Abstract
We report here that gemfibrozil (GFZ) inhibits axenic and intracellular growth of Legionella pneumophila and of 27 strains of wild-type and multidrug-resistant Mycobacterium tuberculosis in bacteriological medium and in human and mouse macrophages, respectively. At a concentration of 0.4 mM, GFZ completely inhibited L. pneumophila fatty acid synthesis, while at 0.12 mM it promoted cytoplasmic accumulation of polyhydroxybutyrate. To assess the mechanism(s) of these effects, we cloned an L. pneumophila FabI enoyl reductase homolog that complemented for growth an Escherichia coli strain carrying a temperature-sensitive enoyl reductase and rendered the complemented E. coli strain sensitive to GFZ at the nonpermissive temperature. GFZ noncompetitively inhibited this L. pneumophila FabI homolog, as well as M. tuberculosis InhA and E. coli FabI.
The advent of AIDS and the emergence of many multidrug-resistant bacterial species have led to renewed efforts to find new antibiotics. The most commonly used antibiotics act by blocking bacterium-specific DNA, RNA, or protein synthesis. Mycobacterium tuberculosis is a major exception to this generalization. While streptomycin, an inhibitor of bacterial protein synthesis, was the first antibiotic to be used successfully to treat M. tuberculosis, isoniazid (INH), an inhibitor of mycobacterial lipid synthesis, is presently the drug most commonly used to treat infections with this organism (2, 43). The differential sensitivity to INH of M. tuberculosis versus mammalian cells reflects the fact that most bacterial fatty acid synthases (type II synthases) are comprised of discrete, separable enzymes encoded by separate genes, while mammalian fatty acid synthases (type I) are dimeric proteins in which a single polypeptide catalyzes the seven enzymatic activities of fatty acid synthesis (21, 52).
In previous studies (45), we reported that gemfibrozil (GFZ), a commonly prescribed and well-tolerated hypolipidemic drug, inhibits the export of various organic anions, including penicillin and fluoroquinolones, from murine macrophages, thereby elevating the intracellular concentration of these antibiotics and enhancing their capacity to block intracellular growth of Listeria monocytogenes. In exploring this system, we discovered that while GFZ has no effect on axenic or intracellular growth of Listeria monocytogenes, it inhibits axenic growth of all Legionella pneumophila strains tested and of 5 wild-type and 22 multidrug-resistant strains of M. tuberculosis and inhibits intracellular growth of L. pneumophila Philadelphia-1 and M. tuberculosis H37RV in human and mouse macrophages, respectively.
Both M. tuberculosis and L. pneumophila are facultative intracellular pathogens that enter host macrophages by phagocytosis (25, 26), grow in nonlysosomal membrane-bound cytoplasmic vacuoles (24), have special nutrient requirements (38, 54, 55), and produce a relatively unique spectrum of membrane lipids (7, 57). However, M. tuberculosis is a slow-growing and dangerous organism with which to work. In contrast, L. pneumophila has a relatively short doubling time (120 min) in axenic culture medium and requires no special biohazard precautions. Therefore, we explored the mechanism(s) responsible for GFZ's antibiotic activity in L. pneumophila, in the expectation that a similar mechanism(s) would be operative in M. tuberculosis.
We report here that GFZ noncompetitively inhibits L. pneumophila and M. tuberculosis enoyl reductases and provide genetic evidence consistent with the hypothesis that GFZ blocks growth of these bacteria by inhibiting their enoyl reductases. These findings, coupled with our inability to select a highly GFZ-resistant strain of L. pneumophila, the sensitivity to GFZ of all 22 drug-resistant M. tuberculosis strains tested, the emerging threat of extensively drug-resistant M. tuberculosis (51), and the paucity of new chemical entities for the treatment of tuberculosis, have prompted us to describe GFZ's antibiotic activities.
MATERIALS AND METHODS
Materials.
GFZ, bezafibrate, clofibric acid, 3-(p-hydroxyphenyl)-propionic acid, hydrochloric acid, propanol, benzoic acid, 3-hydroxy butyrate, dichloroethane, crotonyl coenzyme A (crotonyl-CoA [CCA]), and dodecyl coenzyme A (DCA) were from Sigma-Aldrich, St. Louis, MO.
Bacterial strains and growth conditions.
L. pneumophila Philadelphia-1 (27) and L. pneumophila JR32 (46) were grown in AYE broth (28) lacking bovine serum albumin at 37°C with aeration or on ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered charcoal yeast extract agar (12). The bacterial concentration was measured by determining the absorbance with a Klett colorimeter. M. tuberculosis H37Rv (Pasteur Institute, Paris, France) was grown to mid-log phase in 7H9 broth (Difco, Detroit, MI) containing albumin-dextrose-catalase, glycerol, and 0.25% Tween 80 (Sigma, St. Louis, MO). Mycobacterial suspensions were probe sonicated (Dismembrator 60; Fisher Scientific, Pittsburgh, PA) for 20 s at 1.5 W to disperse clumps. Stocks were frozen in aliquots and stored at −70°C. Aliquots were thawed and plated to determine the number of bacilli, as previously described (34, 35).
L. pneumophila growth in human MDM.
Human mononuclear cells were purified as described previously (50). A total of 4 × 106 monocyte-derived macrophages (MDM)/ml and 1 × 103 CFU/ml log-phase L. pneumophila organisms were suspended in RPMI containing 2 mM glutamine and 10% heat-inactivated human serum (HIHS). One-hundred-microliter aliquots of this mixture were placed in a 96-well microtiter plate. The plates were centrifuged at ∼50 × g to cosediment MDM and L. pneumophila and then incubated for 2.5 h at 37°C. One hundred microliters of fresh medium was added, with or without twice the indicated concentration of GFZ, clofibrate, or bezafibrate. The plates were incubated at 37°C for the indicated times, the cells and medium were harvested and plated on ABCYE agar plates, and the number of L. pneumophila CFU was determined.
M. tuberculosis growth in resident mouse peritoneal macrophages.
Macrophages were harvested from BALB/c mice, as described previously (14), and maintained in antibiotic-free medium unless otherwise stated. Stock vials of M. tuberculosis H37Rv were thawed, sonicated for 20 s at 1.5 W to disperse clumps, diluted in D10 medium, and used to infect duplicate cultures of macrophages at a multiplicity of infection (MOI) of 1 CFU per cell (1:1). The MOI was confirmed by plating serial dilutions of the inoculum on Middlebrook 7H10 agar plates (Difco) containing glycerol and oleic acid-albumin-dextrose-catalase. The viability of the cultured cells was assessed at various time points by either trypan blue exclusion or staining of cells with TP-3 cyanine nucleic acid stain (Molecular Probes, Eugene, OR) as described by Freeman et al. (15). Intracellular bacillary numbers were determined by sonicating infected macrophages four times in phosphate-buffered saline containing 0.25% Tween 80 at 1.5 W for 5 s each time. Tenfold serial dilutions of the mycobacterial suspension were plated on Middlebrook 7H10 agar plates as described above and incubated at 37°C for 2 to 3 weeks before counting of CFU.
Antimicrobial susceptibility.
MICs of L. pneumophila were determined by incubating triplicate samples of logarithmic-phase bacterial suspensions containing ∼2 × 106 CFU/ml in AYE broth containing serial dilutions of GFZ for 48 h at 37°C. Growth was assessed by measuring the A600. For determination of M. tuberculosis susceptibility, axenic cultures of different strains were suspended to a McFarland density standard of 1 (3 × 108 CFU/ml) and then diluted 100-fold. One-hundred-microliter samples of these dilutions were spotted in duplicate onto each of the four quadrants of oleic acid-albumin-dextrose-catalase-enriched Middlebrook agar plates; each quadrant contained 0, 0.2, 0.4, or 0.8 mM of GFZ. The plates were incubated at 37°C for 21 days, at which time the quantity of growth was assessed.
Transmission electron microscopy.
