ± 0.03 | 0.57 ± 0.02 | Y |
| Aspartate | 0.92 ± 0.05 | 0.41 ± 0.01 | Y |
| Serine | 0.51 ± 0.02 | 0.01 ± 0.00 | Y or N |
Y, yes; N, no.
FIG. 3.
TCA cycle of C. jejuni. Substrates in rectangles are intermediates that did not have an effect on the growth of the frdA::cat+ strain. Substrates in ellipses are intermediates that increased the terminal optical density of the frdA::cat+ strain. CoA, coenzyme A.
Investigation of possible polar effects.
The insertion of an antibiotic cassette into the chromosome can affect the transcription of downstream genes (a “polar effect”) (Fig. 1). Immediately 3′ of the sdh operon is Cj0440c, which encodes a TenA/Thi-4 family protein; however, this gene is on the opposite DNA coding strand, and transcription should not be affected (29). 3′ of the frd operon is Cj0411, which encodes a putative GTP-binding protein. Quantitative RT-PCR was employed to measure the transcription of Cj0411 in both the wild type and the frdA::cat+ strain. Polar effects were seen as the relative levels of transcription of Cj0411 in the frdA::cat+ strain were 0.15 ± 0.08 times those in the wild-type strain. To confirm that the phenotype of the frdA::cat+ strain was a result of fumarate reductase disruption and not due to reduced transcription of the GTP-binding protein, a strain with a mutation in this gene was constructed. The Cj0411::cat+ strain behaved like the wild type, with two distinct growth phases and with growth rates in both phases similar to those of the wild type (data not shown), and it exhibited 94% of the succinate dehydrogenase activity (DCPIP reduction) of the wild-type strain. The growth phenotype associated with the frdA::cat+ strain can be attributed solely to the disruption of the frdA gene.
Fumarate reductase and succinate dehydrogenase activities.
Fumarate reductase activity was measured in wild-type, frdA::cat+, and sdhA::cat+ strain cell extracts. In all three extracts reduced benzyl viologen was used to reduce fumarate (Table 3). The activity in cultures was measured at different growth phases (Table 3). The frdA::cat+ strain required addition of pyruvate to the medium for the terminal optical density of the culture to reach an OD600 of 0.5; the greatest OD600 of the unsupplemented cultures was 0.3 (Table 2). The wild type and the sdhA::cat+ strain were grown with pyruvate as controls. Under all of the conditions tested, the frdA::cat+ strain and the sdhA::cat+ strain exhibited fumarate reductase activity, and the sum of these activities equaled the fumarate reductase activity of wild-type C. jejuni (Table 3).
TABLE 3.
Fumarate reductase activity of C. jejuni cell extracts
| OD600 | Fumarate reductase activity (nmol benzyl viologen reduced min−1 mg−1)
|
||
|---|---|---|---|
| Wild type | frdA::cat+ strain | sdhA::cat+ strain | |
| 0.1 | 61 ± 21 | 4 ± 3 | 63 ± 24 |
| 0.2 | 37 ± 13 | 2 ± 0.4 | 40 ± 17 |
| 0.3 | 347 ± 158 | 67 ± 12 | 139 ± 76 |
| 0.5 | 1265 ± 248 | NDa | 386 ± 156 |
| 0.5 (with pyruvate) | 911 ± 254 | 434 ± 236 | 570 ± 188 |
ND, not done.
Succinate dehydrogenase activity was measured for the wild type and the two mutants at different growth phases. Cell extracts were utilized to measure the reduction of the artificial acceptor DCPIP coupled to succinate oxidation. The wild type and the sdhA::cat+ strain had similar succinate dehydrogenase activities under all of the conditions tested (Table 4). The frdA::cat+ strain, however, was unable to oxidize succinate using DCPIP as an artificial electron acceptor under all of the conditions tested (Table 4). Pyruvate was added to the medium so the OD600 of the frdA::cat+ strain cultures could reach 0.5.
TABLE 4.
