Abstract
The rhizobial DctA permease is essential for the development of effective nitrogen-fixing bacteroids, which was correlated with its requirement for growth on C4-dicarboxylates. A previously described dctA mutant of Rhizobium tropici CIAT899, strain GA1 (dctA), however, was unexpectedly still able to grow on succinate as a sole carbon source but less efficiently than CIAT899. Like other rhizobial dctA mutants, GA1 was unable to grow on fumarate or malate as a carbon source and induced the formation of ineffective nodules. We report an alternative succinate uptake system identified by Tn5 mutagenesis of strain GA1 that was required for the remaining ability to transport and utilize succinate. The alternative uptake system required a three-gene cluster that is highly characteristic of a dctABD locus. The predicted permease-encoding gene had high sequence similarity with open reading frames encoding putative 2-oxoglutarate permeases (KgtP) of Ralstonia solanacearum and Agrobacterium tumefaciens. This analysis was in agreement with the requirement for this gene for optimal growth on and induction by 2-oxoglutarate. The permease-encoding gene of the alternative system was also designated kgtP in R. tropici. The dctBD-like genes in this cluster were found to be required for kgtP expression and were designated kgtSR. Analysis of a kgtP::lacZ transcriptional fusion indicated that a kgtSR-dependent promoter of kgtP was specifically induced by 2-oxoglutarate. The expression of kgtPp was found in bacteroids of nodules formed with either CIAT899 or GA1 on roots of Phaseolus vulgaris. Results suggested that 2-oxoglutarate might be transported or conceivably exported in nodules induced by R. tropici on roots of P. vulgaris.
The rhizobia include a number of genera, the majority being members of the Alphaproteobacteria, that form host-specific nitrogen-fixing symbiotic associations with leguminous plants. The development of symbiosis requires specific recognition by root epidermal cells to coordinate nodule morphogenesis in adjacent root cortical cells. During invasion, the bacterium differentiates into a nitrogen-fixing bacteroid upon entry into cells of the newly formed nodules. Within the nodule-invaded cells, bacteroids are surrounded by a peribacteroidal membrane that regulates the exchange of nutrients between the two partners. The plant becomes dependent on the bacteroids for fixed nitrogen, whereas the bacteroids become dependent on the plant for reduced carbon (35). Host recognition and nutrient exchange involve an exchange of molecular signals acting as part of a network to regulate the development and maintenance of symbiosis (7, 20).
Although it has been shown that symbiotic nitrogen fixation is supported by sucrose transferred from leaves to the root nodules, carbohydrates are not directly consumed by bacteroids. It is widely acknowledged that the transport of sugars is driven by passive diffusion across the peribacteroid membrane, and the transport rates are too low to support the energetic requirements needed for nitrogen fixation (36, 38). A current model is that uninfected cells in nodules actively take up and metabolize sucrose to C4-dicarboxylates (DCAs), e.g., malate, which are then discharged to the apoplast or possibly transferred directly to infected cells (38). In agreement with this, rhizobial dctA mutants that are otherwise capable of effective nitrogen fixation form ineffective nodules.
A conserved three-gene cluster required for DCA transport, dct, is found in many bacteria and includes a gene encoding a DctA permease (secondary transporter, family DAACS:2.A.23) (28). The dctA gene is divergently transcribed from the other two genes that encode a two-component signal transduction system, i.e., DctBD. This two-component system specifically recognizes DCAs to activate transcription of dctA for the uptake of DCAs. The two-component dctBD system in Rhizobium is clearly functional in free-living bacteria but is dispensable for Rhizobium-legume symbiosis in that dctBD mutants form effective nodules that are indistinguishable from those of the wild type (39). Factors regulating the expression of dctA in the bacteroid have not yet been identified, although the general transcriptional regulator NifA was proposed to be a possible candidate that might serve to activate dctA in planta (15, 37). The regulation of biological nitrogen fixation by NifA was recently reviewed by Dixon and Kahn (8).
Rhizobium tropici CIAT899 maintains a conserved functional dctABD gene cluster, and a well-defined dctA mutant, strain GA1 (dctA::Gmr), was previously constructed and characterized (5). Surprisingly, unlike other rhizobial dctA mutants, strain GA1 was still able to grow to a limited extent on succinate as a sole carbon source. In addition, strain GA1 was unable to grow on either fumarate or malate as a sole carbon source. These results suggested that an alternative, possibly succinate-specific uptake system might be present in R. tropici (5). Subsequently, Tn5 mutagenesis of strain GA1 was used to identify a cluster of three open reading frames (ORFs) that were required for this strain to grow on succinate as a sole carbon source. Partial DNA sequence analysis of the mutant strains generated indicated that this cluster had remarkable similarity in sequence and organization with a dct cluster. However, the encoded permease of this three-gene cluster had the highest similarity with KgtP permeases, and 2-oxoglutarate was found to be the principal inducing substrate for the alternative system. A role for this kgt system in bacteroids could be suggested in that a permease promoter fusion was expressed in nodules induced either by CIAT899 or by GA1 on Phaseolus vulgaris plants.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in the study are listed in Table 1. R. tropici strains were grown in PY medium (19) or in minimal medium (MM) as described previously (5), with 20 mM NH4Cl as the nitrogen source. Carbon sources were added to a 20 mM final concentration. Escherichia coli strains were grown on LB medium (18). Antibiotics were added to the following final concentrations: nalidixic acid (Nal), 50 μg/ml; kanamycin (Km), 50 μg/ml; tetracycline (Tc), 10 μg/ml; ampicillin (Amp), 50 μg/ml; gentamicin (Gm), 10 μg/ml for R. tropici and 50 μg/ml for E. coli. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added at 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain, plasmid, phagemid, or cosmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| S17-1 | Smr Tpr derivative of E. coli 294 pro recA hsdR [RP4 (IncP):2 kan::Tn7 tet::Mu] in the chromosome | 31 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen Corp. |
| Rhizobium tropici | ||
| CIAT899 | Wild-type strain isolated from Phaseolus vulgaris L.; Nalr | 11 |
| GA1 | CIAT899 dctA::Gmr “cassette” | 5 |
| GA11 | GA1 kgtS::Tn5; Gmr Kmr | This work |
| GA12 | GA1 kgtR::Tn5; Gmr Kmr | This work |
| CIAT899.317 | CIAT899 kgtP::pHRP317; Kmr | This work |
| GA1.317 | GA1 kgtP::pHRP317; Gmr Kmr | This work |
| Plasmids, phagemids, and cosmids | ||
| pBluescript II SK | Phagemid and ColE1 replicon; Ampr | Stratagene |
| pSK-KG | Derived from pBluescript II SK with intergenic region kgtP-kgtS from R. tropici 1.2-kb, EcoRI-SalI site | This work |
| pCR 2.1-TOPO | pUC and f1 origin; Ampr Kmr | Invitrogen Corp. |
| pTH24 | 6-kb HindIII fragment with dctABD genes from S. meliloti cloned into pRK7813 | 39 |
| pRTD1 | Derived from pSUP205 with dct of R. tropici | 5 |
| pMP220 | IncP cosmid with lacZ as reporter gene; Tcr | 32 |
| pRU103 | Derived from pMP220 with intergenic region dctA-dctB from R. leguminosarum 0.9-kb EcoRI; dctA::lacZ; Tcr | 26 |
| pSUP202-1::Tn5 | Derived from pBR325 (ColE1) with oriT region; Kmr | 31 |
| pGA11-10 | Derived from pBluescript II SK with 10-kb fragment of EcoRI including Tn5 from GA11 | This work |
| pGA12-10 | Derived from pBluescript II SK with 10-kb fragment of EcoRI Including Tn5 from GA12 | This work |
| pHRP317 | pMB1 ori; Kmr Spcr Strr Kmr Smr; fQ Sm/Sp cassette from pHRP316 (MscI-EcoRI fragment) in HincII-EcoRI of pK19 | 21 |
| pSB10 | Derived from pCR 2.1 containing a PCR fragment from the internal region of the coding sequence of kgtP | This work |
| pSB11 | Derived from pHRP317 containing an internal fragment of the coding region of kgtP from pCR 2.1::KGT XhoI-SalI | This work |
| pSB12 | Derived from pMP220 with EcoRI-SalI fragment from pGA11-10 containing the presumptive ORF12 | This work |
| pSB13 | Derived from pMP220 with intergenic region kgtP-kgtS from R. tropici 1.5-kb KpnI-PstI fragment | This work |
Genetic and DNA analysis.
