Abstract
Because very little is known about cell division in noncylindrical bacteria and cyanobacteria, we investigated 10 putative cytokinetic proteins in the unicellular spherical cyanobacterium Synechocystis strain PCC 6803. Concerning the eight penicillin-binding proteins (PBPs), which define three classes, we found that Synechocystis can survive in the absence of one but not two PBPs of either class A or class C, whereas the unique class B PBP (also termed FtsI) is indispensable. Furthermore, we showed that all three classes of PBPs are required for normal cell size. Similarly, the putative FtsQ and FtsW proteins appeared to be required for viability and normal cell size. We also used a suitable bacterial two-hybrid system to characterize the interaction web among the eight PBPs, FtsQ, and FtsW, as well as ZipN, the crucial FtsZ partner that occurs only in cyanobacteria and plant chloroplasts. We showed that FtsI, FtsQ, and ZipN are self-interacting proteins and that both FtsI and FtsQ interact with class A PBPs, as well as with ZipN. Collectively, these findings indicate that ZipN, in interacting with FtsZ and both FtsI and FtQ, plays a similar role to the Escherichia coli FtsA protein, which is missing in cyanobacteria and chloroplasts.
The peptidoglycan layer (PG) of bacterial cell wall is a major determinant of cell shape, and the target of our best antibiotics. It is built from long glycan strands of repeating disaccharides cross-linked by short peptides (38). The resultant meshwork structure forms a strong and elastic exoskeleton essential for maintaining shape and withstanding intracellular pressure. Cell morphogenesis and division have been essentially studied in the rod-shaped organisms Escherichia coli and Bacillus subtilis, which divide through a single medial plane (8, 10, 21, 23). These organisms have two modes of cell wall synthesis: one involved in cell elongation and the second operating in septation (2). Each mode of synthesis is ensured by specific protein complexes involving factors implicated in the last step of PG synthesis (2). The complete assembly of PG requires a glycosyl transferase that polymerizes the glycan strands and a transpeptidase that cross-links them via their peptide side chains (35). Both activities are catalyzed by penicillin-binding proteins (PBPs), which can be divided into three classes: class A and class B high-molecular-weight (HMW) PBPs and class C low-molecular-weight (LMW) PBPs (35).
Class A PBPs exhibit both transglycosylase and transpeptidase activities. In E. coli, they seem to be nonspecialized (2), as they operate in the synthesis of both cylindrical wall (cell elongation) and septal PG (cytokinesis). In B. subtilis, PBP1 (class A) is partially localized to septal sites and its depletion leads to cell division defects (31).
Class B PBPs, which comprise two proteins in most bacteria, are monofunctional transpeptidases (35), each involved in longitudinal and septal growth of cell wall, respectively (36). In E. coli, this protein, PBP3, is also termed FtsI, because it belongs to the Fts group of cell division factors whose depletion leads to the filamentation phenotype (11). These at least 10 Fts proteins are recruited to the division site at mid-cell in the following sequential order: FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsL/FtsB, FtsW, FtsI, and FtsN (11). The cytoplasmic protein FtsZ is the first recruited to the division site, where it polymerizes in a ring-like structure (1), which serves as a scaffold for the recruitment of the other Fts proteins and has been proposed to drive the division process (6). Together the Fts proteins form a complex machine coordinating nucleoid segregation, membrane constriction, septal PG synthesis, and possibly membrane fusion.
Unlike the other PBPs, class C PBPs do not operate in PG synthesis but rather in maturation or recycling of PG during cell septation (35). They are subdivided into four types. Class C type 5 PBP removes the terminal d-alanine residue from pentapeptide side-chains (dd-carboxypeptidase activity). Types 4 and 7 are able to cleave the peptide cross-links (endopeptidase activity). Finally, type AmpH, which does not have a defined enzymatic activity, is believed to play a role in the normal course of PG synthesis, remodeling or recycling (for a review, see reference 35).
In contrast to rod-shaped bacteria, less is known concerning PG synthesis, morphogenesis, and cytokinesis, and their relationships, in spherical-celled bacteria, even though a wealth of them have a strong impact on the environment and/or human health. Furthermore, unlike rod-shaped bacteria spherical-celled bacteria possess an infinite number of potential division planes at the point of greater cell diameter, and they divide through alternative perpendicular planes (26, 36, 37, 39). The spherical cells of Staphylococcus aureus seem to insert new PG strands only at the septum, and accordingly the unique class A PBP localizes at the septum during cell division (36). In contrast, the rugby-ball-shaped cells of Streptococcus pneumoniae synthesize cell wall at both the septum and the neighboring region called “equatorial rings” (36). Accordingly, class A PBP2a and PBP1a were found to operate in elongation and septation, respectively (29).
In cyanobacteria, which are crucial to the biosphere in using solar energy to renew the oxygenic atmosphere and which make up the biomass for the food chain (7, 30, 40), cell division is currently investigated in two unicellular models with different morphologies: the rod-shaped Synechococcus elongatus strain PCC 7942 (19, 28) and the spherical-celled Synechocystis strain PCC 6803 (26), which both possess a small fully sequenced genome (http://genome.kazusa.or.jp/cyanobase/) that is easily manipulable (18). In both organisms FtsZ and ZipN/Arc6, a protein occurring only in cyanobacteria (ZipN) and plant chloroplasts (Arc6), were found to be crucial for cytokinesis (19, 26, 28) and to physically interact with each other (25, 26). Also, interestingly, recent studies of cell division in the filamentous cyanobacterium Anabaena (Nostoc) strain PCC 7120, showed that this process is connected with the differentiation of heterocysts, the cells dedicated to nitrogen fixation (34).
In a continuous effort to study the cell division machine of the unicellular spherical cyanobacterium Synechocystis, we have presently characterized its eight presumptive PBPs (22) that define three classes and the putative cytokinetic proteins FtsQ and FtsW, as well as their network of interactions between each other and ZipN. Both FtsI and FtsQ were found to be key players in cell division in interacting with ZipN and class A PBPs. Consequently, ZipN in interacting with FtsZ (26), FtsI, and FtQ, like the FtsA protein of E. coli, could play a role similar to FtsA, which is absent in cyanobacteria and chloroplasts.