Succinate dehydrogenase activity of C. jejuni cell extracts
| OD600 | Succinate dehydrogenase activity (μmol DCPIP reduced min−1 mg−1)
|
||
|---|---|---|---|
| Wild type | frdA::cat+ strain | sdhA::cat+ strain | |
| 0.1 | 11 ± 4 | 2 ± 1 | 18 ± 2 |
| 0.2 | 14 ± 4 | 2 ± 1 | 24 ± 5 |
| 0.3 | 56 ± 17 | 2 ± 1 | 59 ± 18 |
| 0.5 | 87 ± 34 | NDa | 55 ± 21 |
| 0.5 (with pyruvate) | 56 ± 20 | 4 ± 4 | 69 ± 19 |
ND, not done.
Succinate-dependent oxygen uptake is not affected in the sdhA::cat+ strain.
Whole cells of the wild type, the frdA::cat+ strain, and the sdhA::cat+ strain were assayed to determine succinate oxidation as measured by oxygen uptake. Cultures were grown to an OD600 of approximately 0.5. Pyruvate was added to the medium for the frdA::cat+ strain. Addition of pyruvate to the medium had no effect on the wild-type respiration rate (data not shown). The wild type and the sdhA::cat+ strain consumed oxygen at similar rates. The succinate-dependent respiration rate of the wild type was 28 ± 7 nmol O2 consumed/min/108 cells, and the sdhA::cat+ strain respired at a rate of 37 ± 15 nmol O2 consumed/min/108 cells. The frdA::cat+ strain was unable to respire with succinate as the sole electron donor (0.6 ± 1 nmol O2 consumed/min/108 cells). The three strains exhibited similar rates of lactate-dependent respiration, which was used as a positive control.
The frdA::cat+ strain is deficient in host colonization.
At 1 week of age, groups of 10 birds were inoculated with 6.1 × 106 CFU of the wild type or 1.1 × 107 CFU of the frdA::cat+ strain by oral gavage. Two weeks postinoculation, the birds were sacrificed by CO2 asphyxiation, and the cecal contents were collected for enumeration of viable C. jejuni cells. All birds were colonized with C. jejuni at the conclusion of the experiment. The frdA::cat+ strain colonized chickens at significantly lower levels than the wild type (P = 0.001) (Fig. 4). A second host colonization trial was performed with groups of 1-week-old birds inoculated with 2.7 × 106 CFU of the wild type or 1.8 × 106 CFU of the sdhA::cat+ strain by oral gavage. Two weeks postinoculation the cecal contents were examined to enumerate the viable C. jejuni cells. The sdhA::cat+ strain and the wild-type strain colonized at similar levels (Fig. 4). No C. jejuni was recovered from the negative control birds inoculated with PBS in both colonization trials (data not shown).
FIG. 4.
Abilities of C. jejuni strains to colonize chickens. ▪ and ▾, CFU of wild-type C. jejuni/g cecal contents; ▴, CFU of frdA::cat+ strain/g cecal contents for one colonization trial; ⧫, CFU of sdhA::cat+ strain/g cecal contents for second independent colonization trial. The horizontal bars indicate the median values. *, P = 0.001 compared to the wild type.
DISCUSSION
The enzyme annotated as a fumarate reductase is the sole succinate dehydrogenase of C. jejuni. Mutations in this enzyme have serious and previously unsuspected implications for the growth and metabolic flexibility of this important pathogen. Although fumarate reductase activity has been measured in C. jejuni (37, 38), this organism is unable to respire anaerobically using fumarate as a terminal electron acceptor (37, 41), leaving the physiological role of the fumarate reductase in doubt. We constructed mutants with mutations in both the fumarate reductase and succinate dehydrogenase in order determine the in vivo functions of these two enzymes.
Our first indication that the fumarate reductase has a central role in the microaerobic physiology of C. jejuni came from the unusual growth characteristics of the fumarate reductase mutant. The frdA::cat+ strain grew like the wild type until mid-log phase (OD600, approximately 0.3) and then stopped growing (Fig. 2). Wild-type cultures, on the other hand, continued to grow, but the growth rate was lower than the initial growth rate until the terminal optical density was approximately 1.0 (Fig. 2). The sdhA::cat+ strain grew like the wild type, with two distinct growth phases (data not shown).
