Abstract
Efflux pumps function to rid bacteria of xenobiotics, including antibiotics, bile salts, and organic solvents. TolC, which forms an outer membrane channel, is an essential component of several efflux pumps in Escherichia coli. We asked whether TolC has a role during growth in the absence of xenobiotics. Because tolC transcription is activated by three paralogous activators, MarA, SoxS, and Rob, we examined the regulation of these activators in tolC mutants. Using transcriptional fusions, we detected significant upregulation of marRAB and soxS transcription and Rob protein activity in tolC mutants. Three mechanisms could be distinguished: (i) activation of marRAB transcription was independent of marRAB, soxR, and rob functions; (ii) activation of soxS transcription required SoxR, a sensor of oxidants; and (iii) Rob protein was activated posttranscriptionally. This mechanism is similar to the mechanisms of upregulation of marRAB, soxS, and Rob by treatment with certain phenolics, superoxides, and bile salts, respectively. The transcription of other marA/soxS/rob regulon promoters, including tolC itself, was also elevated in tolC mutants. We propose that TolC is involved in the efflux of certain cellular metabolites, not only xenobiotics. As these metabolites accumulate during growth, they trigger the upregulation of MarA, SoxS, and Rob, which in turn upregulate tolC and help rid the bacteria of these metabolites, thereby restoring homeostasis.
Bacteria use efflux pumps to export a variety of xenobiotics (37). Some of these pumps have major clinical significance because they export multiple antibiotics (41). Recently, it has been noted that these pumps also export substances, such as bile salts and steroids, which occur in the environment of enteric bacteria (11; for a review, see reference 42).
An essential component of several efflux systems is TolC. TolC forms a multifunctional outer membrane channel (for a review, see reference 23) with roles in colicin uptake and secretion, bacteriophage adsorption, efflux of multiple antibiotics, detergents, dyes, and organic solvents, and export of hemolysin, heat-stable enterotoxin II (61), microcin J25 (9), and enterobactin (6). Export through the TolC channel requires interaction with two other proteins, an inner membrane transporter (e.g., AcrB) and a periplasmic membrane fusion protein (e.g., AcrA) that links the transporter to TolC. By means of this tripartite structure, xenobiotics or cellular products are pumped directly out of the cell from the cytosol or inner membrane. Basal levels of the AcrAB-TolC pump are important in providing the intrinsic resistance of Escherichia coli to many xenobiotics. Upregulation of the AcrAB-TolC pump engenders a multiple-antibiotic-resistance phenotype which is clinically significant. However, at least seven other sets of proteins in E. coli, such as AcrEF, EmrAB, and MacAB, form similar tripartite pumps with TolC, but they have different substrate specificities. The structures of TolC, AcrB, and AcrA have been solved, and a docking mechanism for AcrAB-TolC has been proposed (12, 32).
tolC and acrAB are members of the marA/soxS/rob regulon, which includes over 40 genes that promote resistance to multiple antibiotics, to numerous other xenobiotics, and to superoxides (3, 13, 24, 27, 38; for comprehensive reviews, see articles cited in reference 54). These genes are transcriptionally activated by three paralogous proteins, MarA, SoxS, and Rob, that bind a sequence upstream of the regulon promoter called the marbox.
Each of these transcriptional activators is regulated in a distinct manner. MarA and SoxS are transcriptionally regulated. The marRAB operon is repressed by MarR and autoactivated by MarA (the role of MarB is unknown). The operon can be derepressed by treating cells with salicylate and related phenolics which decrease the affinity of MarR for its binding sites (1, 28). However, a “mar-independent effect” of salicylate that increases the transcription of marRAB and of inaA, another member of the marA/soxS/rob regulon, has also been described (7, 49). The effect on marRAB transcription was found even in strains with combined deletions or null mutations of marRAB, soxRS, rob, and emrAB (29), indicating the existence of an additional mechanism for activating the regulon. soxS transcription is activated by SoxR after SoxR is activated by exposure to superoxides or nitric oxide (43). Rob is a very abundant and stable protein in E. coli (∼10,000 molecules per cell) but has very little activity in vivo (2, 19, 46). Its activity is increased posttranslationally by treatment with 2,2′-dipyridyl, 4,4′-dipyridyl, bile salts, or decanoate (45, 46). Thus, each activator is activated in response to different environmental signals.
Upregulation of these transcriptional activators engenders a low but significant level of multiple antibiotic, superoxide, and organic solvent resistance. The antibiotic resistance and solvent resistance are due primarily to the AcrAB-TolC pump (4, 13, 55). tolC has four known promoters, two of which (p3 and p4) are activated by MarA, SoxS, and Rob via a single, uniquely configured marbox (10, 25, 62). The acrAB promoter is also activated by MarA, SoxS, and Rob (24).
Here, we examined the effects of tolC on the regulation of MarA, SoxS, and Rob during growth in standard laboratory media. We found elevated levels of transcription of marA and soxS and elevated activity of the Rob protein in tolC efflux mutants. From these findings, we infer that the following homeostatic loop occurs in wild-type bacteria: (i) normal metabolism results in the generation of certain intracellular metabolites that trigger the upregulation of the transcriptional activators MarA, SoxS, and Rob; (ii) these activators, in turn, upregulate tolC, increasing the capacity for excretion of the metabolites via TolC; and (iii) the resulting reduction in the concentrations of the trigger metabolites (TMs) restores the basal levels of the activators. In tolC mutants, the metabolites are not as effectively excreted, and the activator levels remain elevated.
MATERIALS AND METHODS
Bacterial strains.
All strains used in this study are derivatives of E. coli K-12. Their construction and relevant genotypes are given in Table 1. Transduction was performed using bacteriophage P1 clr-100(Ts) as described previously (47). Donor phage for the tolC210::Tn10-48 mutation (35) (referred to below as tolC::Tn10) were obtained by thermal induction of a P1 lysogen of strain LBB735. Transcriptional fusions to lacZ were made in λRS45 as described previously (25, 51, 62) and were assayed as single-copy prophages.
TABLE 1.