MATERIALS AND METHODS
Bacterial strains, growth, plasmids, and gene transfer procedures.
Synechocystis strain PCC 6803 was grown and transformed at 30°C in BG11 medium (33) enriched with 3.78 mM Na2CO3, under continuous white light of standard fluence (2,500 lx, 31.25 μE m−2 s−1) as described in reference 4. E. coli strains TOP10 (Invitrogen), CM404, and DHM1, which were grown on LB (Difco) with or without 1% glucose at 30°C or 37°C, were used for either gene manipulation (TOP10) or the two-hybrid assay (DHM1) (16). The final concentrations of selective antibiotics were as follows: for E. coli, ampicillin at 100 μg ml−1, kanamycin at 50 μg ml−1, nalidixic acid at 20 μg ml−1, and spectinomycin at 100 μg ml−1 and for Synechocystis, kanamycin at 50 to 300 μg ml−1, streptomycin at 5 μg ml−1, and spectinomycin at 2.5 to 10 μg ml−1.
Gene cloning and manipulation.
All Synechocystis genes surrounded by their flanking regions (about 300 bp), for homologous recombinations mediating targeted gene replacement (20), were amplified by PCR from wild-type (WT) DNA using specific primers. After cloning in the appropriate plasmids (see Table 2), site-directed mutagenesis was performed and disruptions were made through standard PCR-driven overlap extension (13). We used deletion cassettes that carry the antibiotic-resistant marker inserted in the same orientation as the gene to be inactivated and verified their DNA sequences (Big Dye kit; ABI Perkin Elmer).
TABLE 2.
Characteristics of the plasmids used in this study
| Plasmid | Relevant feature(s)a | Source or reference |
|---|---|---|
| pGEMT | AT overhang Ampr cloning vector | Promega |
| pUC4K | Source of the Kmr marker gene | Pharmacia |
| pHPΩ45 | Source of the Smr Spr marker gene | 32 |
| ppbp1 | pGEMT with the Synechocystis pbp1 gene and its flanking sequences, where part of the PBP1 CS (from bp 84 to 2424) was replaced by a SmaI site | This study |
| pΔpbp1::Kmr | ppbp1 with the Kmr marker inserted in the unique SmaI site | This study |
| pΔpbp1::SmrSpr | ppbp1 with the Smr Spr marker inserted in the unique SmaI site | This study |
| ppbp2 | pGEMT with the Synechocystis pbp2 gene and its flanking sequences, where part of the PBP2 CS (from bp 111 to 2037) was replaced by a SmaI site | This study |
| pΔpbp2::Kmr | ppbp2 with the Kmr marker inserted in the unique SmaI site | This study |
| pΔpbp2::SmrSpr | ppbp2 with the Smr Spr marker inserted in the unique SmaI site | This study |
| ppbp3 | pGEMT with the Synechocystis pbp3 gene and its flanking sequences, where part of the PBP3 CS (from bp 180 to 1734) was replaced by a SmaI site | This study |
| pΔpbp3::Kmr | ppbp3 with the Kmr marker inserted in the unique SmaI site | This study |
| pΔpbp3::SmrSpr | ppbp3 with the Smr Spr marker inserted in the unique SmaI site | This study |
| pftsI | pGEMT with the Synechocystis ftsI gene and its flanking sequences, where part of the FtsI CS (from bp 129 to 1701) was replaced by a SmaI site | This study |
| pΔftsI::Kmr | pftsI with the Kmr marker inserted in the unique SmaI site | This study |
| ppbp5 | pGEMT with the Synechocystis pbp5 gene and its flanking sequences, where part of the PBP5 CS (from bp 129 to 1326) was replaced by a SmaI site | This study |
| pΔpbp5::Kmr | ppbp5 with the Kmr marker inserted in the unique SmaI site | This study |
| ppbp6 | pGEMT with the Synechocystis pbp6 gene and its flanking sequences, where part of the PBP6 CS (from bp 78 to 1,098) was replaced by a SmaI site | This study |
| pΔpbp6::Kmr | ppbp6 with the Kmr marker inserted in the unique SmaI site | This study |
| ppbp7 | pGEMT with the Synechocystis pbp7 gene and its flanking sequences, where part of the PBP7 CS (from bp 129 to 1602) was replaced by a SmaI site | This study |
| pΔpbp7::Kmr | ppbp7 with the Kmr marker inserted in the unique SmaI site | This study |
| pΔpbp7::SmrSpr | ppbp7 with the Smr Spr marker inserted in the unique SmaI site | This study |
| ppbp8 | pGEMT with the Synechocystis pbp8 gene and its flanking sequences, where part of the PBP8 CS (from bp 72 to 876) was replaced by a SmaI site | This study |
| pΔpbp8::Kmr | ppbp8 with the Kmr marker inserted in the unique SmaI site | This study |
| pΔpbp8::SmrSpr | ppbp8 with the Smr Spr marker inserted in the unique SmaI site | This study |
| pftsQ | pGEMT with the Synechocystis ftsQ gene and its flanking sequences, where part of the FtsQ CS (from bp 138 to 663) was replaced by a SmaI site | This study |
| pΔftsQ::Kmr | pftsQ with the Kmr marker inserted in the unique SmaI site | This study |
| pftsW | pGEMT with the Synechocystis ftsW gene and its flanking sequences, where part of the FtsW CS (from bp 96 to 1029) was replaced by a SmaI site | This study |
| pΔftsW::Kmr | pftsW with the Kmr marker inserted in the unique SmaI site | This study |
| pKT25 | Kmr plasmid encoding the N-terminal T25 domain (amino acids 1-224) of the B. pertussis AC in frame with a downstream multiple cloning site | 16 |
| pKT25-zip | pKT25 with the leucine zipper domain of the yeast GCN4 activator | 16 |
| pKT25-zipN | pKT25 with the full-length Synechocystis zipN CS | This study |
| pKT25-ftsQ | pKT25 with the full-length Synechocystis ftsQ CS | This study |
| pKT25-ftsW | pKT25 with the full-length Synechocystis ftsW CS | This study |
| pKT25-ftsI | pKT25 with the full-length Synechocystis ftsI CS | This study |
| pKT25-pbp1 | pKT25 with the full-length Synechocystis pbp1 CS | This study |
| pKT25-pbp2 | pKT25 with the full-length Synechocystis pbp2 CS | This study |
