Abstract
Clostridium difficile, a major cause of antibiotic-associated diarrhea, produces highly resistant spores that contaminate hospital environments and facilitate efficient disease transmission. We purified C. difficile spores using a novel method and show that they exhibit significant resistance to harsh physical or chemical treatments and are also highly infectious, with <7 environmental spores per cm2 reproducibly establishing a persistent infection in exposed mice. Mass spectrometric analysis identified ∼336 spore-associated polypeptides, with a significant proportion linked to translation, sporulation/germination, and protein stabilization/degradation. In addition, proteins from several distinct metabolic pathways associated with energy production were identified. Comparison of the C. difficile spore proteome to those of other clostridial species defined 88 proteins as the clostridial spore “core” and 29 proteins as C. difficile spore specific, including proteins that could contribute to spore-host interactions. Thus, our results provide the first molecular definition of C. difficile spores, opening up new opportunities for the development of diagnostic and therapeutic approaches.
Clostridium difficile is a gram-positive, spore-forming, anaerobic bacterium that can asymptomatically colonize the intestinal tracts of humans and other mammals (3, 30, 39). Antibiotic treatment can result in C. difficile overgrowth and can lead to clinical disease, ranging from diarrhea to life-threatening pseudomembranous colitis, particularly in immunocompromised hosts (2, 4, 7). In recent years, C. difficile has emerged as the major cause of nosocomial antibiotic-induced diarrhea, and it is frequently associated with outbreaks (21, 22). A contributing factor is that C. difficile can be highly infectious and difficult to contain, especially when susceptible patients are present in the same hospital setting (13).
Person-to-person transmission of C. difficile is associated with the excretion of highly resistant spores in the feces of infected patients, creating an environmental reservoir that can confound many infection control measures (29, 44). Bacterial spores, which are metabolically dormant cells that are formed following asymmetric cell division, normally have thick concentric external layers, the spore coat and cortex, that protect the internal cytoplasm (15, 42). Upon germination, spores lose their protective external layers and resume vegetative growth (24, 27, 36). Bacillus spores and the spores of most Clostridium species germinate in response to amino acids, carbohydrates, or potassium ions (24, 36). In contrast, C. difficile spores show an increased level of germination in response to cholate derivatives found in bile (40, 41). Thus, spores are well adapted for survival and dispersal under a wide range of environmental conditions but will germinate in the presence of specific molecular signals (24, 36).
While the spores of a number of Bacillus species, such as Bacillus subtilis and Bacillus anthracis, and those of other Clostridium species, such as Clostridium perfringens (15, 20), have been well characterized, research on C. difficile spores has been relatively limited. A greater understanding of C. difficile spore biology could be exploited to rationalize disinfection regimes, molecular diagnostics, and the development of targeted treatments such as vaccines. Here we describe a novel method to isolate highly purified C. difficile spores that maintain their resistance and infectious characteristics, thus providing a unique opportunity to study C. difficile spores in the absence of vegetative cells. A thorough proteomic and genomic analysis of the spore provides novel insight into the unique composition and predictive biological properties of C. difficile spores that should underpin future research into this high-profile but poorly understood pathogen.
MATERIALS AND METHODS
Strains and growth conditions.
Clostridium difficile strain 630 (tcdA+ tcdB+; epidemic strain isolated in 1985 from Zurich, Switzerland; 012 ribotype) was routinely grown either in brain heart infusion (BHI) broth, on BHI agar plates (both from Oxoid Limited, Basingstoke, United Kingdom), or in Wilson's broth (45) at 37°C under anaerobic conditions (Mini-Mac 250; Don Whitley, United Kingdom). To enumerate total C. difficile organisms, samples from cultures were removed from the anaerobic cabinet, serially diluted in phosphate-buffered saline (PBS), plated onto BHI agar plates containing 0.5% taurocholate, and immediately returned to the anaerobic cabinet. To enumerate spores, 0.1-ml samples were mixed with 0.1 ml of 100% ethanol for 1 h at room temperature to kill vegetative cells (5). Samples were pelleted and washed twice in PBS before resuspension in 0.1 ml of PBS. Spores were enumerated by serially diluting in PBS and plating onto BHI agar plates containing 0.5% taurocholate. C. difficile was grown for 24 to 48 h at 37°C under anaerobic conditions.
Spore purification.
