Abstract
Sensor histidine kinases are widely used by bacteria to detect and respond to environmental signals. In Bacillus subtilis, KinA is a major kinase providing phosphate input to the phosphorelay that activates the sporulation pathway upon starvation via the phosphorylated Spo0A transcription factor. KinA contains three PAS domains in its amino-terminal sensor domain, which appear to be involved in the sensing of an unidentified sporulation signal(s) produced upon starvation. Prior biochemical studies have suggested that KinA forms a homodimer as a functional enzyme and that the most amino-terminal PAS domain (PAS-A) plays an important role in sensing the signal(s) to activate an ATP-dependent autophosphorylation reaction to a histidine residue. To analyze the structure and function of the kinase in vivo, we have used a strain in which the synthesis of KinA is under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. In vivo functional studies in combination with domain-based deletion analysis show that the cytosolic KinA forms a homo-oligomer as an active form under both nutrient-rich and nutrient-depleted conditions via its amino- and carboxyl-terminal domains independently. Furthermore, we found that a mutant in which the PAS-A domain was deleted was still able to induce sporulation at a wild-type level irrespective of nutrient availability, suggesting that PAS-BC domains are sufficient to maintain the kinase activity. Based on these results, we propose that the primary role of the amino-terminal sensor domain is to form a stable complex as a functional kinase, but possibly not for the binding of an unidentified sporulation signal(s).
Histidine kinases are found mostly in bacteria, as well as in archaea, eukaryotic microorganisms, and plants, and play a role in cellular adaptation to environmental conditions and stresses (10, 32, 42). Typically, each histidine kinase functions as a sensor molecule for environmental cues and transduces signals for various biological processes (16, 42). When a histidine kinase recognizes a stimulus, usually a small extracellular or intracellular effector molecule, the kinase autophosphorylates on a conserved histidine within its catalytic core (32, 39). This phosphate moiety is subsequently transferred to a conserved aspartate residue on the response regulator, thereby inducing a conformational change that leads to DNA-binding activity to regulate target genes. Therefore, the combination of a sensor kinase and a response regulator is known as a two-component signal transduction system (32). In bacteria, two-component systems regulate pathogenicity, virulence, chemotaxis, and stress responses, such as sporulation (16, 21, 30, 32).
The soil bacterium Bacillus subtilis is one of the well-characterized model systems for sporulation (20, 31, 40). Sporulation occurs in response to nutrient deprivation and involves an asymmetric cell division giving rise to two cellular compartments of unequal size that follow different programs of gene expression but communicate between each other to coordinate the maturation of the smaller compartment into a forespore. Finally, a forespore is released as a dormant spore that can survive until conditions are favorable for germination. Thus, this system couples nutrient deprivation with cell morphogenesis by sensing environmental change and directing the transcription of the genes whose products are required to initiate the sporulation process (21, 30).
The major histidine kinase responsible for activating the sporulation pathway in B. subtilis is KinA (29). KinA is a soluble cytoplasmic protein that appears to be active as a dimer and is composed of an amino-terminal sensor domain and a carboxy-terminal autokinase domain (33, 37, 47, 51). The autokinase domain consists of two subdomains: a dimerization and histidine phosphotransfer (DHp) domain and a catalytic ATP-binding (CA) domain (33, 47). The sensor domain contains three PAS domains (PAS-A, -B, and -C) (26, 37, 47). The PAS domain has been reported as a dimerization domain common to the proteins in which imperfect repeat sequences were first recognized: the Drosophila period clock protein (PER), vertebrate aryl hydrocarbon receptor nuclear translocator (ARNT), and Drosophila single-minded protein (SIM) (8, 28). Numerous PAS domain-containing two-component systems have been identified in bacteria (17). PAS domains appear to be used as sensor modules that sense a wide range of stimuli and regulate an associated histidine kinase (17, 32, 43). Presumably, the mechanism relies on the ability of the PAS domain to sense environmental change via a bound ligand (signaling molecule) moiety and to couple that signal to conformational changes of the PAS domain (17, 32, 43). Therefore, as an intracellular sensor, the PAS domains of KinA could respond to metabolic and/or environmental effectors of unknown origin produced by the cells upon nutrient limitation, such as hypoxia, a decrease in the cellular energy level, or a change in redox potential (43). Once activated by a ligand-induced conformational change and autophosphorylation under starvation conditions, KinA indirectly phosphorylates its cognate response regulator, the transcription factor Spo0A, via a phosphorelay involving two additional proteins, Spo0F and Spo0B (see Fig. 1A) (3). In this process, the phosphate moiety on the histidine of KinA is transferred first to an aspartate residue on Spo0F, then from the Spo0F aspartate to a histidine on Spo0B, and finally from the Spo0B histidine to an aspartate on Spo0A, thereby activating Spo0A. Thus, the result of the phosphorelay is phosphorylation of Spo0A, the master transcription factor for sporulation (3, 21, 22, 30).
FIG. 1.
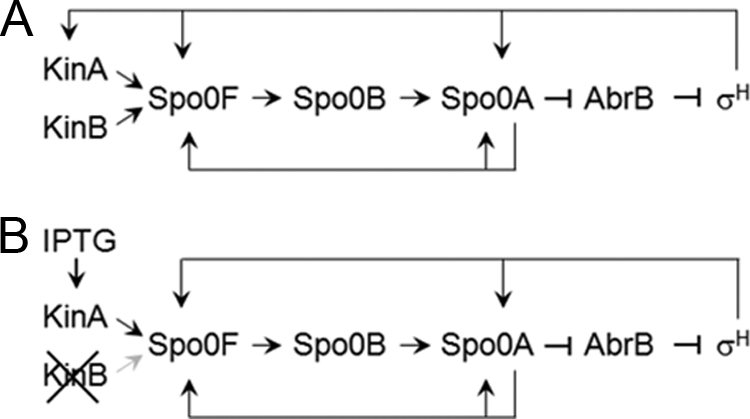
Schematic diagram of the regulatory networks for the three cases used in this study. Arrows and T-shaped bars represent positive and negative actions, respectively. Black and gray symbols represent active and inactive pathways, respectively. (A) Phosphorelay in the wild-type strain (3, 21). Upon nutrient starvation, the KinA and KinB sensor histidine kinases provide phosphate input to the master transcriptional regulator, Spo0A, via two additional regulators, Spo0F and Spo0B. Phosphorylated Spo0A becomes a positive or negative regulator for sporulation genes, including those for Spo0A itself, Spo0F, and the transition state transcription regulator AbrB. AbrB represses the transcription of the sigH (spo0H) gene, encoding the alternative sigma factor σH, which is also essential for sporulation (49). Thus, phosphorylated Spo0A represses abrB, thereby stimulating σH expression. In turn, transcription of the genes for KinA, Spo0F, and Spo0A is activated in the closed-loop system. (B) Artificial phosphorelay system decoupling gene expression from the cell cycle. A strain in which the native promoter of kinA has been replaced by an IPTG-inducible promoter is able to sporulate in response to IPTG addition. A null mutation of kinB is introduced into the system to ensure that the network is dependent solely on KinA or the mutants studied.
Primarily through in vitro biochemical studies of B. subtilis KinA, PAS-B and PAS-C have been implicated in dimerization, and PAS-A has been shown to be important for autophosphorylation activity and to be required for either signal sensing, structural integrity, or both (26, 37, 47). Furthermore, the purified PAS-A domain has been demonstrated to form a dimer by itself (26). Although PAS domains are prevalent in sensor kinases and are known to be involved in signaling in bacteria (43), no signaling molecule(s) that binds directly to the PAS domains has been identified in B. subtilis KinA to date. Thus, there is little direct evidence to support the conclusion that PAS domains in KinA are directly involved in sensing a signal(s) derived from nutrient deprivation.