E. coli strains, plasmids, and phages
| Strain, plasmid, or phage | Relevant genotype | Reference(s) or source |
|---|---|---|
| Strains | ||
| 1411 | lacI3 lacZ118 gyrAa | 24, 39 |
| AG100 | lac+ | 7 |
| AG100AX | AG100 acrAB::Tn903(kan) acrEF::spc | 36 |
| AG100W | AG100 acrAB::spc | H. Nikaido |
| BW1041 | GC4468 λJW1-soxS::lacZ Ampr | 59 |
| CE1 | emrAB::cat | 11 |
| CGSC5634 | tolCΔ(EW1b) | 57c |
| GC4468 | lacΔ4169 | B. Demple |
| LBB512 | thyA derivative of the tolC+ parent of CGSC5634 | J. Fralick |
| LBB735 | MG1655 | J. Fralick |
| LBB801 | MG1655 tolC210::Tn10-48 | 35c |
| M542 | GC4468 (λRS45-rob2::lacZ) | 46 |
| M2561 | N7918 inaA1::lacZ tolC210::Tn10b | This study |
| M2562 | M542 tolC210::Tn10 | This study |
| M2581 | N8453 λRS45-inaA::lacZ | This study |
| M2583 | M2581 tolC210::Tn10 | This study |
| M2605 | 1411 gyrA+inaA1::lacZa,b | This study |
| M2606 | SM1411 gyrA+inaA1::lacZa,b | This study |
| M2676 | N8452 soxR9::cat λJW1-soxS::lacZ | This study |
| M3953 | (mar sad)Δ1738 rob::kan soxR8::cat λRS45-marRAB::lacZ | This study |
| M3954 | rob::kan soxR8::cat λRS45-marRAB::lacZ | This study |
| M4014 | N8452 λJW1-soxS::lacZ Ampr | This study |
| M4110 | M2581/pTA108 | This study |
| M4111 | M2581/pTA:marA | This study |
| M4112 | M2581/pTA:soxS | This study |
| M4113 | M2581/pTA:rob | This study |
| M4114 | M2583/pTA108 | This study |
| M4115 | M2583/pTA:marA | This study |
| M4116 | M2583/pTA:soxS | This study |
| M4117 | M2583/pTA:rob | This study |
| M4141 | M4262 tolC210::Tn10 | This study |
| M4142 | M4263 tolC210::Tn10 | This study |
| M4143 | M4386 tolC210::Tn10 | This study |
| M4165 | M4275 tolC210::Tn10 | This study |
| M4167 | M2581 pyrE60∼Tn10 | This study |
| M4182 | M2676 tolC210::Tn10 | This study |
| M4183 | M4014 tolC210::Tn10 | This study |
| M4187 | M3953 tolC210::Tn10 | This study |
| M4188 | M3954 tolC210::Tn10 | This study |
| M4195 | N7918 emrAB::cat | This study |
| M4196 | N7918 acrEF::spc | This study |
| M4197 | M2561 acrEF::spc | This study |
| M4198 | CGSC5634 inaA1::lacZb | J. Fralick |
| M4199 | LBB512 inaA1::lacZb | J. Fralick |
| M4262 | GC4468 λRS45-tolC(C)::lacZ | This study |
| M4263 | GC4468 λRS45-tolC(B)::lacZ | This study |
| M4275 | GC4468 λRS45-acrAB::lacZ | This study |
| M4386 | GC4468 λRS45-tolC(A)::lacZ | This study |
| M4807 | AG100AX inaA1::lacZb | This study |
| M4820 | AG100W inaA1::lacZb | This study |
| M5572 | M2561/pTrc99A Ampr vector | This study |
| M5573 | M2561/pTrc99A::tolC+ (NcoI site) Ampr | This study |
| M5574 | M2561/pTrc99A:tolC(S257P) (NcoI site) Ampr | This study |
| M5575 | M2561/pTrc99A:tolC(A360T) (NcoI site) Ampr | This study |
| N7881 | AG100 inaA1::lacZb | This study |
| N7918 | GC4468 inaA1::lacZb | This study |
| N8444 | (mar sad)Δ1738 soxRS8::cat | 26 |
| N8452 | (mar sad)Δ1738 rob::kan | 26 |
| N8453 | N8444 rob::kan | This study |
| SM1411 | lacI3 lacZ118 gyrA acrAB::Tn903(kan)a | 24, 39 |
| Plasmids and phages | ||
| pTA108 | Low-copy-number cloning vector | 46 |
| pTA:marA | marA cloned in pTA108 | 46 |
| pTA:soxS | soxS cloned in pTA108 | 46 |
| pTA:rob | rob cloned in pTA108 | 46 |
| pTrc99A Ampr | Expression vector (Pharmacia) | R. Misra |
| pTrc99A::tolC+ (NcoI) | tolC+ cloned in pTrc99A NcoI site | 52 |
| pTrc99A::tolC(S257P) | tolC(S257P) cloned in pTrc99A NcoI site | 52 |
| pTrc99A:tolC(A360T) | tolC(A360T) cloned in pTrc99A NcoI site | 52 |
| λRS45 | Phage used to isolate promoter::lacZ fusions | 51 |
| P1 clr-100(Ts) | Transducing phage | 47 |
The gyrA mutation present in strains 1411 and SM1411 was replaced by the wild-type gyrA+ alleles in M2605 and M2606 during the P1-mediated transduction into these strains of inaA1::lacZ. gyrA and inaA are about 11 kb apart.
The inaA1::lacZ fusions have been described previously (56).
Via J. Fralick.
Culture media and chemicals.
LB (Lennox) media contained (per liter) 10 g Bacto tryptone (Difco, Detroit, MI), 5 g Bacto yeast extract, and 5 g NaCl, and the pH was adjusted to 7.5 with NaOH. M9 minimal medium (33) was supplemented with 0.2% glucose, 1 ng/ml thiamine, and, where indicated, 0.2% Vitamin Assay Casamino Acids (Difco, Detroit, MI). The antibiotics used for genetic selection in tolC+ and tolC strains were ampicillin (100 and 50 μg/ml, respectively), chloramphenicol (25 and 12.5 μg/ml, respectively), tetracycline (15 and 5 μg/ml, respectively), and kanamycin (30 μg/ml). MacConkey-lactose plates (Difco) contained 1% lactose. Since tolC efflux mutants do not grow on MacConkey medium (because it contains bile salts and crystal violet), all strains were routinely checked on this medium.
Growth of cells and β-galactosidase assays.
Bacteria were grown in two ways, unless otherwise indicated. (i) For assays of cells in early log phase to late stationary phase (quasi-growth curve), overnight cultures in LB broth were diluted 1,000-fold, and then nine serial threefold dilutions were made. After growth for 10 to 12 h at 32°C, the A600 of the cultures usually ranged from 0.02 to over 3.0. The cultures were placed on ice and diluted in Z-buffer, and β-galactosidase activity was assayed, as described previously (33). (ii) For assays of cells in early log phase, cells were grown overnight in LB medium at 32°C, diluted 1,000-fold in fresh medium, aerated, grown to an A600 of about 0.2, placed on ice, and diluted, and β-galactosidase activity was assayed as described above. For experiments with cells grown in M9 minimal medium, procedures similar to those described above were used, except that the initial dilution was only 100-fold and the cells were grown for longer times at 32°C. To test posttranscriptional activation of inaA::lacZ fusions by Rob, cells were grown to an A600 of about 0.1, diluted twofold into LB medium with 0 or 5 mM (final concentration) 2,2′-dipyridyl (Sigma Chemical, St. Louis, MO), and aerated for 1 h at 32°C, and β-galactosidase activity was assayed. Expression of tolC cloned in the NcoI site of pTrc99A plasmids was accomplished by addition of 0.4 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) to LB broth.