| pKT25-pbp3 | pKT25 with the full-length Synechocystis pbp3 CS | This study |
| pKT25-pbp5 | pKT25 with the full-length Synechocystis pbp5 CS | This study |
| pKT25-pbp6 | pKT25 with the full-length Synechocystis pbp6 CS | This study |
| pKT25-pbp7 | pKT25 with the full-length Synechocystis pbp7 CS | This study |
| pKT25-pbp8 | pKT25 with the full-length Synechocystis pbp8 CS | This study |
| pUT18 | Ampr plasmid encoding the T18 domain (amino acids 225-339) of the B. pertussis AC in frame with an upstream multiple cloning site | 16 |
| pUT18-zip | pUT18 with the leucine zipper domain of the yeast GCN4 activator | 16 |
| pUT18m1 | pUT18 derivative encoding the AC T18 domain in frame with a downstream multiple cloning site identical to that of pKT25 | This study |
| pUT18m1-zipN | pUT18m1 with the full-length Synechocystis zipN CS | This study |
| pUT18m1-ftsQ | pUT18m1 with the full-length Synechocystis ftsQ CS | This study |
| pUT18m1-ftsW | pUT18m1 with the full-length Synechocystis ftsW CS | This study |
| pUT18m1-ftsI | pUT18m1 with the full-length Synechocystis ftsI CS | This study |
| pUT18m1-pbp1 | pUT18m1 with the full-length Synechocystis pbp1 CS | This study |
| pUT18m1-pbp2 | pUT18m1 with the full-length Synechocystis pbp2 CS | This study |
| pUT18m1-pbp3 | pUT18m1 with the full-length Synechocystis pbp3 CS | This study |
| pUT18m1-pbp5 | pUT18m1 with the full-length Synechocystis pbp5 CS | This study |
| pUT18m1-pbp6 | pUT18m1 with the full-length Synechocystis pbp6 CS | This study |
| pUT18m1-pbp7 | pUT18m1 with the full-length Synechocystis pbp7 CS | This study |
| pUT18m1-pbp8 | pUT18m1 with the full-length Synechocystis pbp8 CS | This study |
CS, protein coding sequence; Δ, deletion.
Microscopy.
A total of 1.25 × 105 Synechocystis cells from mid-log-phase culture were placed on microscope slides and immobilized by a 5- to 10-min incubation at room temperature. Images were captured with a Leica DMRXA microscope equipped with a ×100 oil immersion lens, a Ropper Scientific Micromax cooled charge-coupled device camera, and Metamorph software (Universal Imaging). The final processing of images for presentation was done using Adobe Photoshop.
FACS analysis.
Cells from mid-log-phase liquid cultures were harvested, washed twice, and resuspended in phosphate-buffered saline (Sigma-Aldrich) to a final optical density at 580 nm of 0.3 (1.5 × 107cells ml−1). Then, 2 × 104 cells were analyzed by using a FACSCalibur fluorescence-activated cell sorting (FACS) cytometer (Becton Dickinson) with the following settings: forward scatter (FCS), E01 log; side scatter, 350 V. Results were collected with CellQuest software, version 3.1 (Becton Dickinson). Data were plotted on a two-dimensional graph (x axis, FSC; y axis, number of cells). Then histograms from WT and mutant strains were superimposed. Experiments were repeated twice.
The E. coli BACTH assay.
The bacterial two-hybrid (BACTH) system (16) is composed of two replication-compatible plasmids, pKT25 and pUT18, encoding the intrinsically inactive N-terminal T25 domain and C-terminal T18 domain of the adenylate cyclase (AC) from Bordetella pertussis, as well as an E. coli reporter strain, DHM1, deficient in the endogenous cyclic AMP-producing enzyme. When the coding sequences for physically interacting proteins are cloned in pKT25 and pUT18 and subsequently coexpressed in DHM1, their interaction restores AC activity. This turns on the production of the β-galactosidase (β-Gal) reporter enzyme, leading to the blue-colored colonies on LB indicator plates containing 40 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Eurobio), 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Invitrogen), ampicillin, kanamycin, and nalidixic acid. To reduce the number of cloning experiments, we constructed a slightly modified variant of pUT18 we termed pUT18m1, in which the T18 domain is followed by the same multiple cloning site as the one that follows the T25 domain of AC in pKT25. All Synechocystis open reading frames, were cloned as PstI-BamHI restriction fragments (or NsiI and BglII when they possessed internal PstI or BamHI sites) in the PstI-BamHI sites of the reporter plasmids pKT25 and pUT18m1. DHM1 cells doubly transformed with pKT25- and pUT18m1-derived plasmids (Table 2) were incubated for 2 days at 30°C on LB supplemented with glucose 1% (to allow repression of the lac promoter), ampicillin, and kanamycin. Production of the β-Gal reporter enzyme was monitored as follows. Cells grown overnight (16 h) at 30°C in LB-0.5 mM IPTG were harvested by centrifugation (10 min at 5,000 rpm), washed with lysis buffer (50 mM Tris HCl [pH 8.0], 50 mM NaCl), resuspended in ice-cold lysis buffer containing 2 mM phenylmethylsulfonyl, and homogenized for 5 min on ice. They were then incubated with 1 mg ml−1 lysozyme for 1 h on ice and disrupted by sonication (three 10-s pulses at power 6 on a Microson apparatus; Misonix). The lysates were cleared by centrifugation at 12,000 × g for 30 min at 4°C, and proteins of the soluble extracts were quantified by Bradford assay (Bio-Rad). Then, aliquots (1 to 10 μl) of soluble proteins were added to 0.8 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4 [pH 7.5], 1 mM MgSO4, 50 mM β-mercaptoethanol), and the β-Gal reaction was started by adding 0.2 ml of o-nitrophenol-β-galactoside (ONPG) at 4 mg ml−1 in Z buffer lacking β-mercaptoethanol. The reaction was stopped after 3 min with 0.5 ml of 1 M Na2CO3 and A420 was recorded. One β-Gal unit = 1 nmol of ONPG min−1 mg−1 protein.