Here we report in vivo studies designed to characterize the role of the PAS domains in the regulation of the sensor kinase activity under normal sporulation conditions. We used an artificial phosphorelay network that triggers sporulation in response to isopropyl-β-d-thiogalactopyranoside (IPTG) (13). In this artificial system, sporulation is initiated by inducing the synthesis of KinA, at a level comparable to the wild-type level under natural sporulation conditions, with IPTG as a sporulation initiation signal. Thus, we constructed a series of truncated PAS domain mutants and introduced each construct into the IPTG-inducible system. We demonstrated that the PAS-A deletion mutant is still functional under normal sporulation conditions, whereas PAS-AB and PAS-ABC deletion mutants are unable to carry out the sporulation process successfully. In addition, an in vivo coimmunoprecipitation assay in combination with a blue native gel demonstrates for the first time that KinA forms an active complex, irrespective of the nutrient conditions, through its N and C termini independently. Finally, we constructed coexpression systems for wild-type KinA and its N- and C-terminally truncated mutants, and we demonstrated a dominant-negative effect caused by the mutants. We also used a unique genetic system to demonstrate that the mutant consisting of the C terminus of KinA (the KinAC mutant) is involved in a reverse phosphotransfer reaction.
MATERIALS AND METHODS
Strain construction.
The parent strain for all experiments was B. subtilis strain PY79 (52). Details regarding the full genotypes of strains are provided in Table S1 in the supplemental material.
Plasmid construction.
All plasmid constructions were performed in Escherichia coli DH5α using standard methods. The plasmids used in this study are listed in Table S2 in the supplemental material. Oligonucleotides used for plasmid construction are listed in Table S2 in the supplemental material. To construct pMF416 (kinA spc), the entire kinA gene (bp 1 to 1818, corresponding to amino acid residues 1 to 606) was amplified from PY79 chromosomal DNA by PCR with primers oP1 and oP4. The PCR fragment was digested with HindIII and SphI and was then cloned into pDR111 (2) digested at the same sites. To construct pMF369 (kinAΔPAS-A spc), the PAS-A deletion mutant of kinA (bp 406 to 1818, corresponding to amino acid residues 136 to 606) was amplified from PY79 chromosomal DNA by PCR with primers oP23 and oP4. The PCR fragment was digested with HindIII and SphI and was then cloned into pDR111 digested at the same sites. To construct pMF433 (kinAΔPAS-AB spc), the PAS-AB deletion mutant of kinA (bp 775 to 1818, corresponding to amino acid residues 259 to 606) was amplified from PY79 chromosomal DNA by PCR with primers oP24 and oP4. The PCR fragment was digested with HindIII and SphI and was then cloned into pDR111 digested at the same sites. To construct pMF362 (kinAC spc), the PAS-ABC deletion mutant of kinA (bp 1162 to 1818, corresponding to amino acid residues 387 to 606) was amplified from PY79 chromosomal DNA by PCR with primers oP3 and oP4. The PCR fragment was digested with HindIII and SphI and was then cloned into pDR111 digested at the same sites. To construct pMF446 (kinAN spc), the portion of kinA encoding the N-terminal end of the protein (bp 1 to 1182, corresponding to amino acid residues 1 to 394) was amplified from PY79 chromosomal DNA by PCR with primers oP1 and oP47. The PCR fragment was digested with HindIII and SphI and was then cloned into pDR111 digested at the same sites. To construct pMF448 (kinAN gfp spc), the portion of kinA encoding the N-terminal end of the protein was amplified from PY79 chromosomal DNA by PCR with primers oP1 and oP48. The PCR fragment was digested with HindIII and XhoI. The gfp coding sequence was amplified from pMF139 (kinAC gfp kan) DNA with primers oP43 and oP44 and was digested with XhoI and SphI. The kinAN DNA fragment was cloned into HindIII- and SphI-digested pDR111 in a three-way ligation with the DNA fragment of gfp. The gfp coding sequence (GFPmut2) contains the codon substitutions S65T, V68L, and S72A (7). pMF397 (3× flag-kinA spc) was constructed by amplifying kinA with oP34 and oP4 as described by Ellermeier and Losick (11). The resulting amplified DNA product was then amplified with oP35 and oP4. The resulting DNA product was again amplified with oP36 and oP4 and was then digested with HindIII and SphI. The resulting PCR product was cloned into pDR111 digested at the same sites. To construct pMF451 (kinA-3× flag spc), primers oP1 and oP42 were used to generate a PCR fragment that amplified kinA without a stop codon from PY79 chromosomal DNA; this fragment was then digested with HindIII and XhoI. The DNA containing the 3× flag sequence with a stop codon was amplified from pMF397 plasmid DNA using primers oP51 and oP52. The PCR fragment was digested with XhoI and SphI and was then cloned into pDR111 cut with HindIII and SphI in a three-way ligation with the DNA fragment of kinA. To construct pMF452 (kinA-3× flag erm), pMF451 was digested with EcoRI and BamHI, and the smaller fragment was purified from the agarose gel. The purified fragment was cloned into pDG1664 (20) digested at the same sites.
Media and culture conditions.
For sporulation induction conditions, the resuspension method described by Sterlini and Mandelstam (38) was employed. In brief, B. subtilis strains were grown in casein hydrolysate (CH) medium at 37°C. At the mid-exponential phase of growth (optical density at 600 nm [OD600], 0.5) in CH medium, cells were induced to sporulate by resuspension in Sterlini-Mandelstam (SM) medium. To induce the synthesis of KinA or mutants under the control of the IPTG-inducible hyper-spank promoter (Phyper-spank), IPTG (final concentration, 10 μM) was added just after the cells were suspended in the SM medium. Luria-Bertani (LB) medium was used as the rich medium where indicated. An l-threonine supplement (40 μg/ml) was added to the culture for strains harboring reporter genes at the thrC locus in CH and SM media.
Sporulation efficiency and β-galactosidase assays.
Sporulation efficiency was measured as CFU per milliliter after incubation at 80°C for 10 min. Assays of β-galactosidase activity were performed as described previously (14).
Immunoblot analysis.
Cells grown in LB or SM medium as described under “Media and culture conditions” above were harvested, and cell extracts for immunoblot analysis were prepared by sonication. Immunoblot analysis was performed as described previously (14). Polyclonal antibodies against green fluorescent protein (GFP) (14, 34) and against σA (12), and a monoclonal anti-FLAG antibody (Sigma), were used to detect corresponding proteins or tagged proteins.
Coimmunoprecipitation assay.
Strains expressing both GFP-tagged and FLAG-tagged proteins were examined for the assay in both rich and SM media. A strain expressing both GFP- and FLAG-tagged KinA was used as a control. Cells were induced to synthesize the proteins by IPTG addition. After 2 h of IPTG (final concentration, 10 μM) addition, cells were harvested, and cell extracts were prepared by sonication. A 0.5-ml volume of soluble cell extracts from a 20-ml culture was transferred to a microcentrifuge tube and mixed with 20 μl of anti-FLAG monoclonal antibody-coupled agarose beads (Sigma). The sample mixture was incubated for 2 h at 4°C on a roller shaker. The antibody beads were centrifuged at 1,400 × g at 4°C and were washed five times with 1× wash buffer (Sigma). Finally, 100 μl of 2× sodium dodecyl sulfate (SDS) sample buffer was added to the pellet, and the mixture was heated at 90°C for 5 min. The samples were then centrifuged at room temperature for 10 min at 5,900 × g, and the supernatant was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (16%), followed by immunoblot analysis with polyclonal anti-GFP antibodies (14, 34) and a monoclonal anti-FLAG antibody (Sigma) for the detection of tagged proteins.
BN-PAGE assay.
The blue native PAGE (BN-PAGE) assay was performed according to the protocol provided by Invitrogen. In brief, the proteins examined were purified using coimmunoprecipitation as described in the preceding section but were eluted by competitive elution with a buffer containing FLAG peptide (Sigma) in order to maintain their native forms. The sample eluates were subjected to 4 to 16% BN-PAGE (Invitrogen), followed by immunoblot analysis with polyclonal anti-GFP antibodies (14, 34).
Protein cross-linking.