Plasmids were transferred to R. tropici strains by conjugation using E. coli S17-1 as the donor (31). Plasmids were also transferred into R. tropici strains by electroporation with a MicroPulser (Bio-Rad, CA) using the preprogrammed setting for Agrobacterium tumefaciens. Cloning techniques, transformation of E. coli, and gel electrophoresis were done according to protocols described previously by Ausubel et al. (3). Genomic DNA purification and isolation of plasmid DNA were done using the GenomicPrep blood DNA isolation kit/GenomicPrep cells and tissue DNA isolation kit and the FlexiPrep kit, respectively (Amersham Pharmacia Biotech Inc., Piscataway, NJ). Restriction endonucleases and T4 ligase were used according to the manufacturer's instructions (Promega Corporation, Madison, WI, and Invitrogen Corporation, Carlsbad, CA). DNA/DNA hybridizations were done as described in the NEBlot-Phototope kit (New England Biolabs, Beverly, MA). DNA fragments were isolated from agarose gels using the Sephaglas BandPrep kit (Amersham-Pharmacia, Piscataway, NJ).
DNA sequence analysis was done using an ABI Prism 377 automated DNA sequencer (CTAG Center, UdelaR, Montevideo, Uruguay). Nucleotide sequences were translated and searched using the nonredundant database and the Tblastx program (http://www.ncbi.nlm.nih.gov/BLAST/) (1). Primers were designed using the Primer Premier 5.0 program (Premier Biosoft, Palo Alto, CA). The DNA sequence of the primer specifying the ends of transposon Tn5 used to characterize GA11 and GA12 mutations, 5′-GGTTCCGTTCAGGACGGCTAC-3′, was obtained from Invitrogen (Carlsbad, CA). Other oligonucleotides for DNA sequence analysis and PCR amplification (KGF [5′-CACGGATCCTCGTGTGCAACGCCATCG-3′] and KGR [5′-GGAATTCAACAGCTGCGCCGTCT-3′]) were obtained from Integrated DNA Technologies (Coralville, IA).
The construction of the kgtP mutant strain was done by cointegrating a plasmid containing a segment that spans an internal region of the kgtP ORF. The internal gene fragment was amplified by PCR and cloned into pHRP317 (21), a narrow-host-range vector that does not replicate in rhizobial strains. PCR amplification was done using a 25-μl mixture including primers KGF and KGR by using a Perkin-Elmer GeneAmp PCR system 2400 with 5 min of denaturation at 94°C, followed by 5 cycles of 30 s at 94°C, 30 s at 53.8°C, and 30 s at 72°C and then 25 cycles of 30 s at 94°C, 30 s at 52.8°C, and 30 s at 72°C, followed by a final extension step of 4 min at 72°C.
Determination of succinate uptake activity.
Succinate transport activity was measured for cells grown on the indicated medium to mid-exponential phase, harvested, and suspended in MM without a carbon source at an optical density at 620 nm of 7.0. The succinate transport assay mixture containing [2,3-14C]succinic acid (159.5 μM; 8 μCi/μmol) and unlabeled succinate (3.22 mM) in 0.37 ml MM was equilibrated in a water bath at 30°C. The reaction was initiated by adding 0.25 ml of cell suspension to the mixture, and samples were taken at different times and analyzed as described previously by Batista et al. (4).
Since the presence of glucose in the growth medium was not considered to be ideal for measurements of succinate transport in dctA mutant strains, the assay was modified for GA1, GA11, and GA12 to include a preincubation period of 6 h for the specific induction of the system by adding succinate to MM. In these assays, cells were first centrifuged, washed with MM, and suspended in MM with 20 mM succinate for 6 h, a condition under which growth would not be expected for GA11 or GA12.
Enzyme activity determinations.
β-Galactosidase activity was determined according to a method described previously by Miller (18), with modifications described by Poole et al. (22).
Plant assays.
P. vulgaris seeds were surface sterilized and germinated (33). Following germination, seedlings were transferred to sterile plant growth pouches, and root tips were inoculated with R. tropici cells suspended in inoculation buffer (50 mM phosphate buffer [pH 7.0]) at a concentration of 104 to 106 cells ml−1. Plants were grown in a growth chamber maintained at 65 to 75% relative humidity, 23°C in light and 19°C in dark, with a 16 h photoperiod with N-free Jensen medium (14). Determinations of shoot dry weight were done as previously described (5).
Histochemical localization of β-galactosidase activity.
Whole roots were collected and fixed with a solution containing 1% paraformaldehyde, 0.3 M mannitol, and 10 mM 2-N-morpholino-ethanesulfonic acid (pH 5.6) for 1 h at room temperature under a gentle vacuum. Portions of nodulated roots (0.8 to 1 cm long) were excised with a razor blade and embedded in 4% agarose. Sections (50 to 80 μm) were obtained using a Leica (Nussloch, Germany) VT1000 S vibrating-blade microtome. Nodulated root fragments or nodule sections were washed three times with 50 mM phosphate buffer (pH 7.2) and immersed in a staining solution containing 10 mM phosphate buffer (pH 7.2), 150 mM NaCl, 1 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.03% Triton X-100, and 1 mM X-Gal. The samples were incubated at 37°C overnight in the dark and then briefly cleared with sodium hypochlorite, washed in phosphate buffer, and photographed as described below.
Preparation of fixed, sectioned material.
For histological analysis, nodules were excised and fixed in 50 mM potassium phosphate buffer (pH 7.4) containing 4% paraformaldehyde, 3% glutaraldehyde, and 4% sucrose. Samples were subjected to short pulses of gentle vacuuming until they sank, and after 3 h, they were rinsed in phosphate buffer. After fixation, the samples were rinsed three times with the same buffer, dehydrated by passage through a series of graded ethanol, and embedded in PolyBed 812 resin (Polyscience). Semithin sections (1 to 2 μm thick) were obtained with an Ultracut UCT apparatus (Leica); for optical observation, samples were stained with toluidine blue (0.04% in distilled water) and photographed with a Zeiss (Jena, Germany) Axiophot microscope equipped with a digital camera (Coolpix 995; Nikon).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been submitted to GenBank under the accession numbers GQ249912, GQ249913, GQ249914 and GQ249915.