RESULTS
Construction of Synechocystis mutants lacking or depleted in PBPs.
As very little is known concerning PBPs in noncylindrical bacteria and cyanobacteria, we investigated the role of the eight proteins of the spherical-celled cyanobacterium Synechocystis strain PCC 6803 that share sequence homology with E. coli PBPs (22). These Synechocystis proteins are namely the class A-related PBP1, PBP2, and PBP3; the unique class B member, PBP4 (also termed FtsI); the class C type 4 homologs PBP5 and PBP8; and the class C PBPs type AmpH members PBP6 and PBP7 (Table 1). We constructed pbp gene deletion cassettes (Table 2) (see Materials and Methods), in which each protein coding sequence has been replaced by a transcription terminatorless marker gene for selection, while preserving the flanking DNA regions for homologous recombinations mediating targeted gene replacement in Synechocystis (20). After transformation in Synechocystis, which harbors about 10 chromosome copies per cell (20), we verified through PCR and DNA sequencing that the marker gene had been properly inserted in the Synechocystis chromosome, in place of the studied gene, and we assayed whether the segregation between WT and mutant chromosome copies was complete (the studied gene is dispensable to cell growth) or not (the studied gene is essential to cell viability). All three Δpbp1::Kmr, Δpb2::Kmr, and Δpbp3::Kmr mutants retained no WT chromosome copies (data not shown) and grew as fit as the WT strain (Table 3 and Fig. 1) showing that each of the three class A PBPs is dispensable for Synechocystis viability.
TABLE 1.
Presumptive PBPs in Synechocystis
| Type | Namea | Gene identification no.b |
|---|---|---|
| HMW | ||
| Class A | PBP1 | sll0002 |
| Class A | PBP2 | slr1710 |
| Class A | PBP3 | sll1434 |
| Class B | PBP4/FtsI | sll1833 |
| LMW | ||
| Class C type 4 | PBP5 | slr0646 |
| Class C type 4 | PBP8 | slr0804 |
| Class C type AmpH | PBP6 | sll1167 |
| Class C type AmpH | PBP7 | slr1924 |
Proposed in reference 22.
Gene identifier in CyanoBase.
TABLE 3.
Characteristics of the mutants constructed in this study
| Gene inactivated | Dispensability | Doubling time of growing cells (h) |
|---|---|---|
| None | 10 ± 1 | |
| ftsQ | No | 16 ± 1 |
| ftsW | No | 15 ± 1 |
| pbp1 | Yes | 10 ± 1 |
| pbp2 | Yes | 10 ± 1 |
| pbp3 | Yes | 10 ± 1 |
| pbp4 (ftsI) | No | 15 ± 1 |
| pbp5 | Yes | 10 ± 1 |
| pbp6 | Yes | 10 ± 1 |
| pbp7 | Yes | 10 ± 1 |
| pbp8 | Yes | 10 ± 1 |
| pbp5 and pbp8 | No | 21 ± 2 |
| pbp6 and pbp7 | No | 19 ± 2 |
FIG. 1.
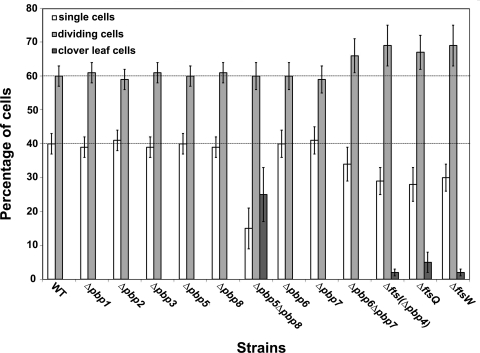
Proportions of various cell types observed in the Synechocystis mutants constructed in this study. In each case, at least 200 randomly chosen cells were classified in the following three categories: single cells, doublets of dividing cells, and cloverleaf-type cell aggregates. Strains are indicated on the x axis, and the percentage of each category is indicated on the y axis (error bars represent the standard deviation). These experiments were performed twice with two independent clones harboring the same mutation.
Synechocystis can survive in the absence of one but not two class A PBPs.
To test whether the three genes possess overlapping functions, we attempted to construct all three double mutants lacking any pair combination of the three class A PBPs. Interestingly, all double mutants invariably died upon restreaking onto selective medium containing both selective antibiotics kanamycin and streptomycin (or spectinomycin), showing that Synechocystis requires at least two HMW PBP class A proteins to survive. Taken together, our results indicate that class A PBPs exhibit crucial and partially overlapping functions.
The absence of PBP2, but not of the other two class A PBPs, PBP1 and PBP3, leads to minicells.