Protein cross-linking with bis-maleimidohexane (BMH; Pierce) was performed as described by Chen et al. (6) with minor modifications. Strains expressing full-length KinA proteins or N-terminal portions of KinA proteins tagged with GFP were processed for the assay. Cells were prepared as described under “Immunoblot analysis” above. The cell pellet was washed in 1× phosphate-buffered saline (PBS; pH 7.2) containing 10 mM EDTA. Cell extracts were prepared by sonication. The cross-linker was then added at a final concentration of 400 μM to the 0.5-ml soluble cell extract prepared from the 10 ml-culture. The mixture was incubated on a roller shaker for 2 h at 4°C. After quenching of the cross-linking reaction by the addition of cysteine at a final concentration of 0.5 M, the samples were subjected to SDS-PAGE (10%), followed by immunoblot analysis with polyclonal anti-GFP antibodies (14, 34).
Dominant-negative and reverse phosphorylation assays.
Strains MF3366 (PspoIIG-lacZ), MF3367 (Phyper-spank-kinAΔPAS-ABC PspoIIG-lacZ), and MF3368 (Phyper-spank-kinAN PspoIIG-lacZ), containing the kinB-null mutation (kinB encodes another sporulation kinase with a transmembrane domain) (44), were used for the dominant-negative assay. Strains MF2678 (PspoIIG-lacZ), MF2831 (Phyper-spank-kinAΔPAS-ABC PspoIIG-lacZ), and MF3251 (Phyper-spank-kinAN PspoIIG-lacZ), containing the kinA-null mutation, were used for the reverse phosphotransfer assay. Expression of the mutant proteins was induced by addition of IPTG at a final concentration of 0.5 mM after suspension in SM medium. Sporulation efficiency and β-galactosidase assays were performed as described above.
Microscope assay.
Cells expressing GFP were examined by fluorescence microscopy. Cells from 0.5 ml of culture were centrifuged and resuspended in 50 μl of 1× PBS. The concentrated cell suspension (3 μl) was placed on an agarose pad containing 1× PBS and was viewed using a fluorescence microscope (Olympus, model BX61). The microscope system control and image processing were performed using SlideBook image analysis software (Intelligent Imaging Innovations, Inc.).
RESULTS
Construction of PAS domain deletion mutants and an assay system to measure kinase activity.
Traditionally, it has been difficult to characterize the phosphorelay network genetically, because the expression of the components is mutually regulated in the closed circuit (Fig. 1A). Thus, decoupling of gene expression in the feedback regulation from the cell cycle is a crucial step for genetic analysis. For this purpose, we have developed an artificial genetic system using a strain harboring an IPTG-inducible kinA gene (Fig. 1B). In this artificial system, addition of the appropriate concentration of IPTG induces protein expression to levels similar to those in the wild-type strain under normal sporulation conditions. When the expression of KinA reaches a certain critical level in the artificial strain in the presence of IPTG, cells undergo sporulation efficiently, even under nutrient-rich conditions (13).
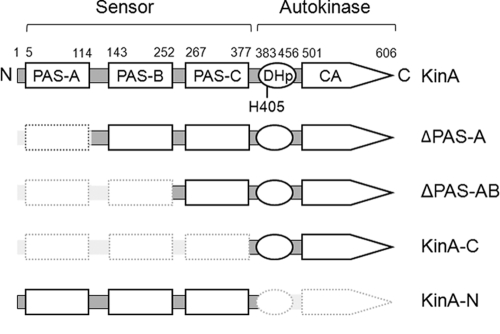
To define the functional role of the sensor domain in KinA, we generated a series of deletion mutants in which each of the three PAS domains (PAS-A, -B, and -C from the N terminus) was deleted stepwise (Fig. 2). Each KinA deletion mutant was fused to the IPTG-inducible promoter (Phyper-spank), and then the fusion was integrated as a single copy at the amyE locus (coding for a nonessential α-amylase used for ectopic integration) by double-crossover homologous recombination in the endogenous kinA-null mutant. To exclude the possibility of involvement of an alternative sporulation kinase pathway, the KinB pathway, in the phosphorelay (22, 25, 44), we introduced a kinB-null mutation into strains tested in this study. We were interested in analyzing the autokinase activity of KinA mutants that were missing PAS domains. To measure the autokinase activity of KinA and its derivatives, we used an indirect reporter system in which the β-galactosidase gene (lacZ) is fused to the Spo0A-directed promoter, PspoIIG. If full-length KinA or the mutant form is active, the protein donates a phosphate to Spo0F, Spo0B, and finally Spo0A. Thus, KinA activity is supposed to be proportional to Spo0A activity, which can be measured by the β-galactosidase assay. For this purpose, the PspoIIG-lacZ reporter gene was integrated at the thrC locus (coding for a nonessential threonine synthase) in the strain harboring our IPTG-inducible expression system for each KinA mutant. These strains were used for subsequent assays.
FIG. 2.
Domain architectures of KinA and truncated KinA mutants. The schematic diagram at the top shows an N-terminal sensor domain and a C-terminal autokinase domain. The sensor domain is further subdivided into three PAS domains (PAS-A, PAS-B, and PAS-C). The autokinase domain is further subdivided into a DHp and a CA domain (33, 47). The catalytic histidine (His405) is located within the DHp domain, as indicated. Amino acid residue numbers for domain boundaries are given above the diagrams.
Functional characterization of PAS domains in the KinA-mediated sporulation pathway.
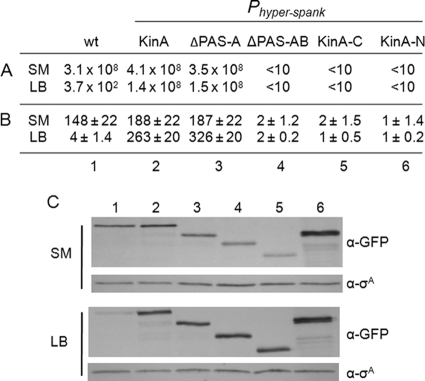
Normally, sporulation occurs during a late phase of growth in nutrient-limited medium. According to the most widely accepted model, the sensor domain is important for signal sensing under these conditions, although no specific signal or ligand for this domain has been identified yet. Thus, we examined KinA-mediated sporulation activity in each deletion mutant under normal sporulation conditions in SM medium (38). In our assays, the ΔPAS-A mutant exhibited normal sporulation efficiency, similar to that of full-length KinA, as measured by the production of heat-resistant CFU, while all other mutants showed reduced sporulation efficiency (Fig. 3A, SM). Consistent with the sporulation efficiency data, the ΔPAS-A mutant exhibited a high level of β-galactosidase synthesis from the PspoIIG-lacZ reporter, while all other mutants showed reduced activities (Fig. 3B, SM). To confirm that all the proteins tested were stably expressed, we constructed a series of strains that expressed GFP-tagged mutant proteins under the control of the IPTG-inducible promoter and then monitored the level of each protein after 2 h of IPTG addition. As shown in Fig. 3C, in SM medium, all the mutant proteins were expressed at levels comparable to those of the functional GFP-tagged full-length KinA (13), showing that the differences in β-galactosidase activity were not due to differences in protein levels. These results indicated that the ΔPAS-A mutant is still able to trigger the phosphorelay under normal sporulation conditions.
FIG. 3.
Quantitative assay for detection of the enzymatic activities of a series of KinA mutants. Genes for full-length KinA and each truncated mutant protein were fused to Phyper-spank and introduced at the amyE locus of cells containing both kinA- and kinB-null mutants. To monitor Spo0A activity, a transcriptional reporter was constructed in which the gene encoding β-galactosidase (lacZ) was fused to the promoter region of the spoIIG operon (PspoIIG) and introduced at the thrC locus of each strain. The wild-type strain (wt; lacking the IPTG-inducible construct) was used as a control. For sporulation conditions (SM), cells in CH medium were grown to the mid-exponential phase (OD600, 0.5), collected by centrifugation, and resuspended in SM medium. Then IPTG was added to a final concentration of 10 μM. For nutrient-rich conditions (LB), cells in LB medium were grown to the mid-exponential phase (OD600, 0.5), collected by centrifugation, and resuspended in fresh LB medium. Then IPTG was added to a final concentration of 10 μM. (A) Sporulation efficiency, expressed in CFU per milliliter, was determined as described in Materials and Methods. (B) The accumulation of β-galactosidase from a PspoIIG-lacZ fusion was monitored at hour 3 after suspension as a measure of Spo0A activity. Data for β-galactosidase activity are expressed in Miner units and are averages of three independent results with standard deviations. Strains used are as follows: MF3366 (wt), MF3316 (full-length KinA), MF3317 (ΔPAS-A), MF3320 (ΔPAS-AB), MF3323 (KinAC), and MF3324 (KinAN). (C) Expression of full-length KinA or KinA mutant protein was examined by immunoblot analysis. Cell extracts of strains expressing GFP fusion proteins were prepared at hour 2 after suspension and were processed for immunoblotting with anti-GFP (α-GFP) antibodies. σA, a constitutively expressed protein, was used as a loading control. Strains used are as follows: MF3593 (wt) (lane 1), MF3352 (full-length KinA) (lane 2), MF3353 (ΔPAS-A) (lane 3), MF3356 (ΔPAS-AB) (lane 4), MF3359 (KinAC) (lane 5), and MF3360 (KinAN) (lane 6).