A possible explanation for the aborted growth of the frdA::cat+ strain is that this mutant is unable to utilize a particular class of substrates for carbon and energy. C. jejuni does not encode a complete glycolytic pathway (29) and must rely on the catabolism of small organic and amino acids for its carbon requirement (10). These acids are incorporated into the TCA cycle through a variety of transport systems and enzymes (15). We added TCA cycle intermediates to test whether such additions could rescue growth. Although none of the added substrates were able to restore biphasic growth to the frdA::cat+ strain (data not shown), addition of certain TCA cycle intermediates did extend the first growth phase and increase the terminal optical density compared to unsupplemented cultures (Table 2). The intermediates that did not extend the primary growth phase of the frdA::cat+ strain include citrate 2-oxoglutarate and succinate, which group together in the TCA cycle immediately preceding (in the oxidative cycle) the succinate-fumarate interconversion. The substrates that were growth stimuli for the frdA::cat+ strain included pyruvate, oxaloacetate, malate, and fumarate, which occur after the succinate-fumarate interconversion. Disruption of Frd results in an inability of C. jejuni to incorporate one-half of the TCA cycle intermediates into biomass, and these intermediates all occur before succinate oxidation in the oxidative TCA cycle (Fig. 3). This is especially detrimental to C. jejuni, which lacks sugar transporters and therefore relies on gluconeogenesis for its carbohydrate requirement (29). Neither oxaloacetate nor pyruvate (starting material for gluconeogenesis) could be produced with the block in the oxidative TCA cycle in the frdA::cat+ strain (Fig. 3).
In addition to direct incorporation of TCA intermediates, C. jejuni can grow on amino acids and has a preference for utilization of specific amino acids (10, 20). Analysis of spent medium from strains grown in MHB showed that the original concentrations of aspartate, glutamate, serine, and proline were reduced by over 50% (10). Therefore, we tested whether these four key amino acids could extend the first growth phase of the frdA::cat+ strain. Both aspartate and serine increased the terminal optical density of the frdA::cat+ strain, while glutamate and proline had no effect (Table 2). Taken together, these data suggest that C. jejuni preferentially uses the amino acids and TCA cycle substrates immediately after the oxidation of succinate in the first growth phase and switches to a less preferred substrate and amino acid class, which includes glutamate and proline, after depletion of the preferred substrates.
In an effort to explain the altered substrate utilization pattern of the the frdA::cat+ strain, we tried to determine the biochemical activities available to each strain. All strains exhibited fumarate reductase activity in all conditions tested. The fumarate reductase activities of the frdA::cat+ strain and the sdhA::cat+ strain were similar, and each of these activities contributed to the total fumarate reductase activity of the wild type (Table 3). Surprisingly, the sdhA::cat+ strain had succinate dehydrogenase activity similar to that of the wild type under all conditions tested, and the frdA::cat+ strain exhibited no succinate dehydrogenase activity (Table 4). Physiologically, succinate oxidation can also be measured using the uptake of oxygen by whole cells. The succinate-dependent respiration rates of whole cells of the wild-type and sdhA::cat+ strain cultures were similar, but the frdA::cat+ strain was unable to respire with succinate. Lactate is an efficient respiratory donor for C. jejuni (12), and the three strains had similar lactate-dependent respiration rates (data not shown), indicating that the respiratory capacity of these strains was not impaired.
The use of succinate as a respiratory chain donor in C. jejuni was initially surprising due to the demonstrated lack of a quinone pool in this organism (4). Typically in facultative anaerobes, succinate oxidation is coupled to ubiquinone, and under anaerobic conditions fumarate oxidation is coupled to menaquinol (42). There is, however, a third functional class of succinate oxidoreductases, which catalyze the oxidation of succinate and the reduction of the low-redox-potential menaquinone (11). C. jejuni contains only menaquinone-6 and a methyl-substituted menaquinone (4). In this class of enzymes the endergonic reaction of succinate oxidation (Em = 30 mV) coupled to menaquinone reduction (Em = −80 mV) has been shown to require a proton gradient (36). Desulfovibrio vulgaris, Geobacter sulfurreducens, and Bacillus subtilis have all been reported to couple succinate oxidation to menaquinone reduction via this mechanism (2, 22, 45).