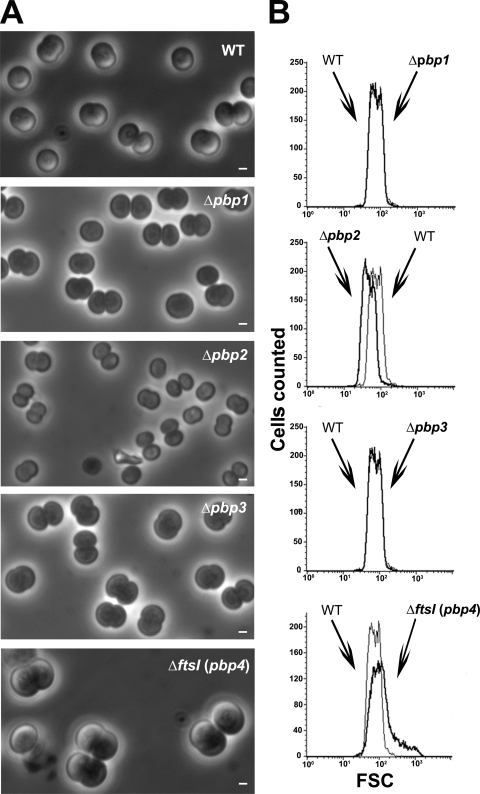
To investigate the impact of each class A PBP on Synechocystis cell morphogenesis, we observed the three Δpbp1::Kmr, Δpbp2::Kmr, and Δpbp3::Kmr single mutants through phase-contrast microscopy. Both Δpbp1::Kmr and Δpbp3::Kmr cells retained normal morphology, whereas Δpbp2::Kmr cells were found to be significantly smaller than WT cells (Fig. 2A). We then confirmed these results using FACS flow cytometry (Fig. 2B), a quantitative evaluation of cell shape (27) that measures the FSC value that is proportional to cell size. In agreement with our microscopy observation, the mean FSC value of Δpbp2::Kmr cells (48.92) appeared to be significantly smaller than those of WT, Δpbp1::Kmr, and Δpbp3::Kmr cells (mean FSC values of 75.94, 74.59, and 73.84, respectively). These findings suggest that PBP2 operates in the synthesis of PG required for normal cell growth of Synechocystis.
FIG. 2.
Morphology of Synechocystis mutant cells depleted in the PBPs of either class A or B. Shown are a phase-contrast image (A; scale bar = 1 μm) and flow cytometry analysis (B) of WT or mutant (Δ) cells totally or partially lacking (Table 3) the indicated PBP. For each mutant, the FSC histogram (bold lines) has been overlaid with that of the WT to better visualize the influence of the mutation on cell size. These experiments were performed twice with two independent clones harboring the same mutation.
FtsI, the unique class B PBP, is indispensable to Synechocystis: its depletion leads to giant cells.
We then studied the role of FtsI, the unique class B PBP (Table 1), using the techniques described above, and found FtsI to be essential to Synechocystis viability. The resulting FtsI-depleted ΔftsI::Kmr/ftsI+ mutant grew more slowly (doubling time of about 15 h) (Table 3) than the WT strain (doubling time of about 10 h). This finding cannot be interpreted in term of a reduced initiation of cytokinesis in the FtsI-depleted mutant, as the fraction of its dividing cells remained similar to, if not higher than, that in the WT strain (71% and 60%, respectively) (Fig. 1). Instead, it is the completion of cytokinesis (i.e., the septation) which seemed to be slower in the FtsI-depleted cells. Indeed, FtsI-depleted cells displayed an increased size (Fig. 2A) and, accordingly, a higher FSC value (152.15) (Fig. 2B) than WT cells (75.94). Furthermore, FtsI depletion led to cloverleaf-like clusters of four large unseparated cells (Fig. 1 and 2) not observed in WT cells. These cloverleaf-like cell clusters likely result from the delayed septation of daughter cells, which was not completed before the initiation of the second round of division. Collectively, our results suggest that FtsI plays a crucial role in the inward synthesis of PG required for completing septation.
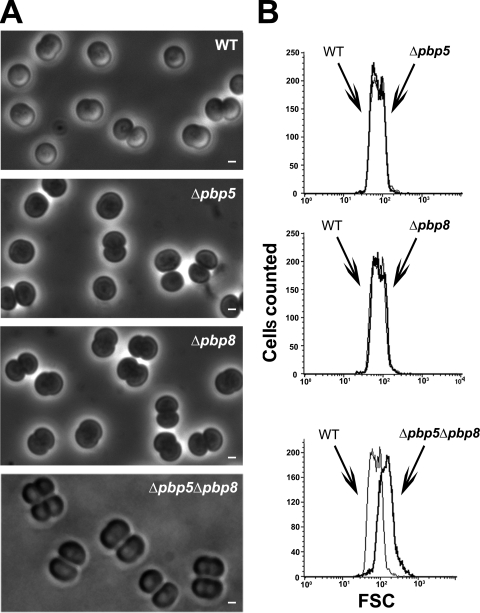
Synechocystis can survive in the absence of one but not the two class C type 4 PBPs, PBP5 and PBP8, operating in septation.
We then studied the role of PBP5 and PBP8, the two class C type 4 PBPs (Table 1). Both appeared to be dispensable to Synechocystis growth (Table 3) and morphology (Fig. 1 and Fig. 3), likely because they have redundant functions. Indeed, it was not possible to obtain the double-deletion mutant lacking both PBP5 and PBP8, irrespective of the sequential order of attempted double deletions, i.e., tentative deletion of either pbp8 in Δpbp5 null recipient cells or pbp5 in Δpbp8 null cells. All resulting cells remained heteroploids in retaining the ability to encode either PBP5 or PBP8 and behaved similarly. They all grew about twice as slowly as WT cells (Table 3), likely because their septation was slow, as suggested by the occurrence of a high proportion (25%) (Fig. 1) of cloverleaf-like four-cell clusters (Fig. 3A) accounting for the observed high FCS value (mean, 148.94) (Fig. 3B), compared to that in WT cells (mean, 75.94). These findings suggest that PBP5 and PBP8, the two class C type 4 PBPs, are required for completion of the septation enabling daughter cell separation.
FIG. 3.
Morphology and size of Synechocystis mutant cells depleted in class C type 4 PBPs. Shown are a phase-contrast image (A; scale bar = 1 μm) and flow cytometry analysis (B) of WT or mutant (Δ) cells totally or partially lacking (Table 3) the indicated PBP. For each mutant, the FSC histogram (bold lines) has been overlaid with that of the WT to better visualize the influence of the mutation on cell size. These experiments were performed twice with two independent clones harboring the same mutation.