L. pneumophila was grown for 2 days on CYE plates, harvested, and suspended at 1010 to 1011 CFU/ml in AYE. A total of 109 to 1010 CFU were plated on CYE agar, without or with 0.12 mM GFZ, and incubated for 3 days at 37°C. The bacteria were harvested, pelleted, fixed by suspension in 2.5% glutaraldehyde in 0.1 M NaPO4 buffer, pH 7.5, for 45 min at 25°C, rinsed overnight in the same buffer, rinsed twice in cacodylate buffer, pH 7.35, postfixed in 1% osmium tetroxide, rinsed successively in cacodylate buffer, pH 7.35, and 0.1 M sodium acetate buffer, pH 6.0, incubated in 1% uranyl acetate in 0.1 M sodium acetate buffer, pH 6.0, for 2.5 h at 4°C, dehydrated through graded alcohols, and embedded in epoxy resin. Six-hundred-angstrom-thick sections were stained with uranyl acetate and lead citrate and examined by transmission electron microscopy, using a JEOL JEM1200EX electron microscope.
Nile blue A staining.
L. pneumophila was grown for 2 days on CYE plates, harvested, and suspended at 1010 to 1011 CFU/ml in AYE. A total of 109 to 1010 bacteria were placed in CYE agar medium without or with 0.12 mM GFZ and incubated for 3 days at 37°C. The bacteria were harvested, suspended in water, smeared on a glass slide, and heat fixed to it. Slides were incubated in a 1% aqueous solution of Nile blue A for 10 min at 55°C, washed with tap water, destained for 1 min in 8% aqueous acetic acid, and air dried. Smears were moistened with water, covered with a no. 1 glass coverslip, and examined by fluorescence microscopy at 460 nm.
GC-MS identification and quantification of 3-HB.
L. pneumophila was grown on CYE agar for 3 days in the absence or presence of 0.12 mM GFZ, harvested, and lyophilized. 3-Hydroxybutyrate (3-HB) propyl esters were prepared for analysis by hydrochloric acid propanolysis of lyophilized bacteria, using benzoic acid as an internal standard, as described previously (40). A standard curve was constructed by converting known quantities of 3-HB and benzoic acid to their propyl esters by hydrochloric acid propanolysis. Prior to analysis, samples were dried under a stream of nitrogen and reconstituted in ethyl acetate. Gas chromatography-mass spectrometry (GC-MS) analysis was performed as described previously (53), using a Hewlett Packard 5987A GC-MS instrument equipped with a DB-1 fused-silica capillary column (30 m by 0.2 mm), using helium as a carrier gas at injector and source temperatures of 220°C and 200°C, respectively. Samples were ionized by electron impact (70 eV). The abundance of ions at m/e 105 was used to measure the quantity of benzoic acid, while the abundance of ions at m/e 87 was used to assess the quantity of 3-HB.
[14C]acetate labeling of L. pneumophila lipids.
L. pneumophila was grown to logarithmic phase in AYE broth at 37°C; 1.0-ml aliquots were pelleted and suspended in fresh medium containing GFZ or other inhibitors at the concentrations indicated. [14C]acetate (specific activity, 48.9 mCi/mmol) (Amersham Biosciences, Piscataway, NJ) was added to a final concentration of 5 μCi/ml, and the cultures were incubated at 37°C. At different time points for up to 1 hour of incubation, lipids were extracted from 100-μl aliquots of bacterial suspension as described by Bligh and Dyer (5). The organic layer was assayed in a scintillation counter, and the rate of [14C]acetate labeling of lipids was determined.
ACP purification.
Acyl carrier protein (ACP) from L. pneumophila was purified as described previously (41). Purified ACP migrated at 20 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described, and Edman degradation of protein eluted from this band gave an eight-amino-acid sequence which was identical to the N-terminal sequence of L. pneumophila ACP (STVEERVR) (lpg 1396) (10). For [14C]acetate incorporation into ACP-linked lipids, logarithmic-phase cultures of L. pneumophila were incubated for 1 h at 37°C with 5 μCi/ml [14C]acetate in AYE, without or with 0.4 mM GFZ or other compounds. ACP was purified, and its specific activity was determined.
Purification of L. pneumophila, E. coli, and M. tuberculosis enoyl reductases.
PCR-amplified DNA encoding L. pneumophila FabI, Escherichia coli EnvM, or M. tuberculosis InhA was cloned into the NdeI and BamHI sites of the pET15 vector (Stratagene, La Jolla, CA) and transformed into E. coli BL21(DE3). Mid-logarithmic-phase bacterial cultures were incubated for 1 h at 37°C in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma-Aldrich, St. Louis, MO), pelleted, suspended in 50 mM sodium phosphate buffer, pH 8, containing 300 mM NaCl, passed twice through a French press at 16,000 lb/in2, and centrifuged to remove insoluble debris. The supernatant was added to a Ni-nitrilotriacetic acid (Ni-NTA) column, and histidine-tagged proteins were eluted with 50 mM sodium phosphate buffer, pH 8, containing 300 mM NaCl and 250 mM imidazole. The identity and purity of all proteins were confirmed by MS of these proteins following their elution from SDS gels (e.g., see Fig. 6b).
FIG. 6.
(A) Aligment of enoyl-CoA reductase amino acid sequences of L. pneumophila (FabI), E. coli (EnvM), and M. tuberculosis (InhA). (B) SDS-PAGE and Coomassie blue staining of purified enoyl reductases from L. pneumophila (lane 1), E. coli (lane 2), and M. tuberculosis (lane 3).
Enoyl reductase activity.
Enoyl reductase activity was assayed, as described previously (3), by measuring the decrease in NADH absorbance at 340 nm at room temperature, using CCA or DCA as a substrate. A standard reaction mix contained 400 μl of 100 μM sodium phosphate buffer, pH 7.5, 100 μm NADH, 2 μg/ml purified protein, and 0.5 mM CCA or DCA. Initial velocity kinetic data were determined at various concentrations of CCA at a fixed concentration of NADH in either the absence or presence of GFZ (4 mM). The data were fitted to the equation that describes noncompetitive inhibition, v = (VA)/Ka(1 + I/Kis) + A(1 + I/Kii), using SigmaPlot 2000, where V is the maximum velocity, Ka is the Michaelis constant for substrate A, and Kis and Kii are the slope and intercept inhibition constants for inhibitor I.
Disc growth inhibition zone assay.
L. pneumophila strain JR32 was transformed with empty pMMB207 plasmid or with pMMB207 chloramphenicol resistance plasmids containing wild-type L. pneumophila fabI, wild-type E. coli fabI, or temperature-sensitive E. coli fabI (from the FT100 strain). The transformed bacteria were grown overnight in AYE broth with 5 μg/ml chloramphenicol (Sigma-Aldrich, St. Louis, MO). The next day, 0.1 ml of culture was layered on CYE agar plates, without or with 1 mM IPTG, and discs containing 1 mM GFZ were added in duplicate to the plates. The plates were incubated at 37°C, and the inhibition zone of bacterial growth was measured after 2 days.
RESULTS
GFZ inhibits growth of L. pneumophila in bacteriological medium and in human MDM.
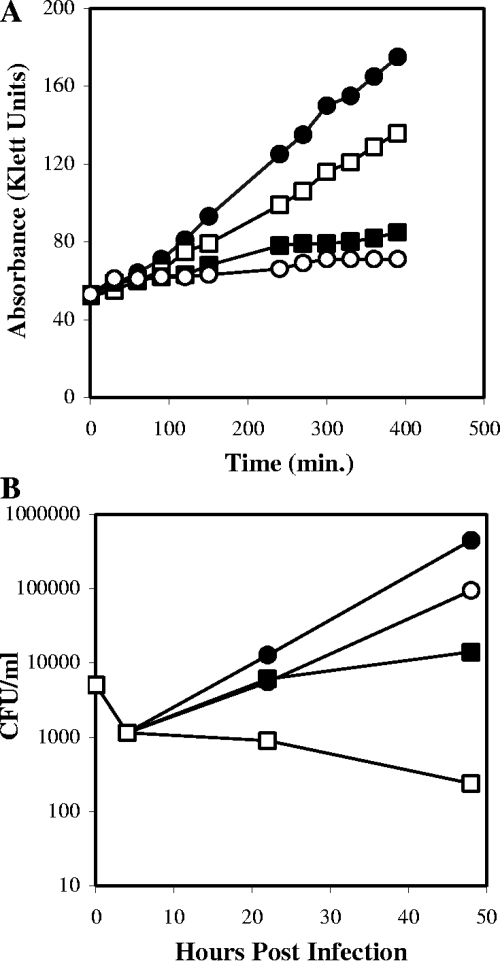
L. pneumophila Philadelphia-1 was grown to logarithmic phase in AYE broth and further incubated in AYE containing 0.1 to 0.4 mM GFZ. Complete inhibition of growth required 0.4 mM GFZ (Fig. 1A). GFZ's MIC90 for L. pneumophila in AYE broth containing 10% heat-inactivated human serum was ∼0.1 mΜ. GFZ also inhibited the growth of 39 other L. pneumophila strains in CYE agar, as measured by a zone inhibition assay (data not shown). Repeated efforts to select spontaneous or ethyl methanesulfonate (EMS)-induced GFZ-resistant mutants or variants of L. pneumophila were uniformly unsuccessful.