We further examined whether these mutants are active under nutrient-rich growth conditions. Thus, we tested KinA-mediated sporulation activity in each deletion mutant in LB medium and obtained essentially the same results as under sporulation conditions (Fig. 3A, B, and C, LB). These results indicated that a PAS-A deletion mutant is still able to induce sporulation to the wild-type level, irrespective of nutrient availability. It should be emphasized that full-length KinA and the ΔPAS-A mutant are active under both nutrient-rich and nutrient-limited conditions once the levels of these proteins reach the sporulating wild-type levels (Fig. 3C, SM). In contrast, the wild-type strain lacking the IPTG-inducible construct did not sporulate under nutrient-rich conditions and served as a negative control (Fig. 3A, B, and C, LB).
KinA forms a homocomplex in vivo.
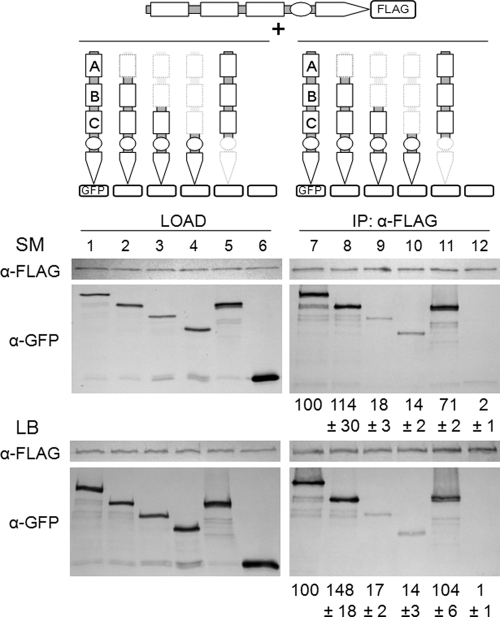
While biochemical studies demonstrate that PAS-A, -B, and -C are involved in dimerization (26, 37, 47), there has been no direct in vivo evidence that KinA exists as a dimer. As an important step toward answering this question, we adopted in vivo coimmunoprecipitation experiments in combination with BN-PAGE and cross-linking analyses. We first constructed a strain harboring C-terminal FLAG-tagged full-length KinA under the control of an IPTG-inducible promoter. To maintain a consistent genetic background throughout this study, we introduced the FLAG-tagged construct at the thrC locus in the kinA kinB double-knockout strain. When IPTG was added to the culture of rich (LB) or SM medium, the resulting strain (MF3503) sporulated with an efficiency similar to that of strains with full-length KinA under normal sporulation conditions, indicating that the FLAG-tagged protein was functional (data not shown). Then the IPTG-inducible GFP fusion of full-length KinA was introduced at the amyE locus into the FLAG-tagged strain to establish the strain coexpressing FLAG-tagged full-length KinA and GFP-tagged full-length KinA, both under the control of the IPTG-inducible promoter. A strain coexpressing only GFP (not fused to KinA) and FLAG-tagged full-length KinA was also constructed and was used as a negative control. Cell extracts were prepared after 2 h of IPTG addition both in rich (LB) medium and under normal sporulation conditions (SM medium) and were then immunoprecipitated with an anti-FLAG monoclonal antibody. Precipitated proteins were analyzed by immunoblotting with anti-FLAG and anti-GFP antibodies.
In Fig. 4, GFP-tagged full-length KinA was detected in a complex with FLAG-tagged full-length KinA (compare lanes 1 and 7). The results confirmed for the first time that KinA exists as a complex in vivo. The complex formation was detected both in rich medium and under normal sporulation culture conditions (Fig. 4, lanes 1 and 7, SM and LB). Furthermore, to identify the domain involved in the formation of the complex, we first constructed GFP fusions of the ΔPAS-A, ΔPAS-AB, ΔPAS-ABC (KinAC [Fig. 2]), and N-terminal domain (KinAN [Fig. 2]) constructs of KinA independently under the control of the IPTG-inducible promoter and then introduced each construct into the strain harboring IPTG-inducible FLAG-tagged full-length KinA. Using each strain, immunoprecipitation experiments with an anti-FLAG monoclonal antibody were performed to detect the formation of a complex. The intensity of each band in the coimmunoprecipitation fraction was quantified by densitometry analysis, and the amount of each protein in the complex was normalized first to the amount of each immunoprecipitated KinA-FLAG protein and then to that of the GFP-tagged proteins or to that of GFP with no tag in the load fraction. Quantification data, expressed as relative ratios, are shown below each gel image in Fig. 4. These results indicated that similar levels of proteins were detected in the complexes with full-length KinA, ΔPAS-A, and KinAN proteins (Fig. 4, lanes 7, 8, and 11), while lower levels (less than 20%) were detected in the complexes with ΔPAS-AB or KinAC proteins (Fig. 4, lanes 9 and 10). Significantly lower levels of protein (less than 5%) were detected in the control GFP lane (Fig. 4, lane 12). Again, essentially the same results were obtained in rich medium and under normal sporulation culture conditions (Fig. 4). Therefore, these results indicated that the N terminus forms a more stable complex than the C terminus with full-length KinA and that at least the PAS-BC domains are sufficient to form a stable complex with full-length KinA, similar to that formed between full-length KinA protomers. To determine whether the complex contains other phosphorelay components, we performed coimmunoprecipitation experiments with cells coexpressing FLAG-tagged full-length KinA and Spo0F-GFP (MF3302). Cells were suspended in SM medium to induce GFP-tagged Spo0F expression from the endogenous promoter, and IPTG was then added to the culture to induce the expression of FLAG-tagged full-length KinA. Cell extracts were processed for immunoprecipitation with an anti-FLAG antibody, followed by immunoblot analysis with anti-GFP and anti-Spo0A antibodies. However, neither Spo0F-GFP nor Spo0A was detected in the immunoprecipitated fraction (data not shown). These results suggest that KinA does not form a stable complex with other phosphorelay components. Nevertheless, these results indicate that KinA forms a homocomplex via its N- and C-terminal domains, irrespective of culture conditions, suggesting that an unidentified signal(s) produced under starvation conditions is not essential for the formation of the homocomplex.
FIG. 4.