Each β-galactosidase assay was carried out in duplicate using the CHCl3-sodium dodecyl sulfate method (33), and all duplicate values were within ±5% of each other. Assays of cells in early log phase were performed at least twice in triplicate. The Kolmogorov-Smirnoff statistic was used to evaluate the probability (PKS) that the (quasi) growth curve data for paired tolC+ and tolC strains were from the same distribution, i.e., indistinguishable from each other (44).
RESULTS
Elevated transcription of marRAB in a tolC mutant.
We considered the possibility that MarA, SoxS, and/or Rob activities may be elevated in tolC mutants because increased transcription of a marA/soxS/rob regulon member, micF (8, 18, 48), had been found in tolC mutants (35). To determine whether marRAB transcription is elevated in tolC mutants, we measured the activity of the marRAB promoter using appropriate lacZ transcriptional fusions in the wild-type and tolC::Tn10 null mutant strains (35). To eliminate possible cross talk between MarA, SoxS, and Rob (30, 31, 50), strains which carry wild-type marRAB but have null mutations in soxS and rob were tested. These strains were diluted in LB broth and grown at 32°C to a range of densities (quasi-growth curve method), and β-galactosidase activity was assayed, as described in Materials and Methods.
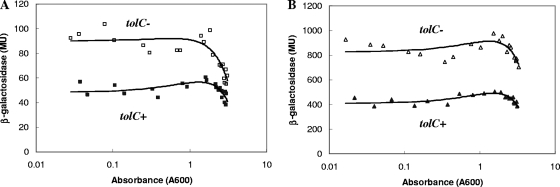
Transcription of marRAB::lacZ was elevated ∼2-fold in the tolC::Tn10 mutant (M4188) compared to the wild-type strain (M3954) in log-phase to early-stationary-phase cells (Fig. 1A). marRAB expression decreased in later stationary phase (A600, >1.6) for both the wild-type and mutant strains, but the decrease was more pronounced in the tolC::Tn10 mutant. Thus, the increased activity of micF observed previously (35) could be due, at least in part, to the increase in the MarA level resulting from the ∼2-fold increase in transcription of marRAB in tolC::Tn10 mutants.
FIG. 1.
β-Galactosidase activities of marRAB::lacZ transcriptional fusions in tolC+ and tolC::Tn10 cells. Cells grown in LB broth to different densities were assayed to determine β-galactosidase activities. (A) ▪, marRAB+ soxS8::cat rob::kan cells (M3954, tolC+); □, M4188 (tolC). The probability (PKS) that the two sets of data are from the same distribution, computed using the Kolmogorov-Smirnov test, was 1.5 × 10−5. (B) ▴, marRABΔ soxS8::cat rob::kan cells (M3953, tolC+); ▵, M4187 (tolC). PKS = 3.4 × 10−5. Note the different scales used. For this and other figures, MS Excel trend lines (second-order polynomials) were fitted to the data only for help with visualization. MU, Miller units.
Activation of marRAB transcription in the absence of marRAB, soxS, and rob.
marRAB transcription can be increased by four distinct mechanisms (54): (i) mutations which prevent MarR repressor synthesis; (ii) treatment with chemicals (e.g., salicylate) which interfere with MarR activity; (iii) transcriptional activation of the promoter by SoxS or Rob binding the marRAB marbox (cross talk); and (iv) a “mar-independent effect” of salicylate on marRAB transcription that has been shown to be independent of marRAB, soxS, rob, and emrAB (7, 29). To determine whether any of these mechanisms played a role in the upregulation of marRAB that was seen in the tolC::Tn10 mutant, we measured the marRAB::lacZ activities in isogenic tolC+ (M3953) and tolC::Tn10 mutant (M4187) strains, both of which have a marRAB deletion in addition to null mutations in soxS and rob (Fig. 1B). Deletion of marR derepressed the levels of marRAB transcription about eightfold, as expected. However, the ratio of the β-galactosidase activity of the tolC mutant to the β-galactosidase activity of the tolC+ strain was similar to ratio found for the marRAB+ strains and was maintained even in late stationary phase. This shows that the effect of the tolC mutation on marRAB transcription is substantially independent of the first three mechanisms described above since it is found even in cells that are defective in MarRAB, SoxS, and Rob. Therefore, the activation of marRAB transcription in tolC mutants resembles the “mar-independent effect” of cells treated with salicylate.
Activation of soxS in tolC::Tn10 mutants depends on soxR.
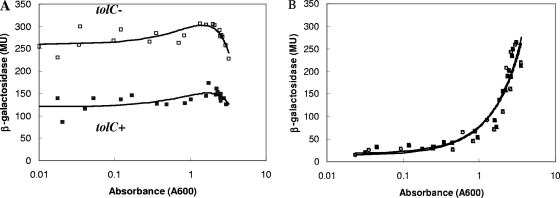
A soxS::lacZ transcriptional fusion was used to monitor soxS transcription (59). The soxS expression in strains with marRAB and rob null mutations also was ∼2-fold higher in the tolC mutant (M4183) than in the tolC+ strain (M4014) (Fig. 2A). In both strains, the β-galactosidase activities decreased somewhat in stationary-phase cells, but the ratio of tolC::Tn10 activity to tolC+ remained relatively constant. Thus, soxS transcription, like marRAB transcription, is also upregulated in tolC mutants.
FIG. 2.
β-Galactosidase activities of soxS::lacZ transcriptional fusions in tolC+ and tolC::Tn10 cells. (A) ▪, soxR+ marRABΔ rob::kan cells (M4014, tolC+); □, M4183 (tolC). PKS = 3.4 × 10−6. (B) ▴, soxR9::cat marRABΔ rob::kan cells (M2676, tolC+); ▵, M4182 (tolC). PKS = 1.0. MU, Miller units.
Superoxides and nitric oxide activate soxS transcription in a two-step manner; they convert SoxR into an active form, which then activates the transcription of soxS (43). In addition to the experiments whose results are shown in Fig. 2A, we tested whether transcriptional activation of soxS in tolC::Tn10 mutants required functional SoxR. Strains M2676 and M4182 carry a wild-type soxS gene but have a soxR9::cat null mutation (59) in addition to marRAB and rob null mutations and the soxS::lacZ fusion. These strains were diluted and grown in LB broth at 32°C, and their β-galactosidase activities were measured (Fig. 2B). As expected, the soxR9::cat tolC+ strain (M2676) had about one-fifth the soxS activity of soxR+ tolC+ strain M4014 in early log phase due to the absence of SoxR, the activator of soxS. Interestingly, the soxS::lacZ activity increased sharply in later growth phases, an effect not seen in the presence of wild-type SoxR. Nevertheless, strain M2676 and its tolC::Tn10 derivative (M4182) had essentially identical β-galactosidase activities regardless of the growth phase. Thus, soxR is necessary for the increased activation of soxS transcription seen in the tolC::Tn10 mutant. This suggests that the mechanism of activation of soxS in tolC mutants is similar to that which occurs when SoxR is activated by superoxides or nitric oxide.
Posttranscriptional activation of Rob in a tolC mutant.