FIG. 1.
GFZ inhibits axenic growth of L. pneumophila. (A) L. pneumophila growth in the absence (•) or presence of 0.1 mM (□) (28% inhibition), 0.2 mM (▪) (76% inhibition), or 0.4 mM (○) (88% inhibition) GFZ, as measured by changes in absorbance using a Klett colorimeter. (B) GFZ inhibits the growth of L. pneumophila in human MDM. CFU of L. pneumophila were measured at the indicated times after infection. Macrophages were maintained in medium alone (•) or in medium containing 0.4 mM GFZ (□) (>100% inhibition), 1.0 mM bezafibrate (○) (86% inhibition), or 1.0 mM clofibric acid (▪) (98% inhibition). Both panels report one experiment representative of three that yielded similar results.
GFZ inhibited growth of L. pneumophila within human MDM (Fig. 1B) and in phorbol myristate acetate-differentiated HL-60 cells (not shown). As in AYE broth, complete inhibition of intracellular growth was observed with 0.4 mM GFZ (Fig. 1B). GFZ at 0.4 mM was not toxic for primary human MDM or macrophage-like cell lines, as measured by trypan blue dye exclusion or MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay, respectively (not shown). We concluded that GFZ is bacteristatic for L. pneumophila in axenic culture and in macrophages.
GFZ, like clofibrate, fenofibrate, and bezafibrate, is a fibric acid. These compounds activate peroxisome proliferator-activated receptors (PPARs), transcriptional activators which affect several metabolic pathways, including cholesterol biosynthesis and β-oxidation of fatty acids (23, 30, 48). To test whether GFZ inhibited L. pneumophila growth in macrophages by affecting host cell functions, we compared the effects of other fibric acids on intracellular growth of L. pneumophila. Clofibrate and bezafibrate, even when used at 1.0 mM, were much less effective inhibitors of L. pneumophila growth in macrophages than GFZ at 0.4 mM (Fig. 1B). Assays in AYE broth confirmed that the MICs for clofibrate and bezafibrate were 0.4 mM and 1.0 mM, 4- and 10-fold higher, respectively, than that found for GFZ. These results, our previous report (45) that GFZ does not inhibit L. monocytogenes growth in macrophages, and our isolation of an EMS-induced variant (F4b) (31) that was fivefold more resistant to GFZ's growth inhibitory effect in both axenic media and macrophages suggest that GFZ inhibits L. pneumophila's growth in macrophages by acting on the bacterium, not on the host cell.
GFZ inhibits the growth of M. tuberculosis in bacteriological medium and in mouse peritoneal macrophages.
The intracellular bacterial pathogen of greatest medical and economic importance is M. tuberculosis. Therefore, we examined GFZ's effect on the growth of 27 M. tuberculosis strains, 22 of which were resistant to one or more antitubercular drugs. GFZ at 0.4 mM completely inhibited growth of 14 of these M. tuberculosis strains, and at 0.8 mM it inhibited growth of all of them, regardless of their profile of resistance to other antibiotics, including INH (Table 1).
TABLE 1.
Susceptibility of various M. tuberculosis strains to GFZ
| Strain | Drug resistance profilea
|
No. of colonies/quadrant
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | E | K | O | C | No GFZ | GFZ at 0.2 mM (50 μg/ml) | GFZ at 0.4 mM (100 μg/ml) | GFZ at 0.8 mM (200 μg/ml) | |
| O80154 | S | S | S | S | S | S | S | 100-500 | 50-99 | 0 | 0 |
| H37RV | S | S | S | S | S | S | S | 100-500 | 100-500 | 1 | 0 |
| CDC K | S | S | S | S | S | S | S | 100-500 | 50-99 | 6 | 0 |
| MH | S | S | S | S | S | S | S | 100-500 | 100-500 | 50-99 | 0 |
| MTB25177 | S | S | S | S | S | S | S | >1,000 | >1,000 | 50-99 | 0 |
| H52578 | R | S | S | S | S | S | S | 100-500 | 100-500 | 0 | 0 |
| CM | R | S | S | S | S | S | S | >1,000 | >1,000 | 50-99 | 0 |
| NM | S | R | S | S | S | S | S | >1,000 | 18 | 0 | 0 |
| CDC P | S | RL | S | S | S | S | S | 100-500 | 100-500 | 0 | 0 |
| CDC T | S | RL | S | S | S | S | S | 100-500 | 100-500 | 0 | 0 |
| CDC D | S | RL | S | S | S | S | S | 100-500 | 100-500 | 0 | 0 |
| S15674 | S | R | S | S | S | S | ND | 100-500 | 50-99 | 10 | 0 |
| F16285 | S | R | S | S | S | S | S | 100-500 | 50-99 | 10 | 0 |
| CDC N | S | S | R | S | S | S | S | 100-500 | 50-99 | 0 | 0 |
| M23294 | S | S | S | R | S | S | S | 100-500 | 50-99 | 4.5 | 0 |
| M41151 | S | S | S | R | S | S | S | 100-500 | 50-99 | 7.5 | 0 |
| CDCL | R | S | R | S | S | S | S | 100-500 | >1,000 | 50-99 | 0 |
| JJ | S | R | R | S | S | S | S | 100-500 | 7 | 0 | 0 |
| F5260 | S | R | R | S | S | S | S | 100-500 | 45 | 0 | 0 |
| O80711 | R | R | R | S | S | S | S | 100-500 | 50-99 | 6 | 0 |
| T30234 | S | R | R | R | S | S | S | 100-500 | 50-99 | 0 | 0 |
| W19521 | S | R | R | R | S | S | S | 100-500 | 50-99 | 0 | 0 |
| W54410 | R | R | R | R | S | S | S | 100-500 | 35 | 0 | 0 |
| RF | R | R | R | R | S | S | S | >1,000 | 100-500 | 0 | 0 |
| O81256 | R | R | R | R | S | S | S | 100-500 | 50-99 | 0 | 0 |
| T45777 | R | R | R | R | R | R | R | 100-500 | 100-500 | 50-99 | 0 |
| AA | R | R | R | R | R | R | R | 100-500 | 100-500 | 50-99 | 0 |
Sensitivity (S) or resistance (R) to antibiotics. Abbreviations: S, streptomycin at 2 mg/ml; I, INH at 1 mg/ml; R, rifampin at 1 mg/ml; E, ethambutol at 5 mg/ml; K, kanamycin at 6 mg/ml; O, ofloxacin at 4 mg/ml; C, ciprofloxacin at 2 mg/ml; RL, low-level resistance to INH at 0.2 mg/ml; ND, not done.