In vivo coimmunoprecipitation assay. Strains coexpressing GFP-tagged full-length KinA, ΔPAS-A, ΔPAS-AB, KinAC, or KinAN and FLAG-tagged full-length KinA, both under the control of the Phyper-spank promoter, were constructed and examined by a coimmunoprecipitation assay. A strain expressing GFP (with no tag) was used as a negative control. (Top) Schematic diagram for each construct and experimental design. (Center and bottom) Cells of each strain were induced to synthesize the tagged proteins by the addition of IPTG (10 μM) just after the cells were suspended in SM sporulation medium or LB medium. FLAG-tagged KinA was immunoprecipitated from crude extracts of cells at hour 2 after suspension by using anti-FLAG beads (Sigma). Samples of crude cell extracts (load) and immunoprecipitates (IP) were subjected to immunoblot analysis using antibodies that recognize FLAG (α-FLAG) and GFP (α-GFP). GFP-tagged proteins were detected after blotting of the resulting IP. The band intensity of each coimmunoprecipitated sample was normalized to the amount of FLAG-tagged KinA in the IP fraction and then to the amount of the GFP-tagged protein (or to that of GFP for the control) in the load fraction. The average of three independent results is shown as a percentage of the full-length KinA level, with error bars indicating the standard deviation (relative ratio). Strains used were MF3373 (KinA-FLAG and KinA-GFP) (lanes 1 and 7), MF3588 (KinA-FLAG and ΔPAS-A-GFP) (lanes 2 and 8), MF3589 (KinA-FLAG and ΔPAS-AB) (lanes 3 and 9), MF3374 (KinA-FLAG and KinAC-GFP) (lanes 4 and 10), MF3375 (KinA-FLAG and KinAN-GFP) (lanes 5 and 11), and MF3376 (KinA-FLAG and GFP) (lanes 6 and 12).
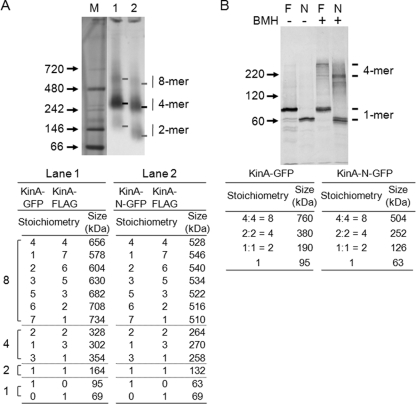
To characterize the homocomplex state of KinA, the complex formed in vivo was isolated in its native conformation by immunoprecipitation and was run on native gels. For this purpose, we used BN-PAGE. Analysis of the KinA complex by BN-PAGE resulted in three bands of different intensities. From migration distances of known molecular masses of marker proteins (Invitrogen) (Fig. 5A, lane M), the three bands found in the full-length KinA complex corresponded to dimeric (∼160-kDa), tetrameric (∼320-kDa), and octameric (∼640-kDa) states, respectively (Fig. 5, lane 1). For further confirmation, we took advantage of a truncated KinA mutant (KinAN) to analyze the formation of complexes, expecting different mobilities. Three bands, corresponding to a dimer (130 kDa), a tetramer (∼260 kDa), and an octamer (∼520 kDa), were detected (Fig. 5A, lane 2). All possible combinations of the complex formations with FLAG- and GFP-tagged proteins and their estimated molecular masses are shown at the bottom of Fig. 5A. The sizes of the complexes calculated from electrophoretic mobilities on the gel were essentially consistent with the estimated values. Nevertheless, the most intense band corresponded to a tetrameric state in both cases.
FIG. 5.
Analyses of the KinA protein complex by BN-PAGE and cross-linking. (A) Strains coexpressing GFP-tagged full-length KinA or KinAN and FLAG-tagged full-length KinA, both under the control of Phyper-spank, were constructed and examined by a coimmunoprecipitation assay. Cell extracts were prepared from strains MF3373 (KinA-FLAG and KinA-GFP) (lane 1) and MF3375 (KinA-FLAG and KinAN-GFP) (lane 2) grown in LB medium in the presence of IPTG (10 μM). The extracts were then processed for coimmunoprecipitation as described in the legend to Fig. 4. Samples of immunoprecipitates were separated by BN-PAGE as described in Materials and Methods. GFP-tagged full-length KinA and KinAN were detected with anti-GFP antibodies. A native protein marker kit (Invitrogen) served as molecular mass standards (lane M). Possible combinations of complex formation are listed, along with the molecular mass of each tagged protein. The individual monomeric forms were not detected, but the sizes are indicated for convenience. (B) Cell extracts were prepared from strains MF3352 (full-length KinA-GFP) (lanes F) and MF3360 (KinAN-GFP) (lanes N) grown in LB medium in the presence of IPTG (10 μM). The extracts were then processed for the BMH cross-linking reaction as described in Materials and Methods. Samples were separated by SDS-PAGE (10%), followed by immunoblot analysis with anti-GFP antibodies. −, no BMH; +, with BMH. Possible combinations of complex formation are listed, along with the molecular mass of each tagged protein.
To further characterize the complex, we performed a protein-protein cross-linking assay. Crude extracts were prepared from cells expressing GFP-tagged full-length KinA or KinAN and were incubated in the absence or presence of BMH, a protein cross-linker specific for free sulfhydryl groups (6). KinA has only five cysteines (C75, C125, C157, C319, and C359) that are localized within the N-terminal sensor domain and are cross-linkable with BMH. While it has been reported that mutation of KinA C75A increases in vivo sporulation efficiency only on a Schaeffer sporulation agar plate (37), the functions of these cysteine residues are not well characterized. After quenching of the cross-linking reactions, samples were resolved by 10% SDS-PAGE, followed by immunoblot analysis. As shown in Fig. 5B, without the cross-linker, full-length KinA and its N-terminal construct migrated as discrete bands with the molecular sizes expected for monomers. In contrast, the cross-linker yielded new sets of larger products that migrated with apparent sizes of 350 kDa for full-length KinA and 220 kDa for the N terminus, which corresponded approximately to the tetramer (Fig. 5B). Other, minor bands detected in the cross-linked samples might be due to inter- or intramolecular cross-linking formations in different ways, depending on the sulfhydryl involved. Nevertheless, these results from gel analyses suggest that KinA preferentially forms a homotetramer complex under physiological conditions.
Expression of the N- or C-terminal domain causes a dominant-negative effect on sporulation.
To extend the characterization of N- and C-terminus-mediated oligomer formation further, we attempted to produce full-length KinA in combination with the N- or C-terminal domain of KinA, predicting a dominant-negative effect. If KinA functions as a homo-oligomer, dominant-negative versions of a hetero-oligomer, composed of full-length and truncated KinA proteins, might be expected to abrogate Spo0A activation via phosphorelay. To examine this effect, we introduced the IPTG-inducible KinA mutant expression system into the kinB-null mutant background (Fig. 6A, bottom). The resulting strain has the IPTG-inducible KinA mutant system in addition to endogenous KinA. Cells were induced to sporulate by suspension in SM medium in the presence of 0.5 mM IPTG for overexpression of the mutant protein. It should be noted that wild-type KinA is expressed at physiological levels under the control of the endogenous promoter, while KinB is not expressed in the kinB-null background. Thus, the mutant form of KinA is present in excess under this culture condition, and its levels are supposed to be sufficient to form the hetero-oligomer and the mutant homo-oligomer predominantly. The results show that the efficiencies of sporulation of the strains overproducing the N or C terminus of KinA were impaired significantly relative to that of the control strain (Fig. 6A). The sporulation process was also monitored by the forespore-specific PspoIIQ-gfp reporter, and the results were consistent with those of the sporulation efficiency assay (Fig. 6A). For quantification of kinase activity, we introduced the PspoIIG-lacZ reporter into each strain. The β-galactosidase activity from the PspoIIG-lacZ reporter in strains overexpressing mutant KinA was significantly decreased (Fig. 6A). These data suggested that the N and C termini of KinA act as dominant-negative inhibitors of KinA function, reducing its signaling activities, likely because of the interaction between full-length KinA and the N- or C-terminal domain to form a hetero-oligomer, as demonstrated in the coimmunoprecipitation and BN-PAGE experiments (Fig. 4 and 5). Alternatively, the catalytic histidine residue (H405) of the C-terminal DHp domain in the mutant homo-oligomer might be phosphorylated by a reverse phosphotransfer reaction from the phosphorylated form of Spo0F, as demonstrated by in vitro experiments (47). This issue was investigated through the experiments described below.
FIG. 6.