In contrast to upregulation of marRAB and soxS transcription in tolC mutants, the transcription of rob (20) in the tolC::Tn10 mutant (M2562) was similar to that in the wild-type strain (M542) (data not shown). However, since the Rob protein can be activated posttranslationally by treatment with various compounds, including 2,2′-dipyridyl and bile salts (45, 46), we tested the effect of the tolC::Tn10 mutation on the activity of Rob using a strategy previously described (46). Strains with null mutations in lacI, marA, soxS, and rob were transformed with a low-copy-number plasmid that carries one of these genes under control of the lacZYA promoter in the absence of LacI repressor. We monitored an inaA::lacZ transcriptional fusion present on λRS74 since the inaA promoter is a member of the marA/soxS/rob regulon (49) and is activated by MarA, SoxS, and Rob. However, ectopic expression of the regulators eliminates their transcriptional activation by stress signals. The moderate overexpression of the activators on the plasmids increased the expression of inaA::lacZ 7- to 14-fold in the tolC+ cells compared to the vector control (Table 2). When inaA transcription was assayed in the tolC::Tn10 mutant, a fivefold increase was seen when the rob plasmid was present (M4117 compared to M4113) but not when the marA (M4115) or soxS (M4116) plasmids were present. This effect of the tolC::Tn10 mutation on Rob is posttranscriptional because (i) rob expression from the plasmid is under control of the plasmid-borne heterologous lacZYA promoter and (ii) the tolC::Tn10 mutation does not affect the transcription of the lacZYA promoter (Table 3). Interestingly, a 1.8-fold increase was seen for the tolC::Tn10 vector strain (M4114) compared to the strain with the vector (M4110). Thus, most of the effect is mediated by Rob, but a portion is mediated by the “mar-independent” activity previously noted for inaA (49).
TABLE 2.
Activation of inaA::lacZ in tolC mutantsa
| Strain
|
Activator controlled by lac promoter | No treatment with 2,2′-dipyridyl
|
Treatment with 5 mM 2,2′-dipyridyl
|
|||||
|---|---|---|---|---|---|---|---|---|
| β-Galactosidase activity (MU)
|
Ratiob | β-Galactosidase activity (MU)
|
Ratiob | |||||
| tolC+ | tolC | tolC+ strain | tolC strain | tolC+ strain | tolC strain | |||
| M4110 | M4114 | None | 6.1 (0.42)c | 11 (1.6) | 1.8 | 12 (0.6) | 16 (1.8) | 1.3 |
| M4111 | M4115 | MarA | 85 (15) | 84 (17) | 1.0 | 63 (17) | 53 (7.4) | 0.84 |
| M4112 | M4116 | SoxS | 80 (24) | 81 (20) | 1.0 | 72 (18) | 61 (9.2) | 0.85 |
| M4113 | M4117 | Rob | 42 (10) | 209 (58) | 5.0 | 675 (139) | 919 (154) | 1.4 |
Strains were grown in LB broth at 32°C to an A600 of 0.1, diluted twofold into LB broth with or without 2,2′-dipyridyl, and aerated for 1 h at 32°C. Cells were placed on ice, and an assay was performed to determine the β-galactosidase activity, which was expressed in Miller units (MU). All strains have chromosomal lacIZYA, marRAB, soxS, and rob null mutations.
Ratio of the activity in the tolC mutant to the activity in the tolC+ strain.
The standard errors of the means are indicated in parentheses.
TABLE 3.
Activities of promoter::lacZ transcriptional fusions in tolC mutantsa
| Promoter::lacZ fusion | Wild type
|
Mutant
|
Mutant/wild-type ratio | |||
|---|---|---|---|---|---|---|
| Strain | β-Galactosidase activity (MU) | Strain | Relevant mutation | β-Galactosidase activity (MU) | ||
| tolC(A)b | M4386 | 331 (16)c | M4143 | tolC::Tn10 | 562 (30) | 1.7 |
| tolC(B)b | M4263 | 133 (6.5) | M4142 | tolC::Tn10 | 250 (2.5) | 1.9 |
| tolC(C)b | M4262 | 76 (5.6) | M4141 | tolC::Tn10 | 74 (3.9) | 1.0 |
| acrAB | M4275 | 50 (2.2) | M4165 | tolC::Tn10 | 128 (3.2) | 2.5 |
| inaA | M2581d | 5.2 (0.17) | M4167d | pyrE∼Tn10 | 6.3 (0.15) | 1.2 |
| M2581d | 5.2 (0.17) | M2583d | tolC::Tn10 | 9.3 (0.03) | 1.8 | |
| inaA1 | M4199 | 43 (1.8) | M4198 | tolCΔ | 77 (0.6) | 1.8 |
| lacZYA | LBB735 | 9.4 (0.14) | LBB801 | tolC::Tn10 | 8.3 (0.89) | 0.88 |
Cells were grown in LB broth at 32°C to an A600 of 0.2 and placed on ice, and an assay was performed to determine the β-galactosidase activity, which was expressed in Miller units (MU).
Fusions A and B contain the tolC marbox; fusion C has a 12-bp deletion of the marbox. See reference 62 for details.
The standard errors of the means are indicated in parentheses.
The strain has a deletion or null mutation of marRAB, soxS, and rob.
We also examined the effect of 2,2′-dipyridyl in conjunction with the tolC::Tn10 mutation to see if the effects were additive or multiplicative (Table 2). 2,2′-Dipyridyl had a dramatic effect only on the strains carrying the pTA:rob plasmid, as previously observed (46). As expected, 5 mM 2,2′-dipyridyl increased inaA expression 16-fold (675/42) in M4113, the tolC+ strain with the pTA:rob plasmid. However, 2,2′-dipyridyl treatment of the tolC::Tn10 mutant (M4117) with the pTA:rob plasmid increased inaA expression only about 4-fold (919/209). Assuming that the cellular concentration of 2,2′-dipyridyl is not greater in the wild-type strain than in the tolC mutant, the separate effects of 2,2′-dipyridyl treatment and of the tolC mutation appear to be additive. This is consistent with the possibility that the tolC mutation leads to activation of Rob in a manner similar to the posttranscriptional activation of Rob by 2,2′-dipyridyl.
Elevated expression of marA/soxS/rob regulon promoters in a tolC mutant.
To determine whether other marA/soxS/rob regulon promoters are upregulated in a tolC mutant, we measured the β-galactosidase activities of wild-type and tolC::Tn10 strains carrying various regulon promoter::lacZ transcriptional fusions. tolC itself has four characterized promoters, p1 to p4, but only p3 and p4 are responsive to MarA, SoxS, and Rob (62). tolC::lacZ promoter fusion A contains all four promoters and showed 1.7-fold-greater β-galactosidase activity in tolC::Tn10 strain M4143 than in parental strain M4386 (Table 3). Similarly, tolC::lacZ promoter fusion B, which contains only promoters p3 and p4, had 1.9-fold-greater activity in tolC::Tn10 strain M4142 than in parental strain M4263. However, no increase was found for fusion C (M4141), which has a partially deleted marbox upstream of the p3 and p4 promoters and is not responsive to MarA, SoxS, and Rob (62). Thus, the effect of the tolC mutation on promoter transcription is highly specific; the transcription of tolC itself is activated via p3 and p4 but not when its marbox is defective.