RESULTS
Isolation and characterization of GA1 derivatives GA11 and GA12 by Tn5 mutagenesis.
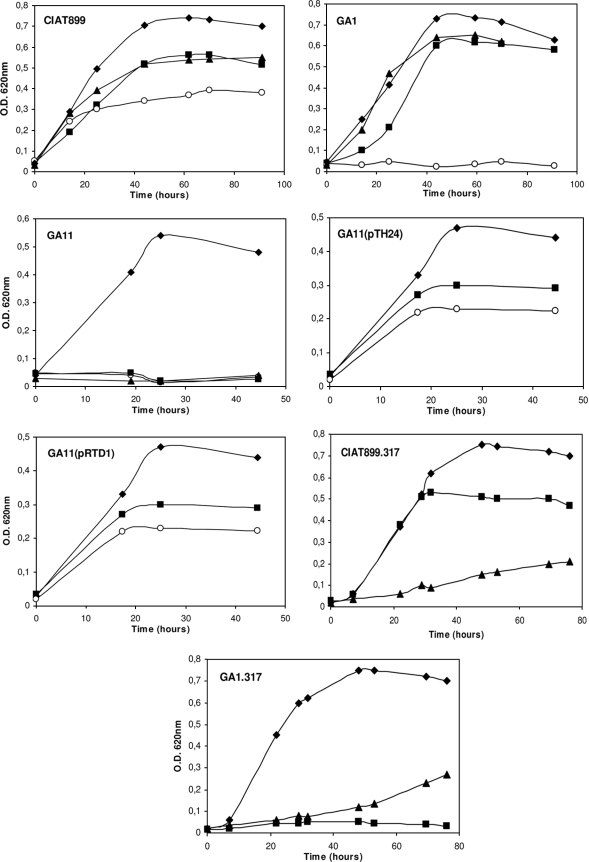
Random insertion mutagenesis of R. tropici GA1 was done by conjugal transfer of pSUP202-1::Tn5 using E. coli S17-1 as the donor (31). Transconjugants were selected on PY solid medium containing Nal, Gm, and Km. From a collection of ca. 1,000 Tn5-derived isolates, four mutant strains that were unable to grow on MM with succinate as a sole carbon source were identified. Two of them, strains GA11 and GA12, were able to grow on MM with glucose as the sole carbon source and were thus used for further characterization as perspective mutants of an alternative succinate transport system. Figure 1 shows the growth profiles for strains GA1 and GA11 and that of their derivatives carrying plasmid pTH24 (39) or pRTD1 (5) with dctABD genes from Sinorhizobium meliloti or R. tropici, respectively. GA12 and derivative strains exhibited the same growth profiles as GA11 and corresponding derivatives grown under the same conditions (data not shown). Transconjugants maintaining the rhizobial dct loci were able to grow on DCAs, suggesting that in strains GA11 and GA12, sequences required for DctA-independent succinate transport had been disrupted by the Tn5 insertions.
FIG. 1.
Growth of R. tropici strains on different carbon sources. Cells were grown in PY medium for 24 h, centrifuged, and suspended in MM supplemented with 10 mM NH4Cl and 20 mM of the indicated carbon substrate. Cultures were incubated in a rotary shaker at 30°C. ⧫, glucose; ▪, succinate; ○, fumarate; ▴, 2-oxoglutarate. O.D., optical density.
The succinate uptake activity of GA11 or GA12 grown in MM with glucose and succinate as a sole carbon source was very low compared with the activity of GA1 grown under the same conditions (Table 2). Assays done using the method that included a 6-h preincubation period in MM with succinate (see Materials and Methods) showed that the succinate uptake activities of strains GA11 and GA12 were indeed very low (1.5 and 0.9 nmol min−1 mg protein−1, respectively) compared with that of GA1 (35.0 nmol min−1 mg protein−1). This confirmed that the insertions inactivated functions for DctA-independent succinate transport.
TABLE 2.
Succinate transport activity in bacteria grown with different carbon sources
| Strain | Mean sp act of succinate uptake (nmol min−1 mg protein−1) ± SDa
|
|||
|---|---|---|---|---|
| Glucose | Glucose + 2-oxoglutarate | Succinate | 2-Oxoglutarate | |
| R. tropici CIAT899 | 0.7 ± 0.2 | ND | 26.3 ± 0.5 | 54.9 ± 0.7 |
| R. tropici GA1 (dctA) | 3.4 ± 0.2 | ND | 60.1 ± 0.6 | 51.7 ± 0.7 |
| R. tropici GA11 (dctA kgtS) | ND | 1.1 ± 0.2 | ND | ND |
| R. tropici GA12 (dctA kgtR) | ND | 1.2 ± 0.3 | ND | ND |
Succinate transport activity was measured in cells grown in MM with 20 mM of each carbon source for about 24 h, harvested, and suspended in MM without a carbon source at an optical density at 620 nm of 7.0. Each value represents the mean ± SD of data from two separate experiments. ND, not determined.
DNA sequence analysis.
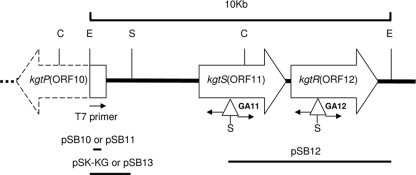
DNA/DNA (Southern) hybridization analysis of total DNA from strains GA11, GA12, CIAT899, and GA1 after digestion with restriction enzymes not recognizing Tn5 (EcoRI or ClaI) was done using EcoRI-digested pSUP202-1::Tn5 as a probe (see Fig. S1 in the supplemental material). Single hybridization bands were evident using EcoRI or ClaI to digest GA11 or GA12 total DNA, indicating that a single copy of Tn5 had inserted into the genome of each mutant strain. Hybridization of genomic DNA of the two mutants digested with EcoRI identified an approximately 10-kb fragment of the same size but with different smaller ClaI fragments. This hybridization profile suggested that Tn5 of both mutants might have inserted into the same ca. 10-kb EcoRI fragment but into different smaller ClaI fragments within or overlapping portions of the 10-kb EcoRI fragment. This possibility suggested that the two mutations were linked but were separated by a genomic ClaI site (see below). The objective, then, was to clone the 10-kb EcoRI genomic fragments from each mutant for sequence analysis of the regions bordering the sites of the Tn5 insertion into GA11 and GA12. To this end, total genomic DNA from each mutant was digested with EcoRI and separated by electrophoresis. Genomic fragments of about 10 kb were purified and ligated into EcoRI-digested pBluescript II SK. Chemically competent E. coli TOP10 cells were transformed with the ligation products, and clones containing a DNA fragment with the Tn5 cloned into pBluescript II SK were isolated as white colonies that were able to grow on LB medium with X-Gal, Amp, and Km. Plasmids containing these insertions (pGA11-10 and pGA12-10), constructed from GA11 and GA12, were then digested with SalI (there is a single central SalI site in Tn5) to clone both the right and left border fragments of the Tn5 insertions including the adjacent genomic sequences in each case.