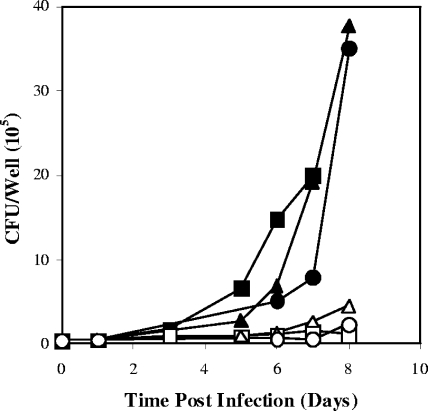
In contrast, 1 to 1.2 mM GFZ was required to block the growth of M. tuberculosis in mouse peritoneal macrophages by ∼80% (Fig. 2). GFZ's inhibitory effect persisted for as long as the drug was maintained in the medium. Removal of GFZ from the medium resulted in resumption of M. tuberculosis growth (not shown), confirming both the viability of macrophages in medium containing this concentration of GFZ and that GFZ is bacteristatic for M. tuberculosis. In contrast, INH at 10 and 1 μg/ml (0.07 μM and 0.007 μM, respectively) killed >99% of M. tuberculosis cells, INH at 0.1 μg/ml held the M. tuberculosis concentration relatively constant, and INH at 0.01 and 0.005 μg/ml had little or no inhibitory effect. Moreover, GFZ at 1 to 1.2 mM neither enhanced nor reduced the capacity of 1 to 0.005 μg/ml INH to inhibit M. tuberculosis growth in macrophages when the two drugs were used in combination (not shown).
FIG. 2.
GFZ inhibits the growth of M. tuberculosis in mouse peritoneal macrophages. Macrophages were infected with M. tuberculosis and incubated in D10 medium without or with GFZ. Data are shown for controls (no GFZ) (▪), cells receiving 1.2 mM GFZ for 8 days (□), controls (no GFZ) receiving fresh medium without GFZ on day 3 (▴), cells receiving 1.2 mM GFZ, with a switch to fresh medium without GFZ on day 3 (▵), controls (no GFZ) receiving fresh medium without GFZ on day 5 (•), and cells with 1.2 mM GFZ, with a switch to fresh medium without GFZ on day 5 (○). Data are the averages for duplicate cultures from one experiment representative of three that yielded similar results.
GFZ distinguishes Nocardia sp. from atypical mycobacteria.
In addition to L. pneumophila and M. tuberculosis, 0.4 mM GFZ inhibited the growth of other human pathogens (e.g., Nocardia sp., Staphylococcus epidermidis, and Staphylococcus aureus) (data not shown). However, it had little or no effect on the growth of atypical mycobacteria, suggesting that it might be used in clinical microbiology to distinguish Nocardia sp. from atypical mycobacteria.
GFZ stimulates accumulation of PHB in L. pneumophila.
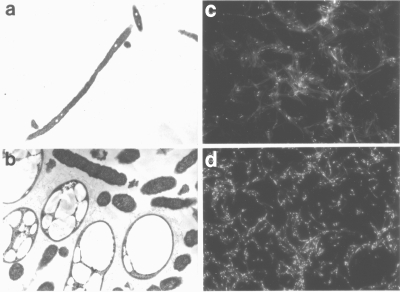
L. pneumophila maintained on CYE agar containing 0.12 mM GFZ developed very large, often irregularly shaped electron-lucent inclusions (Fig. 3b) compared to the relatively small round inclusions found in L. pneumophila maintained in the absence of GFZ (Fig. 3a). Similar electron-lucent inclusions have been reported to contain polyhydroxybutyrate (PHB) (9, 42), a polymer of 3-HB (29, 36). Nile blue A, a dye that stains PHB-containing granules (39), stained these inclusions (Fig. 3d), and GC-MS analysis of extracts of L. pneumophila maintained on CYE without or with 0.12 mM GFZ confirmed that GFZ-treated L. pneumophila contained ∼50-fold more 3-hydroxybutyryl-propyl ester (3-HBPE) than untreated bacteria (42.5 ± 7.5 mg 3-HBPE/mg protein versus 0.75 ± 0.75 mg 3-HBPE/mg protein). Since 3-HB, the precursor of PHB, is an intermediate in fatty acid synthesis (1, 21), these results suggested that GFZ inhibits a step in fatty acid synthesis and led us to investigate this possibility.
FIG. 3.
GFZ promotes PHB accumulation in L. pneumophila. Transmission electron microscopy was performed on L. pneumophila Philadelphia 1 grown on CYE agar in the absence (a) or presence (b) of 0.12 mM GFZ. (c and d) Fluorescence micrographs of Nile blue A-stained L. pneumophila grown on CYE agar in the absence (c) or presence (d) of 0.12 mM GFZ. Magnification, ×10,000 (a), ×20,000 (b), or ×1,000 (c and d).
GFZ inhibits [14C]acetate incorporation into L. pneumophila lipids.
L. pneumophila cells incubated in medium containing [14C]acetate incorporate the acetate primarily into lipids (55). Therefore, we tested GFZ's effect on [14C]acetate incorporation into a chloroform-methanol (5) extract of log-phase (∼5 × 108 CFU/ml) L. pneumophila cells incubated in AYE medium containing [14C]acetate and various concentrations of GFZ. [14C]acetate incorporation into the chloroform-methanol extract was inhibited 19%, 38%, and 76% by GFZ at 0.05 mM, 0.1 mM, and 0.2 mM, respectively, and was almost completely blocked by 0.4 mM GFZ (Fig. 4), the same concentration that completely blocked L. pneumophila growth in this medium (Fig. 1A). We compared the effectiveness of GFZ with similar concentrations of known inhibitors of fatty acid synthesis. Cerulenin (0.45 mM), a β-ketoacylsynthase inhibitor, and INH (0.5 mM), an enoyl reductase inhibitor, inhibited [14C]acetate incorporation into L. pneumophila lipids ∼99% and ∼76%, respectively. Thin-layer chromatographic analysis of these extracts confirmed these results and showed that GFZ decreased labeling of all lipids to approximately the same extent (data not shown). GFZ's inhibitory effect on lipid synthesis is consistent with the hypothesis that it blocks fatty acid synthesis.
FIG. 4.
GFZ inhibits acetate incorporation into L. pneumophila lipids. L. pneumophila was incubated with [14C]acetate in AYE medium in the absence or presence of 0.04 to 0.4 mM GFZ. At different time points, [14C]acetate incorporation into the chloroform-methanol-soluble fraction was measured. Data are the averages for duplicate cultures from one experiment representative of three that yielded similar results.
GFZ inhibits fatty acid elongation on L. pneumophila ACP.
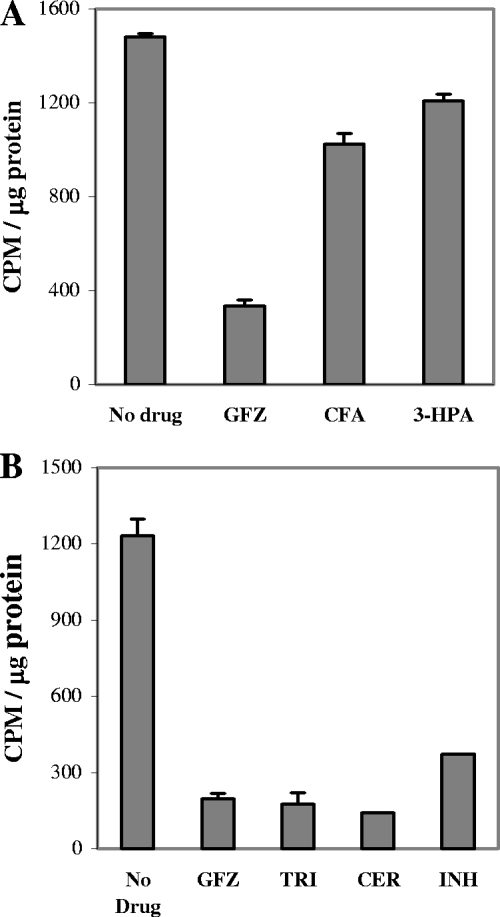
To determine GFZ's effect on assembly of fatty acids on ACP, we purified ACP from L. pneumophila cultures that were incubated for 1 h in [14C]acetate-containing medium without or with 0.4 mM GFZ. GFZ inhibited [14C]acetate association with ACP ∼80% (Fig. 5). Parallel analyses of ACP purified from L. pneumophila incubated in [14C]acetate-containing medium with compounds that share high structural similarity to GFZ, such as 3-(p-hydroxyphenyl)-propionic acid (3-HPA) and clofibric acid, showed that these compounds had only modest (21% and 31%, respectively) inhibitory effects on [14C]acetate association with ACP (Fig. 5A). Other known fatty acid synthesis inhibitors, such as INH, cerulenin, and the commonly used enoyl-ACP reductase inhibitor triclosan, inhibited [14C]acetate association with ACP by 70%, 89%, and 86%, respectively (Fig. 5B).