Because acrAB encodes two components of the AcrAB-TolC pump and is also a member of the marA/soxS/rob regulon, we tested the effect of a tolC mutation on acrAB transcription. The promoter fusion acrAB::lacZ had 2.5-fold-higher β-galactosidase activity in the tolC::Tn10 mutant M4165 than in wild-type strain M4275 (Table 3). We also tested the marA/soxS/rob regulon gene inaA, whose function is not known. The inaA::lacZ promoter fusion (49) had 1.8-fold-higher β-galactosidase activity in tolC::Tn10 strain M2583 than in tolC+ parental strain M2581. Thus, in the absence of TolC functions, the transcription of several regulon promoters (including tolC itself) is upregulated, but not when the marbox is defective.
We also tested whether the activating effect of tolC::Tn10 is specific to regulon promoters by examining the β-galactosidase activity of a chromosomal wild-type lacZYA promoter. We found no significant difference in activity for lacZYA between a wild-type strain (LBB735) and its tolC::Tn10 derivative LBB801 (Table 3). Thus, the effect of tolC::Tn10 is specific for regulon promoters.
The Tn10 transposon is not responsible for the activation of inaA.
We tested the possibility that the Tn10 transposon used here to disrupt tolC was responsible for the activator upregulation seen in our experiments since it carries the tetD gene, which encodes a paralog of MarA, SoxS, and Rob. Since tetD is repressed by TetC, which is also encoded by Tn10 (40), significant amounts of TetD should not be made in strains with Tn10 insertions. However, Griffith and coworkers (16) have shown that when tetD is cloned and overexpressed on a plasmid in a strain (N8453) in which marRAB, soxS, and rob are deleted, it can activate some marA/soxS/rob regulon promoters. Therefore, we tested whether a Tn10 insertion linked to (but not disrupting) pyrE could activate inaA::lacZ in a derivative of strain N8453 (Table 3). We found no significant difference in inaA::lacZ activity between the strain carrying the pyrE-linked Tn10 insertion (M4167) and its parent without the insertion (M2581). A similar result has been obtained for an ara::Tn10 insertion (K. L. Griffith and R. E. Wolf, Jr., personal communication). Furthermore, the expression of inaA1::lacZ in a strain (M4198) which has a deletion of the 5′ end of tolC (tolCΔEW1b) (57) was 1.8-fold greater than that in the tolC+ parental strain (M4199). Thus, it is the disruption of tolC, not the presence of Tn10, that is responsible for the transcriptional activation of inaA and, presumably, the other regulon promoters.
Wild-type and receptor-defective tolC mutants complement tolC::Tn10 with regard to inaA upregulation.
TolC has multiple functions. As an outer membrane protein, it serves as a receptor for the adsorption of phage TLS and for internalization of colicin E1. TolC also serves as a pore for export of xenobiotics, proteins, and enterobactin (23). Since the tolC::Tn10 mutation results in complete loss of both outer membrane activities and export activities, we wished to narrow the possibilities of which function is responsible for the upregulation described here. Accordingly, we examined tolC(S275P) and tolC(A360T) mutants (52), which are defective as receptors for phage TLS and colicin E1 and have a defect in export of hemolysin but behave normally with regard to the efflux of xenobiotics.
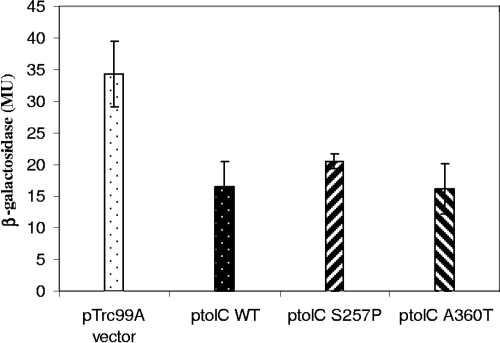
Strain M2561 has a tolC::Tn10 mutation and an inaA1::lacZ reporter gene and expresses about twice as much β-galactosidase as its wild-type parental strain, strain N7918 (Table 4). We transformed strain M2561 with the pTrc99A vector (M5572) or with the vector carrying wild-type (M5573) or mutant tolC under control of the trc promoter, whose expression was derepressed by the presence of 0.4 mM IPTG throughout growth. The presence of the plasmid-borne tolC+ gene in M5573 reduced the activity of inaA ∼2-fold (i.e., to normal tolC+ levels), whereas the vector alone in strain M5572 did not reduce the activity (Fig. 3). This clearly shows that defective tolC is responsible for the upregulation of inaA1::lacZ. The result was similar when the tolC plasmid carried the S257P or A360T mutation (M5574 and M5575, respectively). Thus, these strains behaved like wild-type tolC strains with regard to the inaA activity even though they are defective in TLS, ColE1, and certain hemolysin functions. This is consistent with the hypothesis that the efflux function of TolC is the critical function that regulates the inaA promoter and, by implication, marA, soxS, and Rob.
TABLE 4.
acrAB, acrEF, and emrAB mutations did not increase inaA1::lacZ activity
| Strain | Relevant mutations | β-Galactosidase activity (MU) | Ratioa |
|---|---|---|---|
| N7918 | None | 16 (0.2)b | 1.0 |
| M2561 | tolC::Tn10 | 34 (1.3) | 2.1 |
| M4195 | emrAB::cat | 17 (0.3) | 1.1 |
| M4197 | tolC::Tn10, emrAB | 34 (0.2) | 2.1 |
| M4196 | acrEF::spc | 16 (1.2) | 1.0 |
| M2605 | None | 21 (3.1) | 1.0 |
| M2606 | acrAB::Tn903 | 25 (2.2) | 1.2 |
| N7881c | None | 28 (2.4) | 1.0 |
| M4807c | acrAB::Tn903 acrEF::spc | 28 (5.2) | 1.0 |
| M4820c | acrAB::spc | 33 (2.4) | 1.2 |
Ratio of the β-galactosidase activity (expressed in Miller units [MU]) in the mutant to the β-galactosidase activity in the corresponding wild-type strain.
The standard errors of the means are indicated in parentheses.
The strain was grown in LB broth with 0.2% glucose to repress expression of the chromosomal lacZ.
FIG. 3.
β-Galactosidase activities of inaA1::lacZ transcriptional fusions in cells with a chromosomal tolC::Tn10 mutation and plasmids with different tolC alleles. Cells grown to early log phase in LB broth in the presence of 0.4 mM IPTG were assayed to determine the β-galactosidase activity (pTrc99A vector, M5572; ptolC WT, M5573; ptolC S257P, M5574; ptolC A360T, M5575). The error bars indicate the standard errors of the means. MU, Miller units.
Unlike mutations in tolC, single null mutations in either acrAB, acrEF, or emrAB do not affect regulon expression.