The amino terminus of FrdC of C. jejuni is predicted to contain the five transmembrane helices predicted by TMHMM (www.cbs.dtu.dk/services/TMHMM/), and these helices contain the four conserved His residues that ligate the two heme B molecules required for transmembrane electron transfer (36). These helices and residues are also present in W. succinogenes FrdC and B. subtilis SdhC (36). Also conserved are two glutamate residues, which are active sites for menaquinol oxidation. All these elements are predicted to be required for menaquinone-dependent succinate dehydrogenase activity (45). The fumarate reductase of W. succinogenes is capable of both specific enzymatic activity during succinate-dependent methylene blue reduction and benzyl viologen oxidation through fumarate reduction (26).
The FrdC subunit is a transmembrane anchor with four conserved histidines and contains sites of menaquinol oxidation, which are requirements for succinate:menaquinone dehydrogenases, and the SdhC subunit of C. jejuni lacks these traits. Taken together, these data indicate that FrdCAB is the sole succinate dehydrogenase of C. jejuni and that SdhABC has been misannotated as it does not contribute to succinate dehydrogenase activity. Very recently, this has been shown to be the case in W. succinogenes (14). The W. succinogenes enzyme previously annotated as SdhABC (having 63%, 71%, and 69% homology to C. jejuni SdhABC) was determined to instead be a novel methylmenaquninol:fumarate reductase (MFR). Our data indicate the Sdh is probably an MFR in C. jejuni as well. The in vivo role of MFR in both these organisms has yet to be determined; however, our in vitro studies indicate that FrdCAB and SdhABC each contribute to fumarate reductase activity.
The frdA::cat+ strain showed a significant decrease in the ability to colonize poultry compared to both the wild type and the sdhA::cat+ strain (Fig. 4). The cecum of poultry contains fermentative by-products, including lactate, acetate, hydrogen, and formate (33). Amino acids are abundant in the cecum of poultry as a result of a high-cellulose diet and the biosynthesis of amino acids by microbes in the cecum (32). To study the in vivo availability of amino acids, one can compare the excreta from laying hens and cecectomized laying hens. Removal of the cecum significantly reduces the effect of microbes on digestion, and it is believed that microbial metabolism of amino acids in the cecum is largely responsible for the differences between intact and cecectomized birds (30). The total amino acid excretion was greater for cecectomized laying hens than for intact hens. The levels of proline, threonine, and isoleucine were increased significantly in the excreta of cecectomized laying hens, and it was inferred that the microbes in the cecum utilize these amino acids (31). Because the frdA::cat+ strain is unable to metabolize proline in the TCA cycle, it may be at a disadvantage in the cecum of poultry, thus explaining its decreased ability to colonize.
Fumarate reductase and succinate dehydrogenase have been implicated in colonization and virulence studies in other systems. The fumarate reductase of H. pylori is required for colonization of mice (8) and has been studied as a potential drug target (7, 24). The ability of an E. coli fumarate reductase mutant to colonize mice also was significantly decreased (13). In Salmonella enterica serovar Typhimurium, a complete TCA cycle is necessary for full virulence in mice (39), and a fumarate reductase-succinate dehydrogenase double mutant is avirulent in BALB/c mice (25).
Acknowledgments
This work was supported by USDA-NRI grant 2004-04553.
We thank Jesse L. Grimes, Department of Poultry Science, North Carolina State University, for his help with the poultry colonization trials and Dilan Weerakoon for providing the sdhA::cat+ strain.