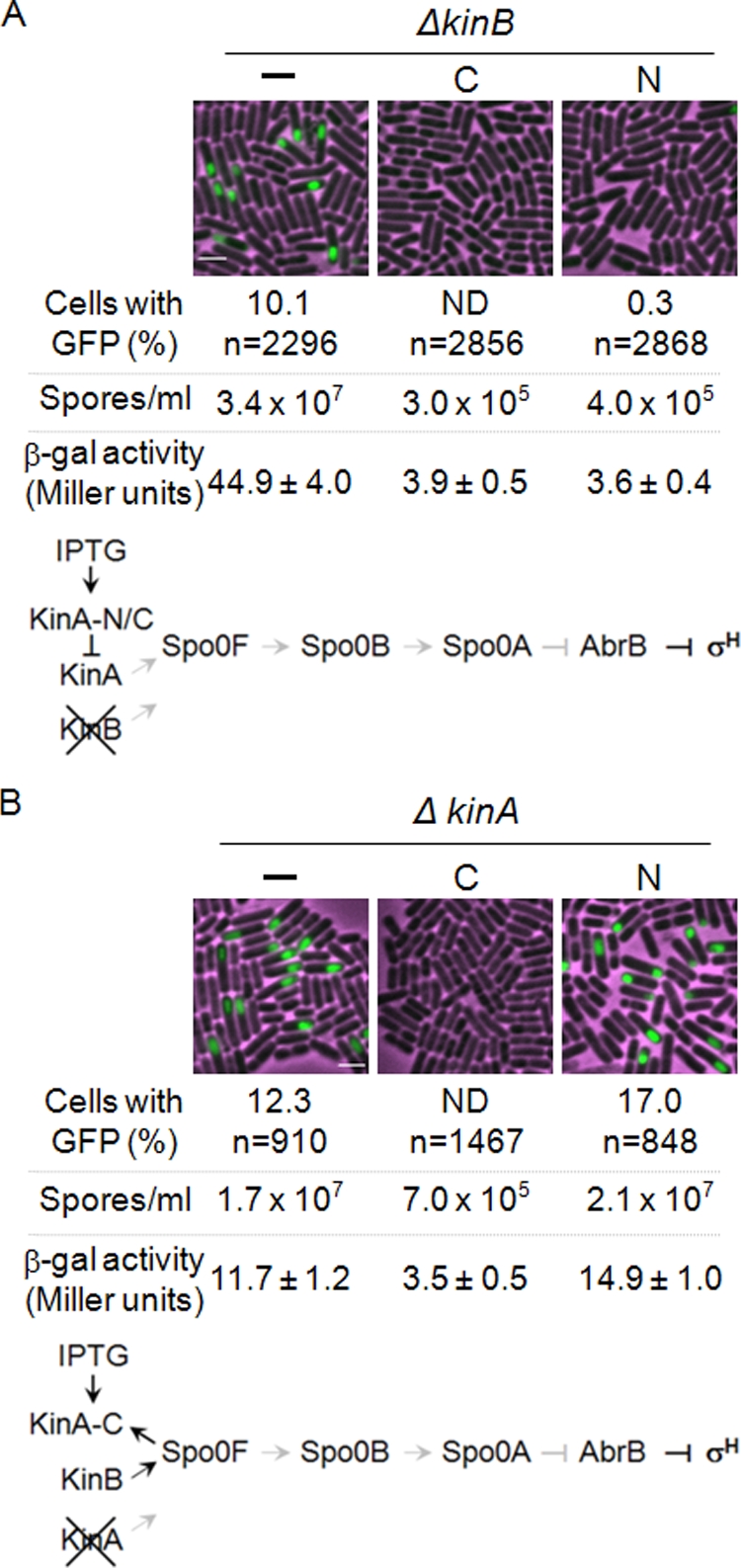
Effects of overexpression of the N- and C-terminal domains of KinA on sporulation. (A) Dominant-negative assay. The gene encoding the truncated KinAN or KinAC protein was fused to Phyper-spank and introduced at the amyE locus into cells containing the kinB-null mutation. The resulting strains have the gene for wild-type KinA at the endogenous locus and the engineered gene for the mutant at the amyE locus in the kinB-null background. Strains with the kinB-null mutation and lacking the IPTG-inducible construct were used as controls (−). C, KinAC; N, KinAN. Transcriptional reporters were constructed in which the genes encoding β-galactosidase (lacZ) and GFP were fused to the promoter region of the spoIIG operon to monitor Spo0A activity and to the promoter region of the spoIIQ operon to monitor forespore-specific σF activity, respectively. Individual reporters were transferred to the thrC locus of each strain and used for the β-galactosidase assay and GFP image analysis, respectively. The percentage of cells with the GFP signal and the total number of cells scored for GFP image analysis (n) are given. Sporulation efficiency was determined as described in Materials and Methods. The accumulation of β-galactosidase from a PspoIIG-lacZ fusion was monitored at hour 3 after suspension as a measure of Spo0A activity. Data for β-galactosidase are averages of three independent results with standard deviations. Strains used were MF3363 (control) (GFP assay), MF3366 (control) (β-galactosidase and sporulation assays), MF3364 (KinAC) (GFP assay), MF3367 (KinAC) (β-galactosidase and sporulation assays), MF3365 (KinAN) (GFP assay), and MF3368 (KinAN) (β-galactosidase and sporulation assays). ND, not detected. The genetic system that allows examination of the dominant-negative effects and the results are diagramed at the bottom. Arrows and T-shaped bars indicate positive and negative actions, respectively. Black and gray symbols represent active and inactive pathways, respectively. (B) Reverse phosphotransfer reaction assay. Each engineered gene construct, as described for panel A, was introduced at the amyE locus into cells containing the kinA-null mutation. The resulting strains have the gene for wild-type KinB at the endogenous locus and the engineered gene for the mutant at the amyE locus in the kinA-null background. Strains with the kinA-null mutation and lacking the IPTG-inducible construct were used as controls. Transcriptional reporters were constructed as described in the legend to panel A. Strains used were MF3386 (control) (GFP assay), MF2678 (control) (β-galactosidase and sporulation assays), MF3387 (KinAC) (GFP assay), MF2831 (KinAC) (β-galactosidase and sporulation assays), MF3388 (KinAN) (GFP assay), and MF3251 (KinAN) (β-galactosidase and sporulation assays). Bars, 2 μm. The genetic system that allows examination of the reverse phosphotransfer reaction and the results are diagramed at the bottom. We note that the sporulation efficiencies and β-galactosidase activities from the PspoIIG-lacZ reporter in the control strain were slightly lower in the presence of 0.5 mM IPTG than in its absence, while the strains shown in Fig. 3 displayed normal levels with or without 10 μM IPTG.
The truncated C-terminal catalytic domain mediates the reverse phosphotransfer reaction in the phosphorelay in vivo.
To distinguish between a dominant-negative effect and reverse phosphotransfer in vivo, we took advantage of the fact that sporulation is impaired but is still maintained at 5 to 10% of the wild-type level in the kinA-null mutant background under normal sporulation conditions in SM medium, as reported elsewhere (25, 44). In this situation, it is known that KinB acts as the alternative sporulation kinase and contributes to the remaining sporulation efficiency (25). Therefore, we constructed a genetic system in which the N- or C-terminal domain of KinA is expressed in the kinA deletion mutant in the presence of endogenous KinB (Fig. 6B, bottom). When the C-terminal domain of KinA was overexpressed in this genetic background, sporulation efficiency was reduced approximately 100-fold (7.0 × 105 spores/ml) relative to that of the control strain (1.7 × 107 spores/ml) (Fig. 6B). In contrast, overexpression of the N-terminal domain of KinA showed little or no inhibitory effect on sporulation efficiency (Fig. 6B). No physical interaction between KinA and KinB has been reported. Therefore, these results suggest that the C-terminal domain of KinA, perhaps as a homo-oligomer, accepts phosphate in a reverse phosphotransfer reaction from Spo0F∼P.
DISCUSSION
The principal contribution of this study is to provide in vivo evidence that (i) the cytosolic sensor kinase KinA forms a homo-oligomer under both nutrient-rich and nutrient-depleted conditions; (ii) the N-terminal sensor domain and C-terminal autokinase domain contribute to complex formation independently, but the former is more stable than the latter; (iii) a PAS-A domain deletion mutant (ΔPAS-A) is still able to induce sporulation to the wild-type level; (iv) the activity of the KinA hetero-oligomer consisting of full-length KinA and the N-terminal sensor domain is impaired in a dominant-negative manner; and (v) the truncated C-terminal autokinase domain accepts phosphate in a reverse phosphotransfer reaction from Spo0F∼P, leading to decreased Spo0A activity and thus impaired sporulation.