The Tn5 border fragments were cloned into pBluescript II SK, thus generating plasmids pGA11L, pGA11R, pGA12L, and pGA12R. DNA sequence analysis of the Tn5 flanking regions (left and right ends) of strain GA11 revealed the presence of an ORF, designated ORF11 (Fig. 2), sharing high sequence similarity with the carboxyl-terminal region of putative integral membrane sensor signal transduction histidine kinases from both Ochrobactrum anthropi strain ATCC 49188 and Symbiobacterium thermophilum IAM strain 14863. A similar DNA sequence analysis revealed the presence of another ORF in the region adjacent to left end of the Tn5 insertion into strain GA12 and was named ORF12 (Fig. 2). ORF12 was likewise highly similar to a presumptive response regulator receiver with a predicted σ54 factor helix-turn-helix DNA-binding motif of O. anthropi strain ATCC 49188. In the genome of O. anthropi ATCC 49188, these two genes were adjacent, showed the same transcriptional organization of a dctBD operon, and could be predicted to specify a similar regulatory system for a transport system (see below). The two O. anthropi ATCC 49188 ORFs also overlapped by 13 nucleotides, indicating that their expression would likely be controlled by a single promoter.
FIG. 2.
Organization of the kgt locus. The triangles indicate the location of theTn5 transposon in each mutant (GA11 and GA12). Restriction sites are abbreviated as follows: E, EcoRI; S, SalI; C, ClaI. The horizontal lines represent the chromosomal fragments cloned into the plasmids indicated above each line. The vectors used to clone the fragments of pSB10 and pSB11 were pCR 2.1 and pHRP317, respectively. For pSB12 and pSB13, the fragments were cloned into pMP220 in both cases.
DNA sequence analysis from the EcoRI cloning site of pGA12-10 and pGA11-10 (primer T7 of the pBluescript II SK vector) (Fig. 2) was done to determine if the same 10-kb EcoRI fragment had been targeted in the two mutants and to provide further evidence that ORF11 and ORF12 were linked. Indeed, the same sequence was found for this end of the two 10-kb EcoRI fragments. The sequence analysis also revealed the presence of another ORF in the two fragments that was divergently oriented with respect to the adjacent ORF11 and ORF12. Restriction mapping also indicated that the orientation of ORF12 was the same as that of ORF11 and that it was tightly linked and just downstream of ORF11 (Fig. 2).
The divergently transcribed ORF upstream of ORF11 was found to be most similar to a general substrate transporter of the major facilitator superfamily MFS_1 (probable MFS dicarboxylate transporter). This ORF, designated ORF10, had the highest similarity with a predicted permease just upstream and divergently oriented from the same predicted dctBD-like two-component system of O. anthropi ATCC 49188 discussed above. ORF10 also had a high level of similarity with the potential permeases for 2-oxoglutarate in Ralstonia solanacearum and A. tumefaciens. ORF10 was thus kgtP-like, and being located adjacent and divergent to ORF11 and ORF12 strongly supported the idea that the three ORFs might function together as a dct-like transport system. The arrangement and sequences of the three ORFs indicated that they specify (principally) a 2-oxoglutarate transport system that also allows for the transport of succinate. This was supported by the observation that mutant strain GA11 was unable to grow on 2-oxoglutarate (Fig. 1). GA12 was also unable to grow on this substrate (data not shown). As such, the ORFs interrupted in GA11 and GA12 were designated kgtS and kgtR, respectively, in R. tropici (Fig. 2). The finding that GA11 and GA12 were also unable to grow on succinate suggested that the remaining ability of GA1 to utilize succinate was dependent on this three-gene system acting as an alternative succinate transport system (Fig. 1). To test this and to determine if the divergently oriented kgtP-like ORF10 upstream of kgtS was a corresponding permease, a well-defined insertion mutation of this ORF was constructed for growth and uptake studies.
Construction of defined mutants of CIAT899 and GA1 in (kgtP-like) ORF10.
Defined mutants of CIAT899 and GA1 in kgtP-like ORF10 were constructed using a suicide plasmid harboring a DNA fragment corresponding to an internal portion of ORF10. The transformation of CIAT899 or GA1 with this plasmid allowed for cointegration of the plasmid into genomic ORF10, resulting in the insertional inactivation of the putative gene. This fragment was isolated by PCR using primers KGF and KGR as described in Materials and Methods. The PCR product was cloned into pCR 2.1-TOPO to construct pSB10 (Table 1 and Fig. 2). A DNA fragment containing the PCR product was isolated from pSB10 after digestion with XhoI and SalI and cloned into pHRP317 (21). The resulting plasmid, designated pSB11, was transferred into R. tropici strains by electroporation. Cointegrate derivatives of CIAT899 or GA1 were isolated on PY solid medium supplemented with either Nal and Km or Nal, Gm, and Km, respectively. The derivatives each contained the same expected cointegrate insertion into ORF10 and therefore appeared to differ only at dctA (data not shown). In correlation, the CIAT899 derivative was able to grow on succinate, indicating that the dctABD system was intact. The GA1 derivative was unable to grow on succinate, indicating that ORF10 was required for the remaining ability of GA1 to grow on succinate. Both strains (CIAT899.317 and GA1.317) also had a greatly reduced ability to grow on 2-oxoglutarate compared with the wild type (Fig. 1). ORF10, as such, was also designated kgtP in R. tropici.
Characterization of defined mutants in kgtP and kgtSR.
GA1.317 was unable to grow on succinate, indicating that the ability of R. tropici to recognize and transport DCAs was specified only by dctA and kgtP. As noted above, both CIAT899.317 (kgtP) and GA1.317 (dctA kgtP) still retained a reduced ability to grow on 2-oxoglutarate as a sole carbon source, which was evident after 40 h of incubation on selective growth medium (Fig. 1). This observation suggested the presence of yet another unidentified alternative transport system but instead for the uptake of 2-oxoglutarate. Also indicated from this result was that the predicted alternative 2-oxoglutarate transport system recognized/transported a C5-dicarboxylate but not DCAs. The reduced ability of CIAT899.317 and GA1.317 to grow on 2-oxoglutarate was not found for GA11 (kgtS) or GA12 (kgtR). Both KgtS and KgtR were absolutely required for residual growth on 2-oxoglutarate, indicating that the potential alternative transport system required the function of KgtSR (Fig. 1). Only the growth profile of GA11 grown on 2-oxoglutarate is shown; the corresponding growth profile of GA12 was identical to that of GA11 (data not shown). Below, we provide evidence that KgtS and KgtR in their normal genomic configuration, as kgtSR, were required to activate a transcriptional promoter for the expression of kgtP. The predicted sensor-regulator proteins encoded by kgtSR had the organization of a standard two-component regulatory system, which consists of a two-gene operon in which an upstream sensor and downstream regulatory gene are coexpressed.
R. tropici sequences of pGA11-10 (Fig. 2) were found to be unable to complement mutant strain GA12 (kgtR) for its ability to grow on 2-oxoglutarate. R. tropici GA11 sequences and a part of the Tn5 of GA11-10 as the corresponding EcoRI-SalI fragment were cloned into pMP220 to construct pSB12 (Table 1). The EcoRI site to which we are referring is downstream of kgtR, as shown in Fig. 2. The cloned R. tropici fragment contained part of kgtS and the entire kgtR gene. Plasmid pSB12 was transferred into GA11 (kgtS), GA12 (kgtR), and GA1 by electroporation, and transformants were isolated on PY solid medium with Gm, Km, and Tc. The growth of GA11 or GA12 with pSB12 in MM with different sole carbon sources showed that neither of these strains was able to grow on either succinate or 2-oxoglutarate, and the transfer of vector pMP220 alone did not confer any growth differences (data not shown). The inability of plasmid pSB12 to complement mutant strain GA12, i.e., kgtR, suggested that the insertion of Tn5 into GA11, i.e., kgtS, had a polar effect on the expression of kgtR, as would be expected if kgtSR were normally cotranscribed as an operon from a promoter region upstream of kgtS. The requirement for kgtSR for the potential alternative 2-oxoglutarate system discussed above also required that these genes be cotranscribed as in an operon.