FIG. 5.
GFZ inhibits fatty acid elongation on L. pneumophila ACP. Logarithmic-phase L. pneumophila cells were incubated for 1 h at 37°C in AYE medium containing [14C]acetate in the absence or presence of 0.4 mM GFZ, 1 mM clofibric acid (CFA), 1 mM 3-HPA, 0.01 mM triclosan (TRI), 0.2 mM cerulenin (CER), or 0.7 mM INH. ACP was purified from these cells, and its protein and radioactive contents were measured, all as described in Materials and Methods.
To confirm that the ACP labeling observed in the absence of GFZ reflected incorporation of [14C]acetate into nascent fatty acids and not its biosynthetic conversion into amino acids subsequently used for ACP synthesis, we blocked L. pneumophila protein synthesis with kanamycin and incubated these kanamycin-treated cells with [14C]acetate. ACP isolated from L. pneumophila incubated in medium containing [14C]acetate and kanamycin had about the same specific activity as ACP isolated from L. pneumophila incubated in kanamycin-free medium containing [14C]acetate (586 cpm/μg protein versus 520 cpm/μg protein). Moreover, GFZ inhibited [14C]acetate incorporation into ACP to about the same extent in kanamycin-treated cells as it did in cells incubated in the absence of kanamycin (148 cpm/μg protein versus 156 cpm/μg protein). These results support the hypothesis that GFZ inhibits fatty acid synthesis.
Identification and purification of L. pneumophila enoyl reductase.
The findings that GFZ stimulates PHB accumulation, blocks fatty acid synthesis, and inhibits [14C]acetate association with ACP in kanamycin-treated L. pneumophila cells strongly suggested that it inhibits one of the enzymes in the fatty acid elongation cycle. Potential targets were β-ketoacylsynthetase, β-hydroxydecenoyl dehydrase, and enoyl reductase. FabI enoyl reductase, the rate-limiting enzyme in E. coli fatty acid synthesis (18), seemed the most likely candidate, since inhibition of this enzyme in E. coli by long-chain fatty acids stimulates accumulation of 3-hydroxybutyryl-ACP (19), and 3-hydroxybutyryl-ACP can be converted to β-hydroxybutyryl-CoA, the precursor of PHB. To examine this possibility, we sought a putative fabI homolog in L. pneumophila.
E. coli strain FT100 (3) carries a mutation in the enoyl reductase homolog gene, envM. FT100 carries a temperature-sensitive (TS) E. coli enoyl reductase (56). We transformed E. coli FT100 with DNA from a wild-type L. pneumophila library by using the shuttle vector pMMB207 and sought colonies that grew at 42°C, the restrictive temperature for E. coli FT100. Since fabI is an essential gene in E. coli, we expected to identify L. pneumophila genes encoding a protein(s) with complementing enoyl reductase activity. Using this strategy, we identified a 1,375-bp fragment from L. pneumophila which complemented the FT100 TS strain for growth at 42°C. Sequence analysis of this 1,375-bp DNA fragment demonstrated that it includes an 804-bp open reading frame (GenBank accession number AE017354; lpg1854) (10) encoding a putative 268-amino-acid protein with 58% identity and 78% similarity to the E. coli EnvM enzyme and 31% identity and 57% similarity to InhA, the INH-sensitive enoyl reductase from M. tuberculosis (Fig. 6A).
Kinetic characteristics of L. pneumophila enoyl-CoA reductase.
The putative L. pneumophila fabI gene was cloned into the pET15b expression vector, generating a protein with an N-terminal six-histidine tag. The protein was purified to homogeneity using nickel-NTA column chromatography (Fig. 6B).
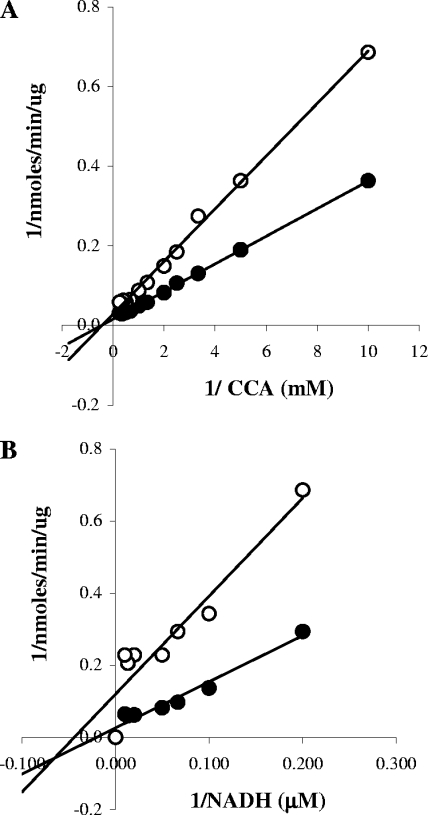
Enoyl reductases catalyze the NADH-dependent reduction of unsaturated fatty acyl groups covalently bound to the pantothenate moiety of ACP. One mole of NAD+ is generated for each mole of crotonyl-ACP reduced. Accordingly, we assessed the enoyl reductase activity of the L. pneumophila protein purified by nickel-NTA chromatography by measuring the decreased absorbance of NADH at 340 nm as it was oxidized to NAD+ (4), and its substrate, CCA, was converted to butyryl-CoA. Kinetic analyses indicated that the L. pneumophila enzyme had a Km of 1.8 ± 0.2 mM (Fig. 7A) and a Vmax of 55 ± 3 nmol/min/μg for CCA in the presence of 100 μM NADH. The Km and Vmax for NADH in the presence of 0.5 mM CCA were estimated to be 18 ± 3 μM and 22 ± 1 nmol/min/μg, respectively (Fig. 7B).
FIG. 7.
Kinetic characteristics of L. pneumophila enoyl-CoA reductase. The activity of purified L. pneumophila FabI was assessed by measuring the decrease in absorbance at 340 nm as NADH was oxidized to NAD+ in the absence (•) or presence (○) of 4 mM GFZ. (A) The dependence of enzyme activity on the concentration of substrate (crotonyl-CoA) was determined in the presence of 100 μM NADH. (B) The dependence of enzyme activity on the concentration of NADH was determined in the presence of 0.5 mM crotonyl-CoA. Data are the averages for duplicate cultures from one experiment representative of three that yielded similar results.
GFZ inhibits L. pneumophila FabI enoyl-CoA reductase.
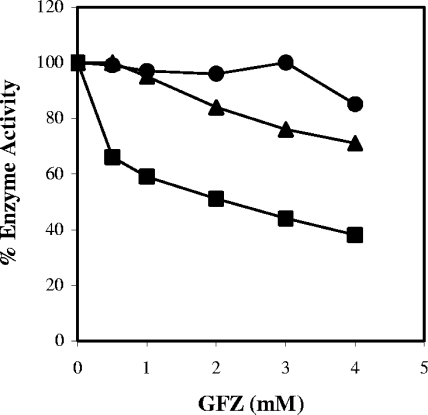
To assess GFZ's effect on L. pneumophila FabI, we measured NADH oxidation in the presence of constant concentrations of enzyme and substrate and concentrations of GFZ varying from 0.5 to 4 mM. All experiments were performed with a submaximal Vmax substrate concentration. The graphs show data from multiple experiments in which 100% enzyme activity ranged from 10 to 20 nmol/minute/μg protein (Fig. 8 and Fig. 9). GFZ inhibited L. pneumophila FabI 35% at 0.5 mM and 60% at 4 mM (Fig. 8). Clofibric acid, which has only a modest inhibitory effect on L. pneumophila growth in macrophages at 1 mM (Fig. 1), inhibited FabI activity by 30% at 4 mM (Fig. 8). 3-HPA did not detectably inhibit L. pneumophila FabI at concentrations of <4 mM, consistent with the result shown in Fig. 5.