Footnotes
Published ahead of print on 12 June 2009.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Butler, J. E., R. H. Glaven, A. Esteve-Nunez, C. Nunez, E. S. Shelobolina, D. R. Bond, and D. R. Lovley. 2006. Genetic characterization of a single bifunctional enzyme for fumarate reduction and succinate oxidation in Geobacter sulfurreducens and engineering of fumarate reduction in Geobacter metallireducens. J. Bacteriol. 188:450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, B. J., and S. C. Cary. 2004. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl. Environ. Microbiol. 70:6282-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlone, G. M., and F. A. Anet. 1983. Detection of menaquinone-6 and a novel methyl-substituted menaquinone-6 in Campylobacter jejuni and Campylobacter fetus subsp. fetus. J. Gen. Microbiol. 129:3385-3393. [DOI] [PubMed] [Google Scholar]
- 5.Carlone, G. M., and J. Lascelles. 1982. Aerobic and anaerobic respiratory systems in Campylobacter fetus subsp. jejuni grown in atmospheres containing hydrogen. J. Bacteriol. 152:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecchini, G., I. Schroder, R. P. Gunsalus, and E. Maklashina. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553:140-157. [DOI] [PubMed] [Google Scholar]
- 7.Ge, Z. 2002. Potential of fumarate reductase as a novel therapeutic target in Helicobacter pylori infection. Expert Opin. Ther. Targets 6:135-146. [DOI] [PubMed] [Google Scholar]
- 8.Ge, Z., Y. Feng, C. A. Dangler, S. Xu, N. S. Taylor, and J. G. Fox. 2000. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb. Pathog. 29:279-287. [DOI] [PubMed] [Google Scholar]
- 9.Ge, Z., Q. Jiang, M. S. Kalisiak, and D. E. Taylor. 1997. Cloning and functional characterization of Helicobacter pylori fumarate reductase operon comprising three structural genes coding for subunits C, A and B. Gene 204:227-234. [DOI] [PubMed] [Google Scholar]
- 10.Guccione, E., R. Leon-Kempis Mdel, B. M. Pearson, E. Hitchin, F. Mulholland, P. M. van Diemen, M. P. Stevens, and D. J. Kelly. 2008. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69:77-93. [DOI] [PubMed] [Google Scholar]
- 11.Hagerhall, C. 1997. Succinate:quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320:107-141. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, P. S., and T. G. Goodman. 1982. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, S. A., F. Z. Chowdhury, A. J. Fabich, A. Anderson, D. M. Schreiner, A. L. House, S. M. Autieri, M. P. Leatham, J. J. Lins, M. Jorgensen, P. S. Cohen, and T. Conway. 2007. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75:4891-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juhnke, H. D., H. Hiltscher, H. R. Nasiri, H. Schwalbe, and C. R. Lancaster. 2009. Production, characterization, and determination of the real catalytic properties of the putative ‘succinate dehydrogenase’ from Wolinella succinogenes. Mol. Microbiol. 71:1088-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly, D. J. 2008. Complexity and versatility in the physiology and metabolism of Campylobacter jejuni. ASM Press, Washington, DC.
- 16.Kortner, C., F. Lauterbach, D. Tripier, G. Unden, and A. Kroger. 1990. Wolinella succinogenes fumarate reductase contains a dihaem cytochrome b. Mol. Microbiol. 4:855-860. [DOI] [PubMed] [Google Scholar]
- 17.Kroger, A., V. Geisler, E. Lemma, F. Theis, and R. Lenger. 1992. Bacterial fumarate respiration. Arch. Microbiol. 158:311-314. [Google Scholar]
- 18.Lancaster, C. R., and J. Simon. 2002. Succinate:quinone oxidoreductases from epsilon-proteobacteria. Biochim. Biophys. Acta 1553:84-101. [DOI] [PubMed] [Google Scholar]
- 19.Lauterbach, F., C. Kortner, S. P. Albracht, G. Unden, and A. Kroger. 1990. The fumarate reductase operon of Wolinella succinogenes. Sequence and expression of the frdA and frdB genes. Arch. Microbiol. 154:386-393. [DOI] [PubMed] [Google Scholar]
- 20.Leach, S., P. Harvey, and R. Wali. 1997. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J. Appl. Microbiol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 21.Lemma, E., C. Hagerhall, V. Geisler, U. Brandt, G. von Jagow, and A. Kroger. 1991. Reactivity of the Bacillus subtilis succinate dehydrogenase complex with quinones. Biochim. Biophys. Acta 1059:281-285. [DOI] [PubMed] [Google Scholar]