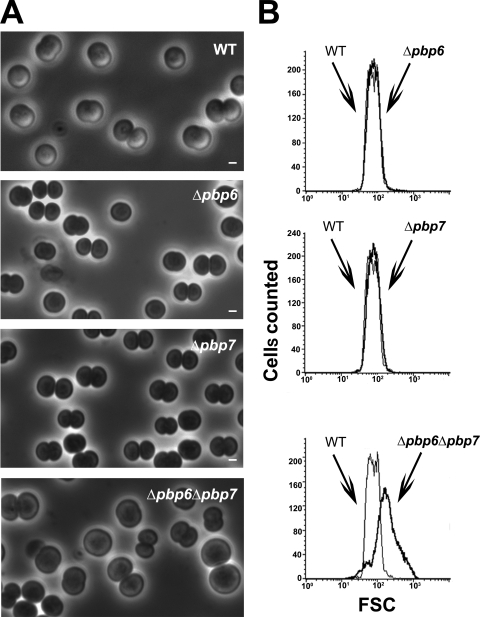
Synechocystis can survive in the absence of one but not both class C type AmpH PBPs, PBP6 and PBP7, the combined depletion of which generates giant cells.
The two Synechocystis class C type AmpH PBPs (Table 1) PBP6 and PBP7 appeared to be dispensable to cell growth (Table 3) and morphology (Fig. 1 and 4), likely because they share redundant functions. Indeed, it was not possible to construct the double-deletion mutant lacking both PBP6 and PBP7, irrespective of the sequential order of the attempted double deletion: i.e., tentative deletion of either pbp7 in Δpbp6 null cells or pbp6 in Δpbp7 null cells. Both resulting heteroploid strains retained the ability to encode either PBP6 or PBP7 and behaved similarly. They grew about twice as slowly as WT cells (Table 3) and generated giant cells (Fig. 4A), likely accounting for the high FCS value (mean, 186.42) of these mutants (Fig. 4B), compared to WT cells (mean, 75.94). We did not observe cloverleaf-like four-cell clusters, unlike the abovementioned cells depleted for the two class C type 4 PBPs.
FIG. 4.
Morphology and size of Synechocystis mutant cells depleted in class C type AmpH PBPs. Shown are a phase-contrast image (A; scale bar = 1 μm) and flow cytometry analysis (B) of WT or mutant (Δ) cells totally or partially lacking (Table 3) the indicated PBP. For each mutant, the FSC histogram (bold lines) has been overlaid with that of the WT to better visualize the influence of the mutation on cell size. These experiments were performed twice with two independent clones harboring the same mutation.
Both FtsQ and FtsW are indispensable to Synechocystis: their depletion leads to giant cells.
As we found FtsI to be indispensable to Synechocystis, we decided to investigate the role of both FtsQ (Sll1632) and FtsW (Slr1267) homolog proteins, because both FtsQ and FtsW were shown in E. coli to interact with FtsI (3) and to be involved in FtsI recruitment to the division site (11). We found that both ftsQ and ftsW are essential to cell viability in Synechocystis (Table 3). The resulting ΔftsQ::Kmr/ftsQ+ and ΔftsW::Kmr/ftsW+ heteroploid mutants grew more slowly than the WT strain (respective doubling times of 16 h, 15 h, and 10 h) (Table 3) and generated giant cells (Fig. 5) (FCS values of 153.74 and 154.46 for ΔftsQ::Kmr/ftsQ+ and ΔftsW::Kmr/ftsW+ cells, respectively), like the ΔftsI::Kmr/ftsI+ mutant (doubling time of about 15 h and FCS value of about 152.15).
FIG. 5.
Morphology and size of Synechocystis mutant cells depleted in proteins FtsQ and FtsW. Shown are a phase-contrast image (A; scale bar = 1 μm) and flow cytometry analysis (B) of WT or mutant (Δ) cells lacking the indicated protein. For each mutant, the FSC histogram (bold lines) has been overlaid with that of the WT to better visualize the influence of the mutation on cell size. These experiments were performed twice with two independent clones harboring the same mutation.
Characterization of the interaction network among PBPs, FtsQ, FtsW, and ZipN.
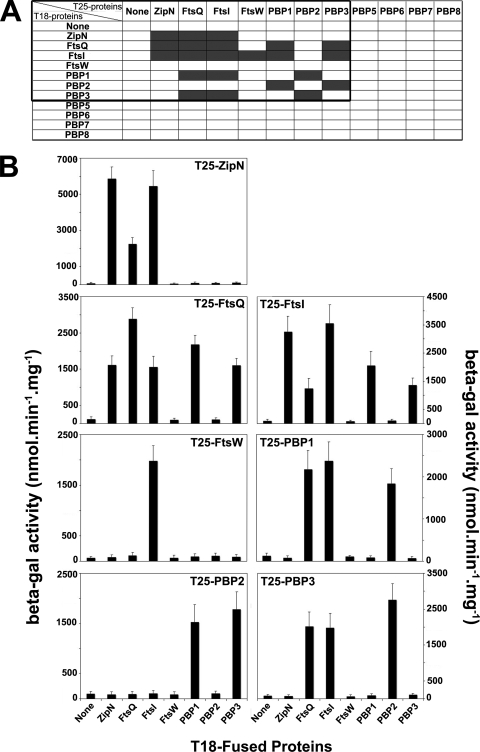
To characterize the interplay between PG synthesis (PBPs) and cytokinesis (Fts proteins), we investigated the pairwise interactions between the presently studied PBPs, FtsQ, FtsW, and ZipN, the previously described cytokinetic factor (26). For this purpose, we used the BACTH system (16), which worked well with cell division proteins from both Synechocystis (26) and E. coli (5, 15), including membrane-associated proteins (17). Full-length coding sequences for the studied Synechocystis proteins were cloned in the two BACTH reporter plasmids pKT25 and pUT18m1 (see Materials and Methods) (Table 2), which were subsequently cotransformed to E. coli to search for pairwise interactions restoring the production of the β-Gal enzyme (Fig. 6). All pairwise interactions were observed in both mutually confirmatory combinations, thereby strengthening their biological relevance. The only exception was FtsW, for which only the T25-FtsW fusion protein, not the T18-FtsW hybrid, led to detection of FtsW partner, as previously reported for the E. coli FtsW protein (15). We found that FtsI, FtsQ, and ZipN are self-interacting proteins (Fig. 6), as observed for E. coli proteins FtsQ (15) and FtsI (24), and Arc6, the chloroplast ortholog of ZipN (25). Also interestingly, FtsI and FtsQ were found to interact with each other (Fig. 6), like their E. coli counterparts (3, 15), as well as with PBP1 and PBP3, which both appeared to interact with PBP2. Furthermore, both FtsI and FtsQ were found to interact with ZipN, the FtsZ-interacting protein occurring only in cyanobacteria (26) and plant chloroplasts (9). Collectively, these findings demonstrate that both FtsI and FtsQ are key players in cyanobacterial cell division. In addition, they suggest (Fig. 7) that ZipN could play a similar role to the E. coli FtsA protein (which is absent from cyanobacteria and chloroplasts), which interacts with FtsZ (11) and both FtsI and FtQ (3, 15). We also found that Synechocystis FtsW interacts with FtsI, as reported in E. coli (15). We observed no interaction with class C PBPs, possibly because these proteins might localize to the periplasm (35), thereby negatively influencing the accumulation and/or activity of the recreated AC reporter enzyme.