TolC interacts with at least eight different pairs of cytoplasmic efflux pumps and membrane fusion proteins to form tripartite transporters with different specificities (23). Prominent among these transporters are the multidrug efflux complexes formed in partnership with AcrA and AcrB, with AcrE and AcrF, and with EmrA and EmrB. If the effects of the tolC mutation on marA, soxS, and Rob described here occurred because the tolC mutation prevented one of these pumps from functioning, a similar effect on marRAB, soxS, and Rob should have been seen when the pump or membrane fusion protein alone was defective even though the TolC protein was the wild-type protein. To identify the putative TolC partners, we constructed strains with single or double null mutations in acrAB, acrEF, and emrAB and used inaA1::lacZ fusions to monitor the effects. However, no upregulation of inaA transcription attributable to the individual pumps was seen (Table 4). The finding that acrAB is not involved in the upregulation is particularly surprising since acrAB is also activated by MarA, SoxS, and Rob and is the major xenobiotic efflux pump in E. coli. In similar experiments, we have examined inaA1::lacZ expression in strains with single null (kan) mutations in tolC, acrA, acrB, acrE, acrF, emrA, and emrB derived from the KEIO Collection (5). With the exception of the tolC strain, which showed three- to fourfold-greater activity than the controls, none of the mutants showed significantly elevated activity (data not shown). Preliminary tests of 32 other mutants from the KEIO Collection that are thought to encode efflux functions have not revealed elevated inaA1::lacZ activities. Thus, we have not identified the relevant efflux pump that partners with TolC. Alternatively, there may be several TolC-associated pumps that must all be made defective before their roles in upregulation of MarA, SoxS, and Rob can be observed.
marRAB and soxS transcriptional effects and Rob posttranscriptional effects in minimal medium.
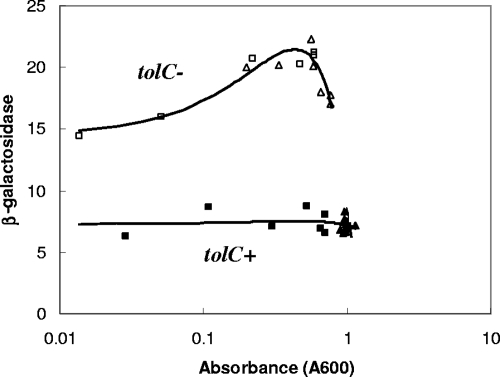
Since LB broth is a rich but poorly defined medium, it may contain trace amounts of xenobiotics that could accumulate inside a tolC mutant and upregulate marRAB, soxS, or Rob. Therefore, we examined the growth of wild-type and tolC::Tn10 strains in minimal M9 salts medium containing glucose, thiamine, and Casamino Acids (Table 5). The tolC::Tn10 mutants grew more slowly than the wild-type parents in this minimal medium (data not shown). marRAB transcription and soxS transcription were increased about twofold in the the tolC mutants, as observed for cells grown in LB medium (Fig. 1 and 2). However, Rob activity was increased only twofold, which was significantly less than the fivefold observed in LB medium. We also tested inaA::lacZ expression in wild-type and tolC::Tn10 cells grown in M9 minimal medium supplemented only with glucose and thiamine (Fig. 4). Clearly, Casamino Acids are not required for the upregulation seen in tolC::Tn10 mutants. Thus, it is unlikely that xenobiotics in the culture medium are responsible for the upregulation of marRAB, soxS, and Rob in the tolC mutants.
TABLE 5.
Effects of tolC::Tn10 on promoter transcription in minimal mediuma
| Strain
|
lacZ fusion | β-Galactosidase activity (MU)
|
Ratiob | ||
|---|---|---|---|---|---|
| tolC+ | tolC | tolC+ strain | tolC strain | ||
| M3954 | M4188 | marRAB | 30 (2.8)c | 57 (12.7) | 1.9 |
| M4014 | M4183 | soxS | 133 (12.3) | 358 (54) | 2.7 |
| M4113 | M4117 | inaA::lacZd | 38 (6.7) | 85 (9.2) | 2.2 |
The strains were grown at 32°C to an A600 of 0.1 to 0.25 in M9 medium supplemented with 0.2% glucose, 1 ng/ml thiamine, and 0.2% Casamino Acids, and an assay was performed to determine the β-galactosidase activity, which was expressed in Miller units (MU).
Ratio of the activity in the tolC mutant to the activity in the wild-type strain.
The standard errors of the means are indicated in parentheses.
As in Table 2, the expression of inaA::lacZ was used to measure the posttranscriptional activation of Rob.
FIG. 4.
Effect of tolC::Tn10 on β-galactosidase activities of inaA1::lacZ transcriptional fusions in cells grown to various densities in M9 minimal medium supplemented with 0.2% glucose and 1 ng/ml thiamine. The results of two experiments (triangles and squares) are combined (filled symbols, N7918 [tolC+]; open symbols, M2561 [tolC::Tn10]). PKS = 2.2 × 10−6.
DISCUSSION
TM hypothesis.
TolC constitutes an outer membrane channel that functions in the export of multiple xenobiotics, enterobactin, peptides, and proteins and in the binding and uptake of colicins and binding of bacteriophage in E. coli and other gram-negative bacteria (23). Misra and Reeves (35) observed that in tolC mutants micF transcription was elevated. We now offer the following explanation for why micF expression is elevated in tolC mutants: the levels of MarA, SoxS, and transcriptionally active Rob are increased in tolC mutants and these proteins transcriptionally activate micF, a member of the marA/soxS/rob regulon. Indeed, other members of the regulon, inaA, acrAB, and tolC itself are also upregulated (Table 3). Furthermore, a tolC promoter (C) lacking a portion of the marbox and the chromosomal lacZYA promoter (not a regulon member) are not upregulated, showing that the upregulation that we have found in tolC mutants is specific for the marA/soxS/rob regulon.
The most likely explanation for the increased activities of the marRAB and soxS promoters and of the Rob protein in tolC mutants is that some products of normal cellular metabolism are not exported as rapidly from tolC mutants as from wild-type cells and therefore accumulate. We propose that these metabolites interact, directly or indirectly, with the marRAB promoter, with SoxR, and with the Rob protein. It seems unlikely that a substance present in the medium is responsible since marA, soxS, and Rob are upregulated even in tolC::Tn10 cells grown in a chemically defined mineral salts medium with only glucose and thiamine added (Fig. 4). Furthermore, the critical TolC function with respect to inaA upregulation is probably efflux and not outer membrane disruption. Ectopic expression of plasmid-borne tolC, including mutations that affect outer membrane properties such as colicin internalization and phage adsorption [tolC(S257P) and tolC(A360T)], prevented (complemented) the upregulation of MarA, SoxS, or Rob seen in tolC mutants (Fig. 3).