FIG. 8.
GFZ is a more potent inhibitor of L. pneumophila FabI than clofibric acid or 3-HPA. The inhibitory effects of 0.5 to 4 mM GFZ (▪), clofibrate (▴), and 3-HPA (•) on enoyl-CoA reductase activity of purified L. pneumophila FabI were assessed by measuring the decrease in absorbance at 340 nm of NADH (100 μM) in the presence of 2 μg/ml purified protein and 0.5 mM CCA. Data are the averages for duplicate cultures from one experiment representative of three that yielded similar results.
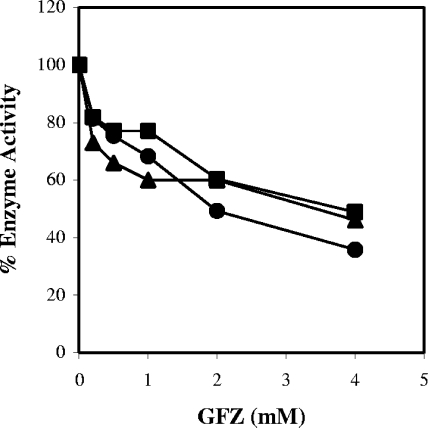
FIG. 9.
GFZ inhibits purified enoyl-CoA reductases from L. pneumophila, E. coli, and M. tuberculosis. The activities of enoyl-CoA reductases from L. pneumophila (FabI) (•), E. coli (EnvM) (▪), and M. tuberculosis (InhA) (▴) were measured by monitoring the rate of decrease in absorbance at 340 nm in the presence of constant concentrations of enzyme and substrate (0.5 mM CCA for FabI and EnvM and 0.5 mM DCA for InhA) at the indicated GFZ concentrations. Data are the averages for duplicate cultures from one experiment representative of three that yielded similar results.
The apparent Km of FabI for CCA did not change much in the presence of 4 mM GFZ, while the apparent Vmax value decreased (Fig. 7A). Very similar results were obtained for NADH saturation (Fig. 7B). For both CCA and NADH, the Kii and Kis were not distinguishable, indicating that FabI binds GFZ with similar affinities in the presence and absence of substrate. The Kii and Kis versus CCA were 4.1 ± 0.8 mM and 5 ± 1 mM, and those versus NADH were 1.4 ± 0.3 mM and 4 ± 3 mM, respectively. Linear transformation of the enzyme activity data suggests that GFZ is a noncompetitive inhibitor of FabI. These findings are reported in preliminary form in U.S. patent applications 5,422,372 and 6,713,043.
GFZ inhibits E. coli and M. tuberculosis enoyl-CoA reductases.
Since E. coli and M. tuberculosis enoyl-CoA reductases are 58% and 31% identical in sequence to L. pneumophila FabI, respectively, we tested GFZ's effect on these enzymes. The E. coli fabI and M. tuberculosis inhA genes were cloned into pET15b, generating enoyl reductase proteins with an N-terminal six-histidine tag, and the proteins were purified as described in Materials and Methods (Fig. 6B). With CCA as a substrate, the apparent Km for E. coli FabI was 2.9 ± 0.2 mM, which is very similar to the Km reported for E. coli FabI by others (4), and the Vmax was 98 ± 4 nmol/min/μg.
GFZ inhibited the E. coli and M. tuberculosis proteins to the same degree as L. pneumophila FabI (Fig. 9). Although E. coli growth was not inhibited by GFZ, the drug behaved as a noncompetitive inhibitor of purified E. coli FabI. The Kii and Kis versus CCA were calculated to be 2.8 ± 0.4 mM and 2.9 ± 0.4 mM, respectively. GFZ also inhibited InhA (Fig. 9). InhA is reported to prefer longer acyl-CoA substrates than those used by E. coli and L. pneumophila enoyl reductases (6, 44) and has a lower specific activity than the E. coli and L. pneumophila enzymes. For InhA enzymatic assays, we used 1 mM DCA as a substrate. Maximum InhA activity was 283 pmol/min/μg protein. Due to its low activity, this enzyme was not characterized further.
GFZ inhibits purified E. coli enoyl reductase but does not inhibit E. coli growth.
Purified E. coli enoyl reductase is as sensitive to GFZ as L. pneumophila FabI. Thus, it was surprising and of interest that GFZ did not inhibit E. coli growth. Ideally, we would have liked to have transfected a FabI-negative E. coli strain with L. pneumophila FabI to explore E. coli's resistance to GFZ. However, E. coli fabI is an essential gene, and it was not possible to knock it out. Hence, we transfected wild-type L. pneumophila with a plasmid containing either L. pneumophila or E. coli fabI under the control of an IPTG-inducible promoter. Discs containing 1 mM GFZ were added in duplicate to CYE agar plates (without or with 1 mM IPTG) containing a wild-type L. pneumophila strain transformed with pMMB207 empty plasmid or pMMB207 plasmid containing either wild-type L. pneumophila fabI, wild-type E. coli fabI, or TS E. coli fabI (from strain FT100). The plates were incubated at 37°C for 2 days, the radius of the growth inhibition zone was measured, and the area of growth inhibition was calculated. The area of growth inhibition for L. pneumophila overexpressing L. pneumophila fabI was 29% smaller than that for the same L. pneumophila strain containing empty vector (Table 2) when IPTG was present but was only 6% smaller in IPTG's absence. The fact that the zone of inhibition decreased to a much greater extent in the presence of IPTG than in its absence confirmed that it resulted from FabI overexpression. E. coli FabI expressed in L. pneumophila also increased the GFZ resistance of transfected L. pneumophila in an IPTG-dependent manner, but the area of inhibition in the presence of IPTG was only half that for L. pneumophila transfected with L. pneumophila FabI (16.5% versus 32%). Whether this reflects less efficient expression or function of E. coli FabI than of L. pneumophila FabI in L. pneumophila cannot be determined from this experiment. What is evident, however, is that E. coli FabI increases L. pneumophila's resistance to GFZ. TS E. coli FabI (FT100), which has only ∼30% of the enoyl reductase activity of wild-type FabI (FT101) at 30°C (3) and is virtually inactive at 37°C, did not enhance the resistance of L. pneumophila to GFZ at 37°C, demonstrating that enzymatically active FabI is required to overcome GFZ's growth-inhibitory effect. These results are consistent with the hypothesis that FabI is a target for GFZ in vivo.
TABLE 2.
Growth inhibition zones of L. pneumophila and E. coli cells overexpressing enoyl reductase
| Gene on plasmid pMMB207 | Inhibition area (mm2)
|
Difference in area of inhibition (1 mM IPTG/no IPTG) | |
|---|---|---|---|
| No IPTG | 1 mM IPTG | ||
| Vector only | 706.5 ± 23.5 | 684.5 ± 69.5 | −3 |
| L. pneumophila fabI | 663.0 ± 91.0 | 452.0 ± 19.0 | −32 |
| E. coli fabI | 660.5 ± 45.5 | 551.0 ± 21.0 | −16.5 |
| TS E. coli fabI | 730.5 ± 24.0 | 754.5 ± 48.5 | +3 |
DISCUSSION
GFZ inhibits axenic growth of 40 different Legionella sp. strains, 27 M. tuberculosis strains (5 antibiotic-sensitive and 22 antibiotic-resistant strains, including 16 strains resistant to INH), and several other human pathogens, such as Nocardia sp. and S. aureus (not shown). It inhibits the growth of L. pneumophila and M. tuberculosis in human and mouse macrophages (Fig. 1 and 2). At submaximal concentrations (e.g., 0.12 mM), GFZ markedly reduces acetate incorporation into L. pneumophila lipids (Fig. 5) and promotes PHB accumulation in this bacterium (Fig. 4). Although it is not the focus of this study, the finding that GFZ stimulates PHB accumulation is noteworthy and suggests that GFZ, and perhaps other compounds with similar activities, shunts intermediates in fatty acid synthesis into PHB. Accordingly, GFZ may be useful for stimulating L. pneumophila, and perhaps other bacteria, to produce commercially valuable polyhydroxyalkanoates (17, 33).