- 22.Lemma, E., G. Unden, and A. Kroger. 1990. Menaquinone is an obligatory component of the chain catalyzing succinate respiration in Bacillus subtilis. Arch. Microbiol. 155:62-67. [DOI] [PubMed] [Google Scholar]
- 23.Leon-Kempis Mdel, R., E. Guccione, F. Mulholland, M. P. Williamson, and D. J. Kelly. 2006. The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60:1262-1275. [DOI] [PubMed] [Google Scholar]
- 24.Mendz, G. L., S. L. Hazell, and S. Srinivasan. 1995. Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori. Arch. Biochem. Biophys. 321:153-159. [DOI] [PubMed] [Google Scholar]
- 25.Mercado-Lubo, R., E. J. Gauger, M. P. Leatham, T. Conway, and P. S. Cohen. 2008. A Salmonella enterica serovar Typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infect. Immun. 76:1128-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mileni, M., F. MacMillan, C. Tziatzios, K. Zwicker, A. H. Haas, W. Mantele, J. Simon, and C. R. Lancaster. 2006. Heterologous production in Wolinella succinogenes and characterization of the quinol:fumarate reductase enzymes from Helicobacter pylori and Campylobacter jejuni. Biochem. J. 395:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mylius, S. D., M. J. Nauta, and A. H. Havelaar. 2007. Cross-contamination during food preparation: a mechanistic model applied to chicken-borne Campylobacter. Risk Anal. 27:803-813. [DOI] [PubMed] [Google Scholar]
- 28.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 30.Parsons, C. M. 1986. Determination of digestible and available amino acids in meat meal using conventional and caecectomized cockerels or chick growth assays. Br. J. Nutr. 56:227-240. [DOI] [PubMed] [Google Scholar]
- 31.Parsons, C. M. 1984. Influence of caecectomy and source of dietary fibre or starch on excretion of endogenous amino acids by laying hens. Br. J. Nutr. 51:541-548. [DOI] [PubMed] [Google Scholar]
- 32.Parsons, C. M., L. M. Potter, and R. D. Brown, Jr. 1983. Effects of dietary carbohydrate and of intestinal microflora on excretion of endogenous amino acids by poultry. Poult Sci. 62:483-489. [DOI] [PubMed] [Google Scholar]
- 33.Salanitro, J. P., I. G. Fairchilds, and Y. D. Zgornicki. 1974. Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl. Microbiol. 27:678-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuel, M. C., D. J. Vugia, S. Shallow, R. Marcus, S. Segler, T. McGivern, H. Kassenborg, K. Reilly, M. Kennedy, F. Angulo, and R. V. Tauxe. 2004. Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996-1999. Clin. Infect. Dis. 38(Suppl. 3):S165-S174. [DOI] [PubMed] [Google Scholar]
- 35.Schirawski, J., and G. Unden. 1995. Anaerobic respiration of Bacillus macerans with fumarate, TMAO, nitrate and nitrite and regulation of the pathways by oxygen and nitrate. Arch. Microbiol. 163:148-154. [Google Scholar]
- 36.Schirawski, J., and G. Unden. 1998. Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential. Eur. J. Biochem. 257:210-215. [DOI] [PubMed] [Google Scholar]
- 37.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, M. A., G. L. Mendz, M. A. Jorgensen, and S. L. Hazell. 1999. Fumarate metabolism and the microaerophily of Campylobacter species. Int. J. Biochem. Cell Biol. 31:961-975. [DOI] [PubMed] [Google Scholar]
- 39.Tchawa Yimga, M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74:1130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 41.Véron, M., A. Lenvoisé-Furet, and P. Beaune. 1981. Anaerobic respiration of fumarate as a differential test between Campylobacter fetus and Campylobacter jejuni. Curr. Microbiol. 6:349-354. [Google Scholar]
- 42.Wallace, B. J., and I. G. Young. 1977. Aerobic respiration in mutants of Escherichia coli accumulating quinone analogues of ubiquinone. Biochim. Biophys. Acta 461:75-83. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 44.Weingarten, R. A., J. L. Grimes, and J. W. Olson. 2008. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaunmuller, T., D. J. Kelly, F. O. Glockner, and G. Unden. 2006. Succinate dehydrogenase functioning by a reverse redox loop mechanism and fumarate reductase in sulphate-reducing bacteria. Microbiology 152:2443-2453. [DOI] [PubMed] [Google Scholar]