Succinate uptake activity in cells grown in the presence of different carbon sources including 2-oxoglutarate was determined (Table 2). As noted above, GA11 and GA12 exhibited a very low succinate uptake activity when they were grown for 24 h in MM with ammonia and a mixture of glucose and 2-oxoglutarate as a carbon source. Succinate transport activity, however, was strongly induced by 2-oxoglutarate in strain CIAT899 and with a higher level of activity than that of cells grown with succinate as the sole carbon source. GA1 exhibited this same high level of succinate uptake when either succinate or 2-oxoglutarate was used as a sole carbon source.
Analysis of dctA and kgtP transcriptional promoter-β-galactosidase fusions in vitro.
The 1.2-kb SalI-EcoRI fragment containing the 5′ end of kgtP as well as upstream sequences where transcriptional promoter sequences should be found was isolated from pGA11-10. This construct was subcloned into pBluescript II SK (and designated pSK-KG) and then transferred into pMP220 as a PstI-EcoRI fragment to construct a kgtP-lacZ promoter fusion, pSB13. Plasmid pSB13 was used to analyze expression from the kgtP promoter region in different genetic backgrounds or under different growth conditions. For comparison, we also analyzed the expression of dctA using plasmid pRU103, which contains a Rhizobium leguminosarum dct-lacZ promoter fusion (26). Table 3 shows the activity of the dctA promoter in different strains. Plasmid pMP220 (Table 1) was included as a vector control. The dctA promoter was specifically induced by succinate, malate, or fumarate in strain CIAT899, but the level of induction was lower than that previously found with other rhizobia. This could be due to the heterologous origin of the dctA promoter region cloned into pRU103. The expression of this promoter in GA1 (dctA), however, was at a level about 3.6-fold higher than that in CIAT899 after growth on glucose and ammonia as C and N sources. Almost the same level of activity was obtained when GA1 (pRU103) was incubated with glucose, succinate, malate, fumarate, or 2-oxoglutarate. This result suggested that, as previously shown (26, 39), in the absence of DctA, the DctB-DctD system loses the ability to be induced only by specific DCAs, and the range of molecules recognized as inducers is increased. In addition, in the absence of DctA, the DctB-DctD system becomes sensitive to cross-induction by various stimuli.
TABLE 3.
β-Galactosidase fusion analysis of the native dctA promoter of R. leguminosarum and kgtP promoter of R. tropicia
| Strain | Genotype | Mean β-galactosidase activity ± SEM under conditions ofb:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glc-NH4
|
Succ-NH4
|
Fum-NH4
|
Mal-NH4
|
Oxo-NH4
|
Oxo-Glut
|
|||||||
| dctAp | kgtPp | dctAp | kgtPp | dctAp | kgtPp | dctAp | kgtPp | dctAp | kgtPp | kgtPp | ||
| CIAT899 | Wild type | 305 ± 34c | 499 ± 50c | 620 ± 54 | 423 ± 54 | 634 ± 66 | 239 ± 45 | 763 ± 62 | 237 ± 22 | 416 ± 60 | 1,380 ± 140 | 2,017 ± 158 |
| GA1 | dctA | 1,086 ± 85 | 889 ± 76 | 1,210 ± 86 | 1,038 ± 120 | 991 ± 45 | 521 ± 65 | 1,184 ± 182 | 847 ± 92 | 1,332 ± 151 | 3,022 ± 253 | 3,304 ± 240 |
| GA11 | dctA kgtS | 710 ± 88 | 80 ± 15 | 2,044 ± 178 | 91 ± 18 | 2,665 ± 279 | 101 ± 29 | |||||
| GA12 | dctA kgtR | 598 ± 83 | 74 ± 23 | 1,317 ± 210 | 94 ± 21 | 1,579 ± 127 | 86 ± 23 | |||||
| CIAT899.317 | kgtP | 114 ± 38 | 3,037 ± 238 | 236 ± 49 | 3,646 ± 229 | 359 ± 49 | 4,160 ± 239 | 3,690 ± 300 | 177 ± 29 | 3,656 ± 230 | 5,531 ± 279 | |
The dctA-lacZ fusion is pRU103, the kgtP-lacZ fusion is pSB13, and the vector control is pMP220. Abbreviations: Glc, glucose; Succ, succinate; Fum, fumarate; Mal, malate; Oxo, oxoglutarate; Glut, glutamate.
The cells were incubated for about 4 h in the presence of the inducer at a 20 mM concentration.
Results are shown as o-nitrophenylgalactoside hydrolyzed (in nanomoles per minute per milligram of protein ± standard errors of the means) and are based on data from three independent cultures assayed in triplicate.
Double mutant strains GA11 (dctA kgtS) and GA12 (dctA kgtR) had higher levels of dctA promoter activity than did the wild type under the same conditions. In contrast with GA1, the dctA promoter in these mutants was regulated the same as in the wild-type strain. Although the levels of expression were consistently higher, these mutants appeared to have a partially restored ability to differentially respond to succinate as well as to 2-oxoglutarate as inducers. These results indicated that KgtS and KgtR can interfere with the ability of DctB and DctD to detect DCAs (succinate) in the absence of DctA. Interference between two-component systems has been termed “cross talk” (17).
A comparative analysis of the kgtP promoter-lacZ fusion, i.e., pSB13, is also shown in Table 3. In the wild type, in contrast with the dctA promoter, a corresponding kgtP promoter was specifically induced by 2-oxoglutarate but not by succinate. The kgtP fusion was strongly induced in the wild type by 2-oxoglutarate using glutamate or ammonia, to a lesser extent, as a nitrogen source. In addition, the kgtP promoter was dramatically induced in the kgtP mutant (CIAT899.317) background with glutamate and again, to a lesser extent, with ammonia as the nitrogen source. The level of expression of kgtP in CIAT899.317, however, was high under all conditions tested, suggesting a hypersensitivity of the KgtSR system in the absence of KgtP. The expression of kgtP in GAI was also uniformly higher than that in the wild type but not as high as that in strain CIAT899.317. A similarly altered expression profile for dctA was also seen with GA1, as noted above. In the absence of DctA, there was a generally higher level of expression of dctA, which may likewise reflect a hypersensitivity of the DctBD system. Note, however, that the level of expression of dctA in the kgtP background was lower than that of the wild type. The level of activity of kgtPp in GA11 or GA12 was very low under the conditions tested, consistent with the absolute requirement of kgtSR for the activity of the kgtP promoter and thus the growth of CIAT899 on 2-oxoglutarate.
These results indicated an interaction between the two-component systems that control the expression of dctA and kgtP. Both permeases were expressed at higher levels in GA1 than in the wild type; however, the expression of dctA was only mildly affected by the absence of KgtP. The kgtPSR system appeared to be more responsive to changes in background or growth conditions and to the dctABD system than vice versa. In comparing the expression levels of the two fusions, however, it should be remembered that the more responsive kgtPp fusion may in part reflect the heterologous nature of the dctAp fusion used in this study.
Plant test results.