We cloned an L. pneumophila protein that is homologous to the E. coli enoyl reductase FabI and that expresses enoyl-CoA reductase activity (Fig. 6). This L. pneumophila enoyl reductase complemented growth of an E. coli strain expressing a temperature-sensitive FabI protein at a nonpermissive temperature. Hence, we termed this L. pneumophila protein Legionella FabI.
Fatty acid synthesis in bacteria is tightly coordinated with membrane phospholipid formation and with other plasma membrane components, and substances that inhibit fatty acid synthesis block bacterial growth (58). Enoyl-ACP reductase executes the final step of fatty acid elongation and is a key regulator of this pathway (18, 21). GFZ is a noncompetitive inhibitor of E. coli, L. pneumophila, and M. tuberculosis enoyl reductases (Fig. 5, 7, 8, and 9). Thus, it is evident why it inhibits L. pneumophila and M. tuberculosis growth. What remains unresolved is the mechanism by which E. coli resists GFZ's growth-inhibitory effects. Aside from the conjecture that E. coli is impermeable to GFZ or highly efficient at pumping it out of the cytoplasm, we have no explanation for its GFZ resistance.
Diazaborines, triclosan, and INH are all enoyl-ACP-reductase inhibitors, but they have different mechanisms of action (20, 21). While mutations in InhA at or near residues involved in NADH binding confer resistance of M. tuberculosis to INH (2, 43), INH-resistant M. tuberculosis strains were as sensitive to GFZ as INH-sensitive strains (Table 1). Moreover, overexpression of L. pneumophila or E. coli FabI in L. pneumophila increased L. pneumophila's resistance to GFZ (Table 2). Similarly, transfection of E. coli with a multicopy vector encoding FabI increased the resistance of the resulting strain to triclosan (22). These findings are consistent with the hypothesis that GFZ inhibits L. pneumophila and M. tuberculosis growth by inhibiting their respective enoyl reductases. The finding that GFZ is equally effective in suppressing growth of INH-sensitive and INH-resistant M. tuberculosis strains suggests that GFZ interacts with InhA at a different site from that for INH.
Both E. coli FabI and FabH are inhibited by elevated intracellular levels of long-chain acyl-ACP (3, 58). GFZ inhibited [14C]acetate incorporation into L. pneumophila ACP by ∼80% (Fig. 5), suggesting that it did not promote accumulation of long-chain acyl-ACP. For this reason, we think it unlikely that GFZ indirectly inhibits L. pneumophila growth by elevating the intracellular concentration of long-chain acyl-ACP.
Several bacterial species metabolize carboxylates by ligating them to CoA (13, 16, 32, 49). Ciprofibrate, which like GFZ is a carboxylate, is converted to ciprofibroyl-CoA in rat and marmoset livers (8, 11). Thin-layer chromatographic analyses of extracts of L. pneumophila incubated with 0.4 mM GFZ and/or [3H]GFZ showed no evidence of any GFZ derivative (e.g., GFZ-AMP or GFZ-CoA) (data not shown). These negative findings, taken together with the findings that GFZ directly inhibits the activity of FabI isolated from L. pneumophila and E. coli and of InhA from M. tuberculosis and that overexpression of L. pneumophila or E. coli FabI in L. pneumophila increases L. pneumophila resistance to GFZ, are most consistent with the hypothesis that GFZ inhibits axenic growth of L. pneumophila and M. tuberculosis by directly blocking FabI- and InhA-mediated fatty acid synthesis, respectively.
GFZ is reported to inhibit fatty acid chain elongation in rat liver microsomes (47). However, it is unknown whether GFZ, like ciprofibrate, is converted to GFZ-CoA in the mammalian liver. If it is, its inhibitory effect on mammalian fatty acid synthesis may be due to GFZ-CoA's effect on mammalian enoyl-CoA reductase, not to GFZ's direct effect on these enzymes. It is also unknown whether GFZ is converted to a CoA derivative by macrophages and whether such CoA derivatives have the capacity to permeate either the cytoplasmic vacuoles in which L. pneumophila and M. tuberculosis reside and grow within macrophages or the walls and membranes of these bacteria when they are resident within their respective membrane-bound compartments. Without resolution of these issues, we cannot be certain whether the mechanism(s) by which GFZ inhibits fatty acid synthesis in L. pneumophila and M. tuberculosis in bacteriological media is the same as that by which it blocks fatty acid elongation in mammalian microsomes and/or growth of L. pneumophila and M. tuberculosis in macrophages. Nonetheless, the findings that GFZ, an approved, widely used, and generally well-tolerated drug, inhibits axenic growth of multiple L. pneumophila strains and of multiple pan-drug-sensitive and multidrug-resistant M. tuberculosis strains and blocks the growth of these pathogens in macrophages maintained in serum-containing medium suggest that it may have similar activity against them in vivo. While the concentration of GFZ required to inhibit growth of these bacteria is greater than that currently used to lower blood lipid levels clinically, it is possible that administration of higher levels of GFZ might be therapeutically beneficial and/or that more potent congeners of this drug could be identified.
The ability of M. tuberculosis to grow in mouse macrophages is reportedly dependent on its capacity to scavenge fatty acids from these cells (37). The findings that GFZ blocks intracellular growth of L. pneumophila and that both GFZ and INH block intracellular growth of M. tuberculosis suggest that these bacteria may shunt host cell fatty acids directly into bacterial membrane phospholipids, in addition to using them as oxidizable substrates for generating ATP and reduced pyridine nucleotides.
Acknowledgments
We thank James C. Sacchettini for his generous gift of the inhA gene and Irina Mozorova and Sergey Pampou from the Columbia Genome Center for their help with this project.
This research was supported by NIH grants AI47694 and AI20516 to S.C.S., AI23549 to H.A.S., AI22616 to G.K., and AI33696 to J.S.B.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Babel, W., J. U. Ackermann, and U. Breuer. 2001. Physiology, regulation, and limits of the synthesis of poly(3HB). Adv. Biochem. Eng. Biotechnol. 71:125-157. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 3.Bergler, H., S. Fuchsbichler, G. Hogenauer, and F. Turnowsky. 1996. The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur. J. Biochem. 242:689-694. [DOI] [PubMed] [Google Scholar]
- 4.Bergler, H., P. Wallner, A. Ebeling, B. Leitinger, S. Fuchsbichler, H. Aschauer, G. Kollenz, G. Högenauer, and F. Turnowsky. 1994. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J. Biol. Chem. 269:5493-5496. [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Bloch, K. 1977. Control mechanisms for fatty acid synthesis in Mycobacterium smegmatis. Adv. Enzymol. Relat. Areas Mol. Biol. 45:1-84. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, P. J. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 83:91-97. [DOI] [PubMed] [Google Scholar]
- 8.Bronfman, M., M. N. Morales, L. Amigo, A. Orellana, L. Nunez, L. Cardenas, and P. C. Hidalgo. 1992. Hypolipidaemic drugs are activated to acyl-CoA esters in isolated rat hepatocytes. Detection of drug activation by human liver homogenates and by human platelets. Biochem. J. 284:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, F. W., R. M. Cole, M. D. Hicklin, J. A. Blackmon, and B. S. Callaway. 1979. Ultrastructure of the Legionnaire's disease bacterium. Ann. Int. Med. 90:642-647. [DOI] [PubMed] [Google Scholar]
- 10.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 11.Drogemuller, C. J., S. Nunthasomboon, and K. M. Knights. 2001. Nafenopin-, ciprofibroyl-, and palmitoyl-CoA conjugation in vitro: kinetic and molecular characterization of marmoset liver microsomes and expressed MLCL1. Arch. Biochem. Biophys. 396:56-64. [DOI] [PubMed] [Google Scholar]