FIG. 6.
Two-hybrid analysis of interactions between PBPs and the cytokinetic proteins FtsQ, FtsW, and ZipN. The occurrence of interaction between the tested proteins coproduced in E. coli DHM1 cells of the BACTH system was ascertained by the production of the β-galactosidase reporter enzyme whose activity (i) turned the corresponding cells (indicated by the gray rectangles in panel A) blue on X-Gal-containing medium and (ii) reached a high value (B; beta-gal). Each bar represents the mean value of two measurements performed on the cellular extracts of three reporter clones originating from independent transformations, and the error bars represent the standard deviation. Cells producing the two interacting ZipN-fusion proteins were used as the positive control (5,900 ± 750 β-Gal units), while those producing a single or no reporter protein served as the negative controls (65 ± 50 U of background β-Gal activity).
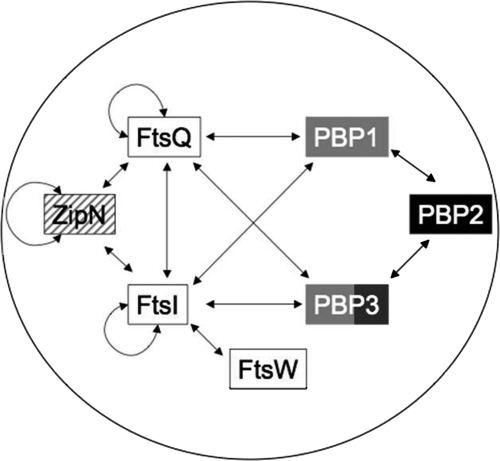
FIG. 7.
Schematic representation of the web of interactions among the 10 presently studied proteins and ZipN, the cytokinetic factor we previously characterized. The spherical morphology of Synechocystis is represented by the circle. The double arrows indicate the presently reported interactions. Black and white letters stand for the proteins respectively indispensable and dispensable to Synechocystis. Black and white squares indicate that the corresponding mutants display decreased and increased cell sizes, respectively. Gray rectangles indicate that the corresponding mutants do not show any morphological defects. The hatched rectangle reminds us that the ZipN-depleted mutant has an aberrant spiral shape (26).
DISCUSSION
As very little is known concerning cell division in noncylindrical bacteria and cyanobacteria, we have investigated several putative cytokinetic proteins in the unicellular spherical cyanobacterium Synechocystis strain PCC 6803. We focused on the eight presumptive PBP-like proteins (22): the three class A PBPs, PBP1, PBP2 and PBP3; the unique class B PBP, FtsI (PBP4); the two class C type 4 PBPs, PBP5 and PBP8; and the two class C type AmpH PBPs, PBP6 and PBP7 (Table 1). We found that Synechocystis can survive without one but not two class A PBPs (Table 3) and that PBP2 is likely involved in the synthesis of PG required for normal cell growth since it leads to minicells. In contrast, both PBP1 and PBP3 appeared to be less important than PBP2 as their absence has no obvious phenotype (Fig. 2).
The unique class B PBP protein, FtsI (PBP4), appeared to be indispensable to Synechocystis. The resulting heteroploid strain (ΔftsI::Kmr/ftsI+) displayed giant cells and cloverleaf-like clusters of four large unseparated cells (Fig. 1 and 2). These cloverleaf-like cell clusters likely result from the delayed septation of daughter cells, which was not completed before the initiation of the second round of division. Collectively, our findings suggest that FtsI might operate in the inward synthesis or incorporation of PG at the septum required for completing separation of daughter cells.
As observed for class A PBPs, we found that Synechocystis can survive without one but not the two class C PBPs of either type 4 (PBP5 and PBP8) or type AmpH (PBP6 and PBP7). Interestingly, the heteroploid mutants resulting from the attempted double deletion of the genes encoding PBP5 and PBP8 on one hand or PBP6 and PBP7 on the other hand grew slowly (Table 3) and displayed giant cells (Fig. 3 and 4), like the ΔftsI::Kmr/ftsI+ heteroploid strain (Table 3 and Fig. 2). Interestingly, the mutants depleted of both PBP5 and PBP8, but not of both PBP6 and PBP7, exhibit a high proportion of cloverleaf-like four-cell clusters (Fig. 1, 3, and 4). These findings suggest that PBP5 and PBP8, but not PBP6 and PBP7, are involved in the completion of the septation allowing daughter cell separation, like FtsI.