Table 4 describes the results obtained from measurements of shoot dry weights of P. vulgaris plants inoculated with each R. tropici strain. In agreement with data from previous studies (5), plants inoculated with GA1 exhibited a low dry-matter weight similar to the values obtained with plants grown in the absence of an externally supplied nitrogen source. In addition, GA11, GA12, or GA1.317 did not promote the growth of the leguminous plant, having dry weights comparable to the values determined for plants inoculated with GA1 or incubated without external sources of nitrogen. As expected, all mutants that were defective in dctA formed ineffective nodules when in association with the plant. The requirement for dctA was evident for plants with nodules formed with GA1 even though a limited acetylene reduction activity was found (5). CIAT899 and CIAT899.317, however, both promoted the growth of the plant to the same level, showing that KgtP was not essential for the formation of effective nodules.
TABLE 4.
Plant test of R. tropici strains
| Strain or condition | Nodulation | Mean shoot dry wt (g plant−1) ± SDa |
|---|---|---|
| Strains | ||
| CIAT899 | + | 0.23 ± 0.08 |
| GA1 | + | 0.14 ± 0.04 |
| GA11 | + | 0.11 ± 0.04 |
| GA12 | + | 0.13 ± 0.04 |
| CIAT899.317 | + | 0.26 ± 0.04 |
| GA1.317 | + | 0.12 ± 0.03 |
| Conditions | ||
| With nitrogen | − | 0.36 ± 0.07 |
| Without nitrogen | − | 0.10 ± 0.02 |
The shoot dry weight was determined using 11 plants of P. vulgaris after 30 days of growth under each condition.
Analysis of β-galactosidase activity in P. vulgaris nodules.
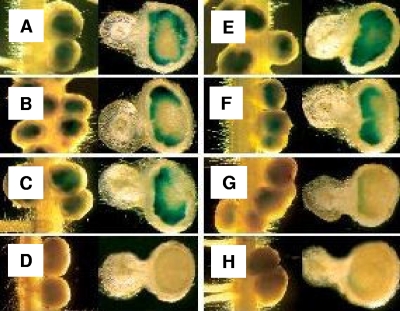
P. vulgaris nodules induced by CIAT899, GA1, GA11, or GA12 carrying plasmid pRU103 or pSB13 were used to monitor dctA and kgtP promoter activity in planta (Fig. 3). The same strains carrying plasmid pMP220 (vector alone) were also included as a control. Analysis of enzymatic activity was done 5 and 10 days after inoculation (see Fig. S2 in the supplemental material and see Fig. 3, respectively).
FIG. 3.
Expression of dctAp (A to D) and kgtPp (E to H) during nodulation. Analysis of β-galactosidase activity was done 10 days postinfection. Images shown in each frame are of whole nodules on the left, and images on the right show internal regions after cross-sections of these nodules. Nodules were induced by CIAT899 (A and E), GA1 (B and F), GA11 (C and G), and GA12 (D and H).
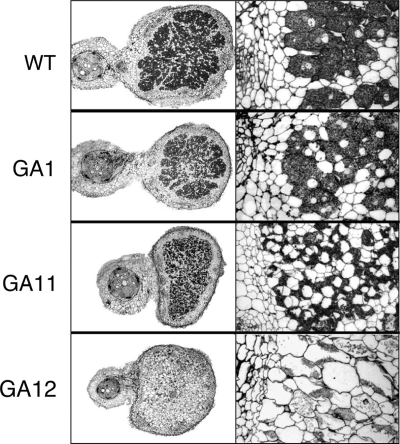
In 5-day-old developing nodules induced by CIAT899, GA1, GA11, and GA12 (see Fig. S2A, S2B, S2C, and S2D in the supplemental material), the dctA promoter was active in bacteroids located in the first invaded cells of nodule primordia. Moreover, in 10-day-old nodules, the dctA promoter was active in bacteroids located in the younger invaded cells of the central tissues of nodules induced by strains CIAT899, GA1, and GA11 (Fig. 3A to C) but was almost inactive in bacteroids of strain GA12 (Fig. 3D). Most likely, bacteroids of strain GA12 underwent early senescence, just after bacteria were released inside the invaded cells, as suggested by the results shown in Fig. 4 (empty invaded cells in GA12-induced nodules). In fact, light microscopy of 20-day-old nodule sections (Fig. 4) revealed that, as described previously for other rhizobial dctA mutants (10, 40), nodules induced by GA11 and GA12 contained a large number of empty, uninfected cells, and/or the infected cells contained few developed bacteroids undergoing senescence. Note that plasmid pMP220 and its derivatives (pRU103 or pSB13) may be lost in the absence of drug selection. However, nodules induced by the same strains carrying empty vector pMP220 showed no β-galactosidase activity. Taken together, these data suggest that the levels of the in planta promoters' activities, although specific, might be underestimated.
FIG. 4.
Histology of P. vulgaris nodules induced by R. tropici strains CIAT899 (wild type [WT]), GA1, GA11, and GA12 examined by light microscopy 20 days postinfection. Images shown on the left are cross-sections of whole nodules, and images on the right show more detail of internal regions of these nodule cross-sections at a higher magnification.
In 5-day-old developing nodules induced by CIAT899 and GA1, the kgtP promoter (see Fig. S2E and S2F in the supplemental material) was active in bacteroids located in the first invaded cells of the nodule primordium, indicating that the kgt system was induced in the symbiosome. In contrast, the activity of the kgtP promoter was almost undetectable in 5-day-old nodules induced by GA11 and GA12 (see Fig. S2G and S2H in the supplemental material). This result, however, is in good agreement with the results obtained with free-living bacteria; i.e., KgtSR was required to induce the kgtP promoter (Table 3). Indeed, in the absence of DctA, bacteroid formation and/or maintenance inside the nodule-invaded cells appeared to be dependent on both KgtR and KgtS, with the requirement of KgtR being more pronounced (Fig. 4).
In 10-day-old nodules induced by strain CIAT899 or GA1, the kgtP promoter was active only in bacteroids present in the younger invaded cells located at the periphery of the central tissue (Fig. 3E and F). In contrast, the kgtP promoter was inactive (or its activity was undetectable) in all bacteroids in nodules induced by GA11 or GA12 at the same day postinfection (Fig. 3G and H). This result indicated that the KgtRS system was essential for the activity of the kgtP promoter during symbiotic interactions.
DISCUSSION
Tn5 mutagenesis of a well-defined dctA mutant of R. tropici CIAT899, i.e., strain GA1, was used to identify a three-gene cluster that is highly similar to a rhizobial dctABD locus that was required for this strain to grow on succinate as a sole carbon source. This alternative succinate transport system, although very similar to DctABD, in fact specified a 2-oxoglutarate transport system that also allowed for succinate recognition, uptake, and subsequent utilization by GA1. Genes encoding the corresponding KgtP permease and KgtSR sensor-regulator were designated such based on sequence similarity and mutational and phenotypic analyses. The encoded kgtP permease had a high levels of similarity with genes encoding the putative MFS permease of Ochrobactrum anthropi ATCC 49188 and permeases for 2-oxoglutarate, i.e., KgtP of R. solanacearum GMI1000 and A. tumefaciens C58.