For discussion, we refer to these accumulated substances that stimulate the upregulation as TMs. What is their nature? It is intriguing that the TMs have three distinct modes of action. (i) Like salicylate, TMs transcriptionally activate the marRAB promoter even in the absence of MarRAB, SoxS, and Rob functions (Fig. 1). Unlike salicylate, there was no evidence that the TMs interact with MarR. (ii) Like superoxides and other triggers of soxS transcription, transcriptional activation of soxS by TMs is mediated by SoxR (Fig. 2). (iii) Like bile salts, decanoate, and 2,2′- and 4,4′-dipyridyl, TMs posttranscriptionally activate the Rob protein, suggesting that there is a direct interaction with Rob (Table 2).
Does one metabolite do all this? An overlap between compounds that can activate marRAB (by derepression) and soxS (via SoxR) has previously been noted (34), and we have observed that high millimolar concentrations of salicylate can activate soxS via SoxR (unpublished data). There is also an overlap between compounds that activate soxS transcription and Rob protein (15; our unpublished data). Thus, it is possible that a single compound or class of compounds activates marRAB, soxS, and Rob, but no such compound has been described yet. Therefore, there could be a number of trigger metabolites. A comparison of the metabolomes of tolC mutant and wild-type cells should help identify the TMs.
What are the TM pumps?
We assume that TolC exports the TMs in conjunction with other components. Generally, TolC seems to interact with an inner membrane-located pump and a membrane fusion protein to form a tripartite complex which extrudes xenobiotics and other molecules into the medium. Among the eight known efflux pump systems that depend upon TolC for function are AcrAB, AcrEF, and EmrAB. However, strains with sets of genes encoding these pumps individually deleted did not show higher activation of inaA1::lacZ transcription, indicating that significant amounts of TMs were not present in these mutants. This suggests either that some other pump interacts with TolC for efflux of the TMs or that a combination of several pumps is involved in their efflux.
It seems reasonable to suppose that high levels of the TMs are toxic, yet tolC mutants grow at rates comparable to those of wild-type strains when they are cultivated in rich media. Thus, there could be a TolC-independent system that exports or detoxifies the TMs. The ability of such a TolC-independent pump or detoxifier to rid the cell of TMs would be expected to be suboptimal; otherwise, we would not have detected the effect of TMs in tolC mutants. Since strains which have null mutations in tolC, marRAB, soxRS, and rob grow very well in rich media, this putative TolC-independent activity might not be regulated by MarA, SoxS, or Rob. It may be possible to identify this activity by isolating chromosomal fragments with activity that is sufficiently elevated that the upregulation observed in tolC mutants is negated.
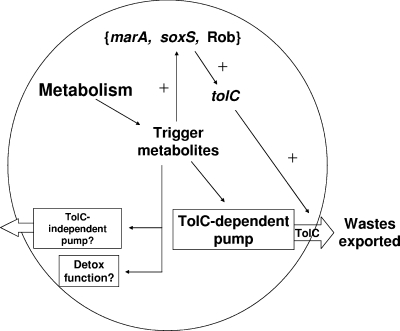
The present results suggest that the MarA, SoxS, and Rob systems are tuned to detect cellular metabolites, not only xenobiotics like salicylate and bile salts. The buildup of these metabolites may then signal the need to excrete them or detoxify them (Fig. 5). In this way, TolC and TMs may regulate each other in wild-type cells; excess TMs would activate marA, soxS, and Rob, which would then increase tolC expression and increase the export of TMs. Other pumps, now known to export xenobiotics, may export other cellular metabolites. If so, it may be that the efflux of xenobiotics evolved from pumps that originally were dedicated to the export of cellular metabolites.
FIG. 5.
Hypothetical components of a waste disposal system in E. coli (for simplicity, only the outer membrane is indicated). Metabolism generates TMs in the cell that are disposed of via an unspecified TolC-dependent pump. If the TolC pump is defective for efflux, TMs accumulate and trigger the activation (+) of marRAB, soxS, and Rob, which then activate the marA-soxS-rob regulon promoters, including the tolC promoters p3 and p4. Since tolC mutants are viable, a suboptimal TolC-independent pump or other detoxification function may also be present.
Export of metabolites.
Helling and coworkers (17) have come to similar conclusions. These authors found that 10% of transposon-generated mutants selected for resistance to low levels of nalidixic acid had mutations in amino acid or adenine biosynthetic genes (icdA, metE, icdA, purB). The increased resistance was accompanied by an increase in acrAB transcription and was dependent on wild-type alleles of soxS plus either marA or rob. Helling and coworkers proposed that the mutational blockage of certain biosynthetic pathways results in accumulation of particular intermediates, which then activate the SoxS and MarA or Rob systems. In effect, this is a “toxic waste disposal” system (17). Our conclusions differ from the conclusions of Helling et al. in one respect. Helling et al. concluded that acrAB and tolC are required to expel the accumulated metabolic intermediate. Since we found that acrAB null mutations did not elevate the expression of an inaA1::lacZ fusion, we concluded that acrAB is not essential for efflux of TMs. This discrepancy is readily accounted for by the fact that Helling and coworkers (17) used a nalidixic acid resistance assay as their end point assay. In the acrAB mutants, as in tolC mutants, nalidixic acid resistance is so compromised that it cannot be used to monitor the presence of a separate pump.
It has been commonly assumed that when metabolites are overproduced by bacteria and other organisms, they simply leak out of the cells. The “feeding” to neighboring cells of biosynthetic intermediates accumulated in certain mutants is often observed, but how the intermediates get out of the cell has not been explored. Kawamura-Sato et al. (21) have shown that AcrEF is important for indole excretion. Franke et al. (14) have shown that YfiK plays a significant role in the excretion of cysteine-cystine, and Yamada et al. (60) have shown that a number of other pumps, including some pumps known to be active in multidrug efflux, also are involved in cystine excretion. TolC, but not AcrAB, has been shown to also have a role in this process (58). Importantly, the aaeAB genes encode an efflux system which exports p-hydroxybenzoate (pHBA), an intermediate in ubiquinone synthesis, and which protects the cell from exogenous pHBA and a few related compounds (53). The system is regulated by AaeR and is inducible by pHBA and salicylate. Van Dyk and coworkers suggested that AaeAB might provide a “metabolic relief valve” for excess pHBA. This is very similar to our conclusions. However, null mutations in aaeA and aaeB did not upregulate inaA1::lacZ fusions (data not shown).
Downstream of tolC are three genes, ygiA, ygiB and ygiC, which may be part of the tolC operon (22). If the tolC mutations which we used in this study are polar on the downstream ygiABC genes, it may be that the latter genes are responsible for the regulatory effects that we have described. We tested this by asking whether a plasmid carrying the ygiABC genes (kindly provided by L. Thomason and D. Court) can complement the tolC::Tn10 mutant with regard to inaA1::lacZ activation. No complementation was observed, indicating that the ygiABC genes are not involved in the upregulation of inaA (data not shown). Furthermore, we have seen that cloned wild-type tolC alone complemented the upregulation due to a tolC::Tn10 chromosomal mutation (Fig. 3). Thus, it is unlikely that the ygiABC genes play a role in the tolC effect on activator regulation.