It has been proposed that the probable signal recognition and regulation region of KinA is within the sensor domain of the N-terminal 400 amino acids (aa) consisting of three PAS domains (PAS-A, -B, and, -C), yet how unidentified sporulation signals are processed and how processing is related to the activation of Spo0A are not well understood (37, 47). The intrinsic phosphorelay feedback loop in the process of initiation of sporulation has both negative and positive aspects. The network is directed by an estimated 10 positive and 5 negative regulations (36). Therefore, characterization of KinA had been difficult to perform by traditional genetic methods and thus relied heavily on in vitro studies (26, 37, 47). Until now, there have been discrepancies in the results of published, mostly in vitro, studies of the structure and function of KinA (26, 37, 47). Thus, to eliminate those ambiguities observed in the biochemical experiments, we developed a genetic system using a strain harboring IPTG-inducible KinA and reported that inducing the synthesis of the sensor kinase during growth triggers efficient sporulation (13). It should be emphasized that our artificial sporulation system does not overexpress the KinA protein but closely mimics the normal levels of KinA protein in the wild-type strain under nutrient deprivation conditions (13). As a useful extension of the previous system, we establish the artificial phosphorelay network in which the addition of the inducer causes efficient sporulation under normal sporulation conditions in the kinB deletion background and the initiation of sporulation is entirely dependent on IPTG. Thus, our artificial genetic system provides a unique and simple experimental setup to reduce the ambiguities in previous experimental procedures, since (i) functional assays for KinA and its mutants can be performed in vivo in the absence of KinB; (ii) the sporulation signal is defined as IPTG; (iii) the quantification of sporulation dynamics is simplified, because it is easy to control the precise time and cell density for the initiation of sporulation with IPTG addition; and (iv) the system is fully functional both with rich medium and under normal sporulation conditions. Therefore, the artificial phosphorelay system that is intended to decouple gene expression from the cell cycle can be useful for further characterization of the molecular interactions between the sensor kinase and the upstream or downstream components of the signaling network.
The traditional view of sporulation signal transduction has been that the sensor kinase(s) receives an unidentified ligand(s) of unknown origin that is produced under nutrient deprivation conditions; the sensor kinase then becomes active through a ligand-induced conformational change resulting in autophosphorylation (19, 21). In brief, the model asserts that the putative ligand(s) appears to be received by the N-terminal sensor domain, presumably PAS-A (26, 37, 47). The major discrepancy between the experimental results reported here and the results published previously (26) is that the ΔPAS-A construct (aa 151 to 606) used for the previous study has an 8-amino-acid deletion of the PAS-B domain (aa 143 to 150) (Fig. 2). In contrast, we use a ΔPAS-A construct comprising aa 136 to 606 in this study (Fig. 2; see also Materials and Methods). Using our system, we demonstrated that the previously published ΔPAS-A mutant (aa 151 to 606) showed decreased Spo0A activity and impaired sporulation efficiency relative to those of our ΔPAS-AB mutant (aa 259 to 606) (data not shown).
Several ligand candidates have been suggested in the literature, while in some cases their actual functions on the phosphorelay components have not been well characterized. These include small biological molecules (e.g., cis-saturated fatty acids) (41) whose levels might change in response to the cellular metabolic status. Furthermore, protein factors have been reported to modulate KinA activity. These include Sda (4, 9, 23), KipI (23, 48), and YheH, which putatively partners with YheI to form an ABC transporter (15). Sda and KipI have been characterized in detail, and they appear to work by binding to the autokinase domain, not the N-terminal sensor domain, and modulating KinA activity (23, 33, 51). The ABC transporter YheH/YheI has been identified as a KinA-interacting protein in a yeast two-hybrid study of B. subtilis protein-protein interactions (15). The disruption of either yheH or yheI, or both, had little or no effect on sporulation efficiency (15). Nevertheless, all those protein factors show inhibitory effects on sporulation only when artificially overexpressed. These results suggest that those protein factors act as fine-tuning devices rather than as primary regulators. Furthermore, the sensor domain of KinA appears not to be involved in sensing or interacting with those protein factors as ligands.
In the absence of the structure of full-length KinA, homology models based on other sensor histidine kinases, the complex consisting of Spo0F and Spo0B, and the complex consisting of Sda and the C-terminal catalytic domain of KinA have been used to predict the dimer structure of KinA and the mechanism of autophosphorylation (23, 33, 45, 51). These models suggest that the ATP binding site (CA domain) on one protomer is located close to the target His405 on the other protomer but distal to the corresponding His on the same polypeptide chain, thus explaining why autophosphorylation occurs in trans. Based on this trans-phosphorylation model, the C terminus of KinA forms a homodimer using the helix bundle of the DHp domain, where the ATP binding domain of one protomer phosphorylates the catalytic histidine H405 of the other protomer, as proposed for other sensor kinases (5, 32, 45, 46). Recently, structural analysis of the complex between Sda and the autokinase domain of the KinB dimer (1:2 stoichiometry) revealed that Sda interacts with a motif (GFXXL) that is highly conserved among all the sporulation kinases and is localized in the DHp domain (1). These results suggest that Sda inhibits all of the sporulation histidine kinases. However, all those structural studies use the C-terminal domain, termed the catalytic core of kinases in the literature, and show that the C-terminal domain forms a dimer (1, 26, 33, 51). Thus, complete structural analysis of kinases would be required to prove whether the dimer or the tetramer represents the natural structure under physiological conditions.
Using the coimmunoprecipitation assay, we confirmed for the first time that KinA exists as a homo-oligomer in vivo under both nutrient-rich and nutrient-depleted conditions (Fig. 4). Furthermore, we showed that both the N terminus (KinAN) and the C terminus (KinAC) of KinA form a hetero-oligomer with full-length KinA when coexpressed, and we found that the former is more abundant than the latter (Fig. 4). These results suggest that the N- and C-terminal domains independently contribute to the formation of the oligomer. Further analyses by use of BN-PAGE and cross-linker experiments show that the complex exists predominantly as a tetramer. We note that other phosphorelay components are not detected in the complex and that KinAN, which does not interact with Sda or KipI, behaves similarly to full-length KinA. These results suggest that the complex is a homotetramer of KinA. In support of our results, the PAS domain found in other sensor kinases in bacteria has been reported to be a tetramer under certain conditions (27, 35). Although BN-PAGE is widely used for the analysis of the molecular masses and oligomeric states of protein complexes, we note that the technique is not completely reliable, since the electrophoretic behavior of the protein species is dependent on the extent of binding with Coomassie brilliant blue and is influenced by the native shape of the protein (24). We also demonstrated that KinAN behaves in a dominant-negative fashion, impairing Spo0A activation (Fig. 6A). These results indicate that the formation of a hetero-oligomer between full-length KinA and the N terminus of KinA impairs sporulation. Thus, our results support the trans-phosphorylation model but as the homotetramer complex. To resolve this issue conclusively, we have to wait for structural information.