The MFS permease-encoding gene family is an extensive and diverse superfamily of secondary transporters found in the three branches of life. Most of these encoded permeases are composed of 400 to 600 amino acids with 12, 14, or 24 predicted transmembrane α-helical regions. These porters are suggested to function by uniport, symport (with solute, H+ or Na+), or antiport and exhibit specificity for a range of organic molecules including sugars, drugs, Krebs cycle intermediates, amino acids, and osmolites, etc. (27). KgtP (2.A.1.6.2) (28) is a proton symporter for 2-oxoglutarate, which is constitutively expressed in E. coli (30). This porter is 432 amino acids long with 12 membrane-spanning regions (29). In contrast with E. coli kgtP, upstream of kgtP in R. tropici CIAT899 was a divergently transcribed sensor-regulator two-component system, kgtSR, which exhibited similarity with the dctBD genes of rhizobia. A transcriptional promoter-lacZ gene fusion of the R. tropici kgtP gene was constructed, and expression studies indicated a promoter in the region upstream of kgtP that was strongly induced by 2-oxoglutarate in the wild-type strain. The kgtSR two-component system was absolutely required for the expression of this 2-oxoglutarate-responsive promoter. Well-defined plasmid cointegration mutants of the kgtP gene were constructed in CIAT899 and GA1, but these mutants retained a limited ability to grow on 2-oxoglutarate. This result suggested the presence of yet another alternative transport system that allowed the transport of this C5-dicarboxylate. This possible alternative transport system also had an absolute requirement for the kgtSR two-component system.
Expression studies using the kgtP-lacZ fusion and a previously constructed dctA-lacZ fusion in the wild-type background yielded the expected expression profile for each gene: dctA was induced by DCAs, succinate, malate, or fumarate, and kgtP was strongly induced by 2-oxoglutarate. As expected, no induction of kgtP was observed in GA11 (dctA kgtS) or GA12 (dctA kgtR). Our results indicated an interaction between the two-component systems that controlled the expression of dctA and kgtP. Both permeases were expressed at higher levels in GA1 than in the wild-type strain. The expression of dctA seemed to be affected by the absence of KgtP, since the level of expression was lower for CIAT899.317 than for CIAT899 under all conditions tested. The kgtPRS system appeared to be more responsive to changes in background or growth conditions and to the dctABD system than vice versa.
Succinate transport activity in CIAT899 was induced by succinate or by 2-oxoglutarate. Consistent with previously determined gene expression profiles, the transport systems induced by these two carbon sources should be DctA and KgtP, respectively. The level of succinate transport by CIAT899 incubated with 2-oxoglutarate was higher than that in cells incubated with succinate and comparable to transport levels determined for GA1 cells incubated in the presence of succinate or 2-oxoglutarate. Taken together, these results suggested that KgtP was mediating the uptake of succinate in CIAT899 or GA1 cells grown on 2-oxoglutarate or GA1 cells grown with succinate. Note that these activities were measured in the presence of high concentrations of succinate, since the KgtP system appeared to have a relatively low affinity for succinate, as noted below (5).
Previous studies with bacteroids of Bradyrhizobium japonicum isolated from soybean nodules and maintained under anaerobic conditions allowed the identification of the active mechanisms for the uptake of succinate, malate, 2-oxoglutarate, and glutamate (25). Values were obtained for the apparent Km and Vmax for the uptake of succinate, malate, and 2-oxoglutarate in these bacteroids. The Vmax values determined for succinate and malate were 2.72 and 1.05 nmol min−1 mg bacteroid−1, respectively, and the Km for both solutes was 0.04 mM. Uptake measurements for 2-oxoglutarate, however, indicated a relatively low affinity for this substrate compared to the Km for the other DCAs, i.e., a Km of 3.88 mM, reaching saturation with 5 mM 2-oxoglutarate. The Vmax for 2-oxoglutarate was also much higher, at 71.90 nmol min−1 mg bacteroid−1 (25). The genome of B. japonicum USDA110 contains an ORF (GenBank accession number NC_004463) with high similarity to the kgtP gene of E. coli, suggesting the presence of a system for the transport of 2-oxoglutarate similar to the one which we describe for R. tropici. This possibility was consistent with measurements made for succinate transport done with R. tropici cells grown in MM with different carbon sources. As noted in Materials and Methods, the determination of succinate uptake activity was done using succinate at concentrations higher than 3 mM, since the activity of the KgtP uptake system (in GA1) likely had a low affinity for this substrate (5).
Results obtained with wild-type strain CIAT899 in bean nodules indicated that the KgtP permease was expressed in bacteroids of R. tropici formed in determinate nodules of P. vulgaris and that this expression was induced by specific compounds present in the symbiosomes. The microaerophilic environment imposed on bacteroids in nodules involves the tight control of the pathways involved in the metabolism of DCAs, including the Krebs cycle. In addition, mature nitrogen-fixing bacteroids are nongrowing cells that must carefully balance the flow of metabolites into and out of the cell. Isolated soybean nodules, e.g., appear to exhibit respiratory inhibition when incubated with excessive levels of DCAs (6). It was further suggested that since aconitase mutants of B. japonicum can form effective bacteroids, a complete functional tricarboxylic acid (TCA) cycle may not be necessary and may in fact be detrimental to mature bacteroids (34). Mutants in α-ketoglutarate dehydrogenase, e.g., are impaired in nodule growth and development but can eventually form effective nodules (12). These observations indicated that some enzymes of the TCA cycle may not be fully expressed or perhaps become unnecessary in mature bacteroids. The participation of bypass pathways (e.g., α-ketoglutarate decarboxylase bypass to succinate) could also be used to maintain carbon balance and energy homeostasis (13). Another way to maintain this balance would be to export specific intermediates from the cell that would tend to overaccumulate (24), e.g., 2-oxoglutarate.
The high ratio of NADH/NAD+ generated under microaerobic conditions could induce the inhibition of α-ketoglutarate dehydrogenase and result in the accumulation of 2-oxoglutarate. If KgtP functioned for the excretion of 2-oxoglutarate and protons (or Na+), then this process could also be used by the bacteroid to capture reductive energy as well as a pathway to diminish the internal levels of 2-oxoglutarate. In symporters, the free energy required for the accumulation of an organic solute is provided from the electrochemical gradient of the other solute, i.e., inorganic cations, H+ or Na+. These reactions are reversible, and in some cases, the concentration gradient of the organic solute or metabolite could control the transport of the cation (23). Under fermentative conditions, when the concentration gradient of a corresponding metabolite exceeds the electrochemical gradient of the cation, the metabolites synthesized as end products can be coexcreted with the cations.
Encarnación et al. (9) previously analyzed the growths of different Rhizobium strains under fermentative-like conditions. The cells incubated in MM after successive subculture accumulated poly-β-hydroxybutyrate (PHB) and excreted amino acids and organic acids including 2-oxoglutarate. Those authors noted that this efflux was not the consequence of the intracellular accumulation of large amounts of these compounds and suggested that a specific transport system should be induced and expressed in these bacteria under fermentative conditions. The metabolic regulation of bacteroids was suggested to share some similarities with that of fermentative bacteria. An illustration that those authors provided in this regard was the proposed regulatory role for PHB biosynthesis in lowering cellular levels of NAD(P)H. An adjustment to lower cellular NAD(P)H levels should allow the TCA cycle to operate or to partially operate. In this regard, those authors pointed out that, e.g., PHB is accumulated by rhizobia in determinate nodules.