We think that the giant morphology of spherical cells results from their septation being slowed down more importantly than their growth. This interpretation is supported by the finding that the specific inhibition of Z-ring assembly by the antibacterial compound PC190723 causes dramatic enlargement of the spherical-celled organism S. aureus (12). In contrast, in a rod-shaped bacterium such as E. coli, when septation is more affected than cell growth (14), the corresponding fts mutants become filamentous (8, 10, 21, 23). To confirm that the giant-cell phenotype in a spherical-celled organism is equivalent to the filamentous phenotype of a rod-shaped organism, we studied the impact on Synechocystis of the depletion of the FtsQ and FtsW homologs, because their depletion in E. coli triggers filamentation. The other reason why we studied the presumptive Synechocystis FtsQ and FtsW proteins is that in E. coli both FtsQ and FtsW were found to interact with FtsI (3). First, we found that both the cyanobacterial FtsQ and FtsW are indispensable to Synechocystis, like FtsI. Then, as expected, we found that both the FtsQ- and FtsW-depleted mutants exhibit similar defects to FtsI: i.e., they grow slowly (Table 3) and display giant cells (Fig. 3 and 5). Furthermore, we used a convenient BACTH system to show that both FtsQ and FtsW physically interact with FtsI, as observed in E. coli. We also used this two-hybrid system to characterize the interplay between PG synthesis (PBPs) and cytokinesis (Fts proteins). We also included the ZipN protein in our study because it is the crucial FtsZ partner in cyanobacteria (26) and plant chloroplasts, where it is termed “Arc6” (9). Interestingly, we found that FtsI, FtsQ, and ZipN are self-interacting proteins (Fig. 6), as observed for E. coli proteins FtsI and FtsQ (24) and chloroplastic protein Arc6 (25). Also interestingly, we found that both FtsI and FtsQ interact with ZipN, as well as with PBP1 and PBP3, which both appeared to interact with PBP2 (Fig. 6). Consequently, both FtsI and FtsQ appeared to be key players in cyanobacterial cell division. Collectively, our findings (Fig. 7) indicate that ZipN, in interacting with FtsZ (26) and both FtsI and FtQ (this study), plays a role similar to the E. coli FtsA protein, which is absent from cyanobacteria and chloroplasts. The E. coli FtsA protein assembles at the Z ring early and participates in the sequential recruitment of the Fts proteins, including both FtsI and FtsQ (6), which interact directly with FtsA (3, 15).
Acknowledgments
We thank Daniel Ladant (Pasteur Institute, Paris, France) for the gift of the BACTH system.
This work was supported in part by French Scientific “ANR Biosys06: SULFIRHOM.” M.M. and C.S. were recipients of fellowships from the CEA (France), and K.M. was the recipient of thesis fellowships from the Algerian Ministry for Education.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Addinall, S. G., and B. Holland. 2002. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 2.den Blaauwen, T., M. A. de Pedro, M. Nguyen-Disteche, and J. A. Ayala. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321-344. [DOI] [PubMed] [Google Scholar]
- 3.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 4.Domain, F., L. Houot, F. Chauvat, and C. Cassier-Chauvat. 2004. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53:65-80. [DOI] [PubMed] [Google Scholar]
- 5.Ebersbach, G., E. Galli, J. Moller-Jensen, J. Lowe, and K. Gerdes. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68:720-735. [DOI] [PubMed] [Google Scholar]
- 6.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris, M., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226-228. [Google Scholar]
- 8.Gerdes, K., J. Moller-Jensen, G. Ebersbach, T. Kruse, and K. Nordstrom. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 9.Glynn, J. M., J. E. Froehlich, and K. W. Osteryoung. 2008. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20:2460-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 11.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 12.Haydon, D. J., N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington, and L. G. Czaplewski. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321:1673-1675. [DOI] [PubMed] [Google Scholar]
- 13.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924-932. [DOI] [PubMed] [Google Scholar]
- 14.Justice, S. S., D. A. Hunstad, L. Cegelski, and S. J. Hultgren. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162-168. [DOI] [PubMed] [Google Scholar]
- 15.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karimova, G., C. Robichon, and D. Ladant. 2009. Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J. Bacteriol. 191:333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koksharova, O. A., and C. P. Wolk. 2002. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 58:123-137. [DOI] [PubMed] [Google Scholar]
- 19.Koksharova, O. A., and C. P. Wolk. 2002. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 184:5524-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labarre, J., F. Chauvat, and P. Thuriaux. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 171:3449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaver, M., P. Dominguez-Cuevas, J. M. Coxhead, R. A. Daniel, and J. Errington. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849-853. [DOI] [PubMed] [Google Scholar]
- 22.Leganes, F., A. Blanco-Rivero, F. Fernandez-Pinas, M. Redondo, E. Fernandez-Valiente, Q. Fan, S. Lechno-Yossef, and C. P. Wolk. 2005. Wide variation in the cyanobacterial complement of presumptive penicillin-binding proteins. Arch. Microbiol. 184:234-248. [DOI] [PubMed] [Google Scholar]
- 23.Lutkenhaus, J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539-562. [DOI] [PubMed] [Google Scholar]
- 24.Maggi, S., O. Massidda, G. Luzi, D. Fadda, L. Paolozzi, and P. Ghelardini. 2008. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology 154:3042-3052. [DOI] [PubMed] [Google Scholar]
- 25.Maple, J., C. Aldridge, and S. G. Moller. 2005. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43:811-823. [DOI] [PubMed] [Google Scholar]
- 26.Mazouni, K., F. Domain, C. Cassier-Chauvat, and F. Chauvat. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52:1145-1158. [DOI] [PubMed] [Google Scholar]
- 27.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 186:8326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyagishima, S. Y., C. P. Wolk, and K. W. Osteryoung. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126-143. [DOI] [PubMed] [Google Scholar]
- 29.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 30.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 33.Rippka, R., J. Deruelles, J. Waterbury, M. Herdman, and R. Stanier. 1979. Generic assignements, strains histories and properties of pure culture of cyanobacteria. J. Genet. Microbiol. 111:1-61. [Google Scholar]
- 34.Sakr, S., R. Jeanjean, C.-C. Zhang, and T. Arcondeguy. 2006. Inhibition of cell division suppresses heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauvage, E., F. Kerff, M. Terrak, J. A. Ayala, and P. Charlier. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234-258. [DOI] [PubMed] [Google Scholar]
- 36.Scheffers, D.-J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69:585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzagoloff, H., and R. Novick. 1977. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 129:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 39.Westling-Häggström, B., T. Elmros, S. Normark, and B. Winblad. 1977. Growth pattern and cell division in Neisseria gonorrhoeae. J. Bacteriol. 129:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]