It has been reported previously using in vitro experiments that the C terminus of KinA (the autokinase domain) by itself, in the absence of the N-terminal PAS domains, cannot undergo autophosphorylation. However, it has been shown that the C terminus of KinA can accept Pi from Spo0F∼P through the reverse phosphotransfer reaction (47). Under our experimental conditions, shown in Fig. 6B, sporulation is induced by KinB at a slightly reduced efficiency in the kinA mutant, and Spo0F is phosphorylated mainly by the KinB pathway. Consistent with the biochemistry results reported by Wang et al. (47), our genetic data showed that sporulation was impaired by overexpression of KinAC, suggesting that the dominant-negative effect of the C terminus of KinA is due to the reverse phosphotransfer reaction from Spo0F∼P to the KinAC homooligomer (Fig. 6B). Thus, we propose that the oligomer conformation remains “open” in the absence of the N-terminal domain without autophosphorylation and provides room for docking Spo0F∼P to facilitate the reverse phosphotransfer reaction. Therefore, KinAC may accept phosphorylated Spo0F preferentially and transfer the phosphate moiety from Spo0F∼P to itself. As a result of the reverse phosphotransfer reaction, the limited phosphate supply from Spo0F∼P to Spo0B in the phosphorelay may cause decreased phosphorylation of Spo0A and finally result in a sporulation defect. Alternatively, KinAC might be able to interact with KinB by forming a hetero-oligomer, since the autokinase domains (DHp-CA) of the two kinases show 60% similarity (40% identity), and both kinases bind Sda, although no physical interaction between these two kinases has been detected by the systematic study of protein-protein interactions using yeast two-hybrid analysis (15; H. Yoshikawa, personal communication). It should be noted that the N or C terminus of KinA could be used as a potential inhibitor of sporulation. Thus, it will be interesting to see if similar effects occur in pathogenic sporulating bacteria, not only by use of the N-terminal domain, as reported elsewhere (50), but also by use of the C-terminal domain of the kinase.
In summary, we have used the IPTG-inducible KinA system to analyze the structure and function of the sensor kinase both under normal sporulation conditions and under nutrient-rich conditions. Our artificial system, which activates the phosphorelay to trigger sporulation irrespective of culture conditions, provides new insight into the role of the PAS domains in kinase activity. We conclude that the PAS-A domain is not required for kinase activity, whereas at least two PAS domains (PAS-BC) are sufficient to maintain enzyme activity. We also confirm that KinA functions as a homo-oligomer, predominately as a homotetramer. Furthermore, in the in vitro phosphorylation assay, the rate of KinA autophosphorylation is independent of the enzyme concentration (18). This in vitro result indicates that KinA, in a purified form, is constitutively active, excluding the possibility of signal involvement. Thus, based on these results, we propose that the primary role of the N-terminal sensor domain is to form a stable homo-oligomer, which is essential for catalytic activity but not for ligand binding. We therefore speculate that some other mechanism to modulate the level and activity of the kinase might be a critical contributor to the initiation of sporulation. Further experiments to clarify this point are in progress.
Supplementary Material
Acknowledgments
We thank Kottayil I. Varughese, Karon P. Cassidy, and Hirofumi Yoshikawa for valuable comments on the manuscript and Kazuo Kobayashi for the kinB mutant strain. We also thank Jean-Calvin Nguele, Mou Bhattacharya, Ashlee Dravis, Jeff Dinh, Daniel Duan, Esmeralda Ramirez-Pena, Joseph Amaya, and Steve Reyes for technical assistance and discussions.
This work was supported by the Texas Advanced Research Program (003652-0072-2007) and the Welch Foundation (E-1627).
Footnotes
Published ahead of print on 26 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bick, M. J., V. Lamour, K. R. Rajashankar, Y. Gordiyenko, C. V. Robinson, and S. A. Darst. 2009. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 386163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 1844881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64545-552. [DOI] [PubMed] [Google Scholar]
- 4.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104269-279. [DOI] [PubMed] [Google Scholar]
- 5.Cai, S. J., and M. Inouye. 2003. Spontaneous subunit exchange and biochemical evidence for trans-autophosphorylation in a dimer of Escherichia coli histidine kinase (EnvZ). J. Mol. Biol. 329495-503. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. L., J. J. Rosa, S. Turner, and R. B. Pepinsky. 1991. Production of multimeric forms of CD4 through a sugar-based cross-linking strategy. J. Biol. Chem. 26618237-18243. [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 17333-38. [DOI] [PubMed] [Google Scholar]
- 8.Crews, S. T. 1998. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 12607-620. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham, K. A., and W. F. Burkholder. 2009. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol. Microbiol. 71659-677. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34633-640. [DOI] [PubMed] [Google Scholar]
- 11.Ellermeier, C. D., and R. Losick. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 201911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 579-88. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, M., and R. Losick. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 192236-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 4327-38. [DOI] [PubMed] [Google Scholar]
- 15.Fukushima, S., M. Yoshimura, T. Chibazakura, T. Sato, and H. Yoshikawa. 2006. The putative ABC transporter YheH/YheI is involved in the signalling pathway that activates KinA during sporulation initiation. FEMS Microbiol. Lett. 25690-97. [DOI] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 18.Grimshaw, C. E., S. Huang, C. G. Hanstein, M. A. Strauch, D. Burbulys, L. Wang, J. A. Hoch, and J. M. Whiteley. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 371365-1375. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29477-508. [DOI] [PubMed] [Google Scholar]
- 20.Guérout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 18057-61. [DOI] [PubMed] [Google Scholar]
- 21.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47441-465. [DOI] [PubMed] [Google Scholar]
- 22.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3165-170. [DOI] [PubMed] [Google Scholar]
- 23.Jacques, D. A., D. B. Langley, C. M. Jeffries, K. A. Cunningham, W. F. Burkholder, J. M. Guss, and J. Trewhella. 2008. Histidine kinase regulation by a cyclophilin-like inhibitor. J. Mol. Biol. 384422-435. [DOI] [PubMed] [Google Scholar]
- 24.Krause, F. 2006. Detection and analysis of protein-protein interactions in organellar and prokaryotic proteomes by native gel electrophoresis: (membrane) protein complexes and supercomplexes. Electrophoresis 272759-2781. [DOI] [PubMed] [Google Scholar]
- 25.LeDeaux, J. R., N. Yu, and A. D. Grossman. 1995. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177861-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., D. R. Tomchick, C. A. Brautigam, M. Machius, R. Kort, K. J. Hellingwerf, and K. H. Gardner. 2008. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry 474051-4064. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., J. Xu, and M. Li. 1997. The human Δ1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J. Biol. Chem. 272705-708. [DOI] [PubMed] [Google Scholar]
- 28.Nambu, J. R., J. O. Lewis, K. A. Wharton, Jr., and S. T. Crews. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 671157-1167. [DOI] [PubMed] [Google Scholar]
- 29.Perego, M., S. P. Cole, D. Burbulys, K. Trach, and J. A. Hoch. 1989. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J. Bacteriol. 1716187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC.
- 31.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-518. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC.
- 32.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7626-633. [DOI] [PubMed] [Google Scholar]
- 33.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejewski, A. D. Grossman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13689-701. [DOI] [PubMed] [Google Scholar]
- 34.Rudner, D. Z., and R. Losick. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 161007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Söderbäck, E., F. Reyes-Ramirez, T. Eydmann, S. Austin, S. Hill, and R. Dixon. 1998. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol. Microbiol. 28179-192. [DOI] [PubMed] [Google Scholar]
- 36.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3561-566. [DOI] [PubMed] [Google Scholar]
- 37.Stephenson, K., and J. A. Hoch. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9815251-15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 11329-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 40.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30297-341. [DOI] [PubMed] [Google Scholar]
- 41.Strauch, M. A., D. de Mendoza, and J. A. Hoch. 1992. cis-unsaturated fatty acids specifically inhibit a signal-transducing protein kinase required for initiation of sporulation in Bacillus subtilis. Mol. Microbiol. 62909-2917. [DOI] [PubMed] [Google Scholar]
- 42.Szurmant, H., R. A. White, and J. A. Hoch. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trach, K. A., and J. A. Hoch. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 869-79. [DOI] [PubMed] [Google Scholar]
- 45.Varughese, K. I., I. Tsigelny, and H. Zhao. 2006. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J. Bacteriol. 1884970-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldburger, C. D. 2003. His-Asp signal transduction via a monomeric histidine phosphotransfer protein. Structure 111461-1462. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 1832795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., R. Grau, M. Perego, and J. A. Hoch. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 112569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J. Bacteriol. 173521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, A. K., J. A. Hoch, M. Grynberg, A. Godzik, and M. Perego. 2006. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J. Bacteriol. 1886354-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitten, A. E., D. A. Jacques, B. Hammouda, T. Hanley, G. F. King, J. M. Guss, J. Trewhella, and D. B. Langley. 2007. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 368407-420. [DOI] [PubMed] [Google Scholar]
- 52.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 121-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.