Glutamate added to cultures of rhizobia can result in the induction of nitrogen fixation and the release of ammonia. It is also known that nitrogen-fixing bacteroids do not show signs of nitrogen limitation. Glutamine synthetase and glutamate synthase levels remain low, and the high-affinity ammonia uptake system remains off, while such bacteroids continue to excrete ammonia. To account for this behavior, Kahn et al. (16) previously proposed that a malate-aspartate shuttle, similar to that found in mitochondria, could be active in bacteroids. According to this model, the organelle does not have a net incorporation of carbon compounds and instead incorporates reductive power in exchange for ammonia (2, 24, 38). The model proposed that the bacteroid should incorporate malate and glutamate and excrete 2-oxoglutarate and aspartate. Malate is oxidized and forms aspartate after transamination using a molecule of glutamate in the bacteroid. The final products, aspartate and 2-oxoglutarate, should then be excreted from the cell. Consistent with this, KgtP could be operating in bacteroids to ensure the excretion of 2-oxoglutarate as part of a malate-aspartate shuttle.
Supplementary Material
Acknowledgments
This research was supported by a grant from the International Foundation for Science (IFS-Crop Science), Sweden.
We thank Philip Poole, Division of Microbiology, School of AMS, University of Reading, Whiteknights, Reading RG6 6AJ, United Kingdom, for useful discussions, strains, and supplies and for hosting Silvia Batista in his laboratory during an early stage of this study. We are also grateful for helpful comments provided by two anonymous reviewers.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appels, M. A., and H. Haaker. 1991. Glutamate oxaloacetate transaminase in pea root nodules. Plant Physiol. 95:740-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 4.Batista, S., S. Castro, M. Ubalde, and G. Martínez-Drets. 1994. Effect of divalent cations on succinate transport in Rhizobium tropici, R. leguminosarum bv. phaseoli and R. loti. World J. Microbiol. 10:249-255. [DOI] [PubMed] [Google Scholar]
- 5.Batista, S., A. I. Catalán, I. Hernández-Lucas, E. Martínez-Romero, O. M. Aguilar, and G. Martínez-Drets. 2001. Identification of a system that allows a Rhizobium tropici dctA mutant to grow on succinate, but not on other C4-dicarboxylates. Can. J. Microbiol. 47:509-518. [DOI] [PubMed] [Google Scholar]
- 6.Bergersen, F. J., and G. L. Turner. 1990. Bacteroids from soybean root nodules: respiration and N2-fixation in flow-chamber with oxyleghaemoglobin. Proc. R. Soc. Lond. B Biol. Sci. 238:295-320. [Google Scholar]
- 7.Brencic, A., and S. C. Winans. 2005. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69:155-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, R., and D. Kahn. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621-631. [DOI] [PubMed] [Google Scholar]
- 9.Encarnación, S., M. Dunn, K. Willms, and J. Mora. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finan, T. M., J. M. Wood, and D. C. Jordan. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 154:1403-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, P. H., S. E. Viteri, F. Mackie, A. A. T. Vargas, and A. Palacios. 1982. Variation in acid soil tolerance among strains of Rhizobium phaseoli. Field Crops Res. 5:121-128. [Google Scholar]
- 12.Green, L. S., and D. W. Emerich. 1997. Bradyrhizobium japonicum does not require α-ketoglutarate dehydrogenase for growth on succinate or malate. J. Bacteriol. 179:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, L. S., L. Youzhong, D. W. Emmerich, F. J. Bergersen, and D. D. Day. 2000. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 182:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, H. L. 1942. Nitrogen fixation in leguminous plants. I. General characteristics of root nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linnean Soc. New South Wales 66:98. [Google Scholar]
- 15.Jording, D., P. K. Sharma, R. Schmidt, T. Engelke, C. Uhde, and A. Pühler. 1992. Regulatory aspects of the C4-dicarboxylate transport in Rhizobium meliloti: transcriptional activation dependence on effective symbiosis. J. Plant Physiol. 141:18-27. [Google Scholar]
- 16.Kahn, M. L., J. Kraus, and J. E. Sommerville. 1985. A model of nutrient exchange in the Rhizobium-legume symbiosis, p. 193-199. In H. J. Evans, P. J. Bottomley, and W. E. Newton (ed.), Nitrogen fixation research progress. Martinus Nijhoff, Dordrecht, The Netherlands.
- 17.Laub, M. T., and M. Goulian. 2007. Two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 19.Noel, K. D., A. Sánchez, J. Fernández, J. Leemans, and M. A. Ceballos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldroyd, G. E. D., and J. A. Downie. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59:519-546. [DOI] [PubMed] [Google Scholar]
- 21.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram− bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 22.Poole, P. S., A. Blyth, C. J. Reid, and K. Walters. 1994. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv viciae. Microbiology 140:2787-2795. [Google Scholar]
- 23.Poolman, B., and W. N. Konings. 1993. Secondary transport in bacteria. Biochim. Biophys. Acta 1183:5-39. [DOI] [PubMed] [Google Scholar]
- 24.Prell, J., and P. Poole. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 14:161-168. [DOI] [PubMed] [Google Scholar]
- 25.Reibach, P. H., and J. G. Streeter. 1984. Evaluation of active versus passive uptake of metabolites by Rhizobium japonicum bacteroids. J. Bacteriol. 159:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid, C. J., and P. S. Poole. 1998. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 180:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. M. Heijne, S.-C. Huang, D. L. Jack, P. S. Jähn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T.-T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 28.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seol, W., and A. J. Shatkin. 1991. Escherichia coli kgtP encodes an α-ketoglutarate transporter. Proc. Natl. Acad. Sci. USA 88:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seol, W., and A. J. Shatkin. 1992. Escherichia coli α-ketoglutarate permease is a constitutively expressed proton symporter. J. Biol. Chem. 267:6409-6413. [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis of gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 32.Spaink, H. P., J. H. Robert, C. A. Okker, E. P. Wijffelman, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 33.Tatè, R., A. Riccio, M. Iaccarino, and E. J. Patriarca. 1997. A cysG mutant strain of Rhizobium etli pleiotropically defective in sulfate and nitrate assimilation. J. Bacteriol. 179:7343-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thony-Meyer, L., and P. Kunzler. 1996. The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for an effective root-nodule symbiosis. J. Bacteriol. 178:6166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udvardi, M. K., and D. A. Day. 1997. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:493-523. [DOI] [PubMed] [Google Scholar]
- 36.Udvardi, M. K., L.-J. O. Yang, S. Young, and D. A. Day. 1990. Sugar and amino acid transport across symbiotic membranes from soybean nodules. Mol. Plant-Microbe Interact. 3:334-340. [Google Scholar]
- 37.Wang, Y., L. Giblin, B. Boesten, and F. O'Gara. 1989. Genetic analysis and regulation of the Rhizobium meliloti genes controlling C4-dicarboxylic acid transport. Gene 85:135-144. [DOI] [PubMed] [Google Scholar]
- 38.White, J., J. Prell, E. K. James, and P. Poole. 2007. Nutrient sharing between symbionts. Plant Physiol. 144:604-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarosh, O. K., T. C. Charles, and T. M. Finan. 1989. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol. Microbiol. 3:813-823. [DOI] [PubMed] [Google Scholar]
- 40.Yurgel, S. N., and M. L. Kahn. 2004. Dicarboxylate transport by rhizobia. FEMS. Microbiol. Rev. 28:489-501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.