- 12.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrandez, A., B. Minambres, B. Garcia, E. R. Olivera, J. M. Luengo, J. L. Garcia, and E. Diaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 14.Fortier, A. H., and L. A. Falk. 2002. Isolation of murine macrophages. In J. E. Coligan, A. M. Kruisbeek, D. M. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley and Sons, New York, NY [Book online.] 14. Unit 14.1. Basic protocol 2. http://www.mrw2.interscience.wiley.com/cponline/tserver.dll?command=doGetDoc&sUI=&database=CP&useScheme=WIS_Framed.Scheme&getDoc=cp_cpim_fs.html. [DOI] [PubMed]
- 15.Freeman, S., F. A. Post, L. G. Bekker, R. Harbacheuski, L. M. Steyn, B. Ryffel, N. D. Connell, B. N. Kreiswirth, and G. Kaplan. 2006. Mycobacterium tuberculosis H37Ra and H37Rv differential growth and cytokine/chemokine induction in murine macrophages in vitro. J. Interferon Cytokine Res. 26:27-33. [DOI] [PubMed] [Google Scholar]
- 16.Gobel, M., K. Kassel-Cati, E. Schmidt, and W. Reineke. 2002. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: cloning, characterization, and analysis of sequences encoding 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. J. Bacteriol. 184:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hankermeyer, C. R., and R. S. Tjeerdema. 1999. Polyhydroxybutyrate: plastic made and degraded by microorganisms. Rev. Environ. Contam. Toxicol. 159:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Heath, R. J., and C. O. Rock. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J. Biol. Chem. 270:26538-26542. [DOI] [PubMed] [Google Scholar]
- 19.Heath, R. J., and C. O. Rock. 1996. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:1833-1836. [DOI] [PubMed] [Google Scholar]
- 20.Heath, R. J., S. W. White, and C. O. Rock. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58:695-703. [DOI] [PubMed] [Google Scholar]
- 21.Heath, R. J., S. W. White, and C. O. Rock. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467-497. [DOI] [PubMed] [Google Scholar]
- 22.Heath, R. J., Y. T. Yu, M. A. Shapiro, E. Olson, and C. O. Rock. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316-30320. [DOI] [PubMed] [Google Scholar]
- 23.Hemmingway, C. J., K. K. Tey, and M. R. Munday. 1995. Short-term inhibition of fatty acid and cholesterol biosynthesis by the lipid lowering drug gemfibrozil in primary rat hepatocyte cultures and rat liver in vivo. Biochem. Soc. Trans. 23:496S. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 27.Horwitz, M. A. 1983. Symbiotic interactions between Legionella pneumophila and human leukocytes. Int. Rev. Cytol. 14(Suppl.):307-328. [PubMed] [Google Scholar]
- 28.Horwitz, M. A., and S. C. Silverstein. 1983. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J. Clin. Investig. 71:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, B. W., W. S. Mauchline, P. J. Dennis, C. W. Keevil, and R. Wait. 1999. Poly-3-hydroxybutyrate in Legionella pneumophila, an energy source for survival in low-nutrient environments. Appl. Environ. Microbiol. 65:822-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, F. Y., V. S. Kamanna, M. Y. Chuang, K. Morgan, and M. L. Kashyap. 1996. Gemfibrozil stimulates apolipoprotein A-I synthesis and secretion by stabilization of mRNA transcripts in human hepatoblastoma cell line (Hep G2). Arterioscler. Thromb. Vasc. Biol. 16:1052-1062. [DOI] [PubMed] [Google Scholar]
- 31.Kabbash, C. A. 2000. Characterization of gemfibrozil's novel mechanism of antibacterial action. Ph.D. dissertation. Columbia University, New York, NY.
- 32.Kawaguchi, K., Y. Shinoda, H. Yurimoto, Y. Sakai, and N. Kato. 2006. Purification and characterization of benzoate-CoA ligase from Magnetospirillum sp. strain TS-6 capable of aerobic and anaerobic degradation of aromatic compounds. FEMS Microbiol. Lett. 257:208-213. [DOI] [PubMed] [Google Scholar]
- 33.Mahapatra, K., M. S. Kumar, and T. Chakrabarti. 2007. Production and recovery process of polyhydroxybutyrate (PHB) from waste activated sludge. J. Environ. Sci. Eng. 49:164-169. [PubMed] [Google Scholar]
- 34.Manca, C., S. Paul, C. E. Barry III, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauchline, W. S., and C. W. Keevil. 1991. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl. Environ. Microbiol. 57:3345-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niederweis, M. 2008. Nutrient acquisition by mycobacteria. Microbiology 154:679-692. [DOI] [PubMed] [Google Scholar]
- 39.Ostle, A. G., and J. G. Holt. 1982. Nile blue A as a fluorescent stain for poly-β hydroxybutyrate. Appl. Environ. Microbiol. 44:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riis, W., and W. Mai. 1988. Gas chromatographic determination of poly-β-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J. Chromatogr. 445:285-289. [Google Scholar]
- 41.Rock, C. O., J. L. Garwin, and J. E. Cronan, Jr. 1981. Preparative enzymatic synthesis of acyl-acyl carrier protein. Methods Enzymol. 72:397-403. [DOI] [PubMed] [Google Scholar]
- 42.Rodgers, F. G., and M. R. Davey. 1982. Ultrastructure of the cell envelope layers and surface details of Legionella pneumophila. J. Gen. Microbiol. 128:1547-1557. [DOI] [PubMed] [Google Scholar]
- 43.Rozwarski, D. A., G. A. Grant, D. H. Barton, W. R. Jacobs, Jr., and J. C. Sacchettini. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 44.Rozwarski, D. A., C. Vilcheze, M. Sugantino, R. Bittman, and J. C. Sacchettini. 1999. Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate. J. Biol. Chem. 274:15582-15589. [DOI] [PubMed] [Google Scholar]
- 45.Rudin, D. E., P. X. Gao, C. X. Cao, H. C. Neu, and S. C. Silverstein. 1992. Gemfibrozil enhances the listeriacidal effects of fluoroquinolone antibiotics in J774 macrophages. J. Exp. Med. 176:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez, R. M., M. Vinals, M. Alegret, M. Vazquez, T. Adzet, M. Merlos, and J. C. Laguna. 1992. Inhibition of rat liver microsomal fatty acid chain elongation by gemfibrozil in vitro. FEBS Lett. 300:89-92. [DOI] [PubMed] [Google Scholar]
- 48.Schoonjans, K., B. Staels, P. Grimaldi, and J. Auwerx. 1993. Acyl-CoA synthetase mRNA expression is controlled by fibric acid derivatives, feeding and liver proliferation. Eur. J. Biochem. 216:615-622. [DOI] [PubMed] [Google Scholar]
- 49.Schuhle, K., J. Gescher, U. Feil, M. Paul, M. Jahn, H. Schagger, and G. Fuchs. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 185:4920-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sethy-Coraci, I., L. W. Crock, and S. C. Silverstein. 2005. PAF-receptor antagonists, lovastatin, and the PTK inhibitor genistein inhibit H2O2 secretion by macrophages cultured on oxidized-LDL matrices. J. Leukoc. Biol. 78:1166-1174. [DOI] [PubMed] [Google Scholar]
- 51.Shah, N. S., A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martin-Casabona, F. Drobniewski, C. Gilpin, M. Havelkova, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rusch-Gerdes, A. Van Deun, V. Vincent, K. Laserson, C. Wells, and J. P. Cegielski. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, S. 1994. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 8:1248-1259. [PubMed] [Google Scholar]
- 53.Steel, D. J., T. L. Tieman, J. H. Schwartz, and S. J. Feinmark. 1997. Identification of an 8-lipoxygenase pathway in nervous tissue of Aplysia californica. J. Biol. Chem. 272:18673-18681. [DOI] [PubMed] [Google Scholar]
- 54.Tesh, M. J., and R. D. Miller. 1981. Amino acid requirements for Legionella pneumophila growth. J. Clin. Microbiol. 13:865-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tesh, M. J., S. A. Morse, and R. D. Miller. 1983. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J. Bacteriol. 154:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnowsky, F., K. Fuchs, C. Jeschek, and G. Hogenauer. 1989. envM genes of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:6555-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahringer, U., Y. A. Knirel, B. Lindner, J. H. Helbig, A. Sonesson, R. Marre, and E. T. Rietschel. 1995. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res. 392:113-139. [PubMed] [Google Scholar]
- 58.Zhang, Y. M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]