Acknowledgments
We thank the National BioResource Project (NIG, Japan) for providing 39 mutants from the KEIO Collection; J. M. Bostock, M. Cashel, D. Court, B. Demple, C. A. Elkins, J. A. Fralick, R. B. Helling, R. Misra, H. Nikaido, L. Thomason, and B. Weiss for providing various strains and plasmids; I. Botos, G. Hummer, and M. E. Wall for invaluable help with formulating the statistics; and S. Busby, R. Misra, and R. E. Wolf, Jr. for discussions.
This research was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Alekshun, M., and S. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali Azam, T., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleuel, C., C. Grosse, N. Taudte, J. Scherer, D. Wesenberg, G. J. Krauss, D. H. Nies, and G. Grass. 2005. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187:6701-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado, M. A., P. A. Vincent, R. N. Farias, and R. A. Salomon. 2005. YojI of Escherichia coli functions as a microcin J25 efflux pump. J. Bacteriol. 187:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 11.Elkins, C. A., and L. B. Mullis. 2006. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J. Bacteriol. 188:1191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswaran, J., E. Koronakis, M. K. Higgins, C. Hughes, and V. Koronakis. 2004. Three's company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 14:741-747. [DOI] [PubMed] [Google Scholar]
- 13.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke, I., A. Resch, T. Dassler, T. Maier, and A. Bock. 2003. YfiK from Escherichia coli promotes export of O-acetylserine and cysteine. J. Bacteriol. 185:1161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuentes, A. M., J. J. Diaz-Mejia, R. Maldonado-Rodriguez, and C. F. Amabile-Cuevas. 2001. Differential activities of the SoxR protein of Escherichia coli: SoxS is not required for gene activation under iron deprivation. FEMS Microbiol. Lett. 201:271-275. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, K. L., S. M. Becker, and R. E. Wolf, Jr. 2005. Characterization of TetD as a transcriptional activator of a subset of genes of the Escherichia coli SoxS/MarA/Rob regulon. Mol. Microbiol. 56:1103-1117. [DOI] [PubMed] [Google Scholar]
- 17.Helling, R. B., B. K. Janes, H. Kimball, T. Tran, M. Bundesmann, P. Check, D. Phelan, and C. Miller. 2002. Toxic waste disposal in Escherichia coli. J. Bacteriol. 184:3699-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jair, K. W., R. G. Martin, J. L. Rosner, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 177:7100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jair, K. W., X. Yu, K. Skarstad, B. Thöny, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakeda, M., C. Ueguchi, H. Yamada, and T. Mizuno. 1995. An Escherichia coli curved DNA-binding protein whose expression is affected by the stationary phase-specific sigma factor sigma S. Mol. Gen. Genet. 248:629-634. [DOI] [PubMed] [Google Scholar]
- 21.Kawamura-Sato, K., K. Shibayama, T. Horii, Y. Iimuma, Y. Arakawa, and M. Ohta. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179:345-352. [DOI] [PubMed] [Google Scholar]
- 22.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467-489. [DOI] [PubMed] [Google Scholar]
- 24.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 25.Martin, R., W. Gillette, S. Rhee, and J. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 26.Martin, R., W. Gillette, and J. Rosner. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623-634. [DOI] [PubMed] [Google Scholar]
- 27.Martin, R., and J. Rosner. 2003. Analysis of microarray data for the marA, soxS, and rob regulons of Escherichia coli. Methods Enzymol. 370:278-280. [DOI] [PubMed] [Google Scholar]
- 28.Martin, R., and J. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, R., and J. Rosner. 1997. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J. Bacteriol. 179:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, R. G., K. W. Jair, R. E. Wolf, Jr., and J. L. Rosner. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michán, C., M. Manchado, and C. Pueyo. 2002. SoxRS down-regulation of rob transcription. J. Bacteriol. 184:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 35.Misra, R., and P. Reeves. 1987. Role of micF in the tolC-mediated regulation of OmpF, a major outer membrane protein of Escherichia coli K-12. J. Bacteriol. 169:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 38.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill, A. J., J. M. Bostock, A. M. Moita, and I. Chopra. 2002. Antimicrobial activity and mechanisms of resistance to cephalosporin P1, an antibiotic related to fusidic acid. J. Antimicrob. Chemother. 50:839-848. [DOI] [PubMed] [Google Scholar]
- 40.Pepe, C. M., C. Suzuki, C. Laurie, and R. W. Simons. 1997. Regulation of the “tetCD” genes of transposon Tn10. J. Mol. Biol. 270:14-25. [DOI] [PubMed] [Google Scholar]
- 41.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 43.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 44.Press, W. H., S. A. Teulosky, W. T. Vetterling, and B. P. Flannery. 1992. Numerical recipes in Fortran 77—the art of scientific computing, 2nd ed. Cambridge University Press, Cambridge, England.
- 45.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 46.Rosner, J., B. Dangi, A. Gronenborn, and R. Martin. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosner, J. L. 1972. Formation, induction, and curing of bacteriophage P1 lysogens. Virology 48:679-689. [DOI] [PubMed] [Google Scholar]
- 48.Rosner, J. L., T. J. Chai, and J. Foulds. 1991. Regulation of ompF porin expression by salicylate in Escherichia coli. J. Bacteriol. 173:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosner, J. L., and J. L. Slonczewski. 1994. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J. Bacteriol. 176:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneiders, T., and S. Levy. 2006. MarA-mediated transcriptional repression of the rob promoter. J. Biol. Chem. 281:10049-10055. [DOI] [PubMed] [Google Scholar]
- 51.Simons, R., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 52.Vakharia, H., G. German, and R. Misra. 2001. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active alpha-hemolysin. J. Bacteriol. 183:6908-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Dyk, T. K., L. J. Templeton, K. A. Cantera, P. L. Sharpe, and F. S. Sariaslani. 2004. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, D. G., M. N. Alekshun, and P. F. McDermott. 2005. Frontiers in antimicrobial resistance—a tribute to Stewart B. Levy. ASM Press, Washington, DC.
- 55.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, S., F. E. Tuttle, D. Blankenhorn, D. C. Dosch, and J. L. Slonczewski. 1992. pH dependence and gene structure of inaA in Escherichia coli. J. Bacteriol. 174:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitney, E. N. 1971. The tolC locus in Escherichia coli K12. Genetics 67:39-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiriyathanawudhiwong, N., I. Ohtsu, Z. D. Li, H. Mori, and H. Takagi. 2009. The outer membrane TolC is involved in cysteine tolerance and overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 81:903-913. [DOI] [PubMed] [Google Scholar]
- 59.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada, S., N. Awano, K. Inubushi, E. Maeda, S. Nakamori, K. Nishino, A. Yamaguchi, and H. Takagi. 2006. Effect of drug transporter genes on cysteine export and overproduction in Escherichia coli. Appl. Environ. Microbiol. 72:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamanaka, H., H. Kobayashi, E. Takahashi, and K. Okamoto. 2008. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J. Bacteriol. 190:7693-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, A., J. Rosner, and R. Martin. 2008. Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol. Microbiol. 69:1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]