Prior to spore purification, C. difficile was grown statically in 500 ml of Wilson's broth (45) under anaerobic conditions for 7 to 10 days. Cultures were pelleted by centrifugation at 10,000 rpm for 15 min in a Sorvall RC 5C Plus centrifuge using an SLA 3000 rotor and were resuspended in 35 ml of sterile water. Samples were washed in water four to six times, during which the supernatant became clear. Samples were then pelleted and resuspended in 30 ml of PBS prior to sonication for 90 s at 35% amplitude in a VibraCell sonicator (Sonics, Newton, CT) with a tapered probe (model CV33). Then 3 ml of 10% Sarkosyl NL30 (VWR International Ltd., Poole, United Kingdom) was added to the samples before incubation at 37°C with agitation for 1 h. Samples were then pelleted by centrifugation at 4,000 rpm for 10 min in a Sorvall Legend RT centrifuge. Pellets were resuspended in 10 ml of PBS containing 0.125 M Tris buffer (pH 8) and 10 mg/ml lysozyme (Roche, Mannheim, Germany) and were incubated overnight (∼16 h) at 37°C with agitation. Samples were then sonicated as described above, and 1% Sarkosyl was added to the samples prior to incubation for 1 h at 37°C with agitation. Samples were then layered onto 50% sucrose and were centrifuged for 20 min at 4,000 rpm in a Sorvall Legend RT centrifuge. Pellets were resuspended in 2 ml of PBS containing 200 mM EDTA, 300 ng/ml proteinase K (recombinant PCR grade; Roche, Mannheim, Germany), and 1% Sarkosyl NL30 (VWR, Poole, United Kingdom) and were incubated for 1 to 2 h with shaking at 37°C until the samples cleared. Samples were then layered onto 50% sucrose and centrifuged as described above. Pellets were resuspended in water and were washed twice prior to resuspension in sterile water. Spores were stored at 4°C. Typically, the original 500-ml culture contained 5 × 109 to 1 × 1010 viable C. difficile spores, of which ∼108 to 109 were ethanol resistant. The purification protocol typically recovered >90% of the input spores.
Endospore staining.
C. difficile cultures or pure spores were stained according to the directions of the Schaeffer and Fulton spore stain kit (Sigma, United Kingdom). Stains were viewed on a Zeiss Axiovert 200m microscope equipped with a charge-coupled device camera.
Spore inactivation and germination assays.
The concentration of the spore preparation was determined by visual enumeration of serial dilutions using a hemocytometer. The spore preparations were devoid of vegetative cells, so the spores were easily recognized by their characteristic phase-bright appearance (23).
Spore germination and outgrowth.
Pure spores were serially diluted and plated onto BHI agar plates (germination background control). Preliminary experiments demonstrated that C. difficile spores germinated equally well on BHI, Wilson's, and Brazier's agar plates. For experiments, the BHI agar plates were supplemented with either cholate, taurocholate, or glycocholate at a concentration of 0.1%. C. difficile was grown for 48 h before counting.
Heat and chemical inactivation.
To test for heat resistance, pure spores were placed in water in a total volume of 0.5 ml. Samples either were placed in a heating block set to 60°C or 70°C or were left at room temperature. At 0.5, 3, and 24 h, samples were removed and enumerated by culturing as described above. To test for chemical resistance, pure spores were placed in either 70% ethanol (VWR, United Kingdom) or 1% Virkon (a strong oxidizing agent with the active compound potassium peroxymonosulfate) (Antec International, Suffolk, United Kingdom) in a total volume of 0.5 ml for 20 min. Treated spores were pelleted by centrifugation and were washed in sterile water before enumeration as described above.
Electron microscopy.
Pelleted spores were resuspended in a freshly prepared primary fixative that contained 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.42) with added magnesium and calcium chloride at 0.1% and 0.05%, respectively, at 20°C for 10 min before transfer to an ice bath for the remainder of 2 h. The spores were pelleted again at 3,000 rpm and were rinsed three times, for 10 min each time, in sodium cacodylate buffer with added chlorides on ice. Secondary fixation with 1% osmium tetroxide in sodium cacodylate buffer only was carried out at room temperature for 1 h. All subsequent steps were performed at room temperature. Spores were rinsed three times in cacodylate buffer over 30 min and were mordanted with 1% tannic acid for 30 min, followed by a rinse with 1% sodium sulfate for 10 min. The samples were dehydrated through an ethanol series of 20%, 30% (staining en bloc with 2% uranyl acetate at this stage), 50%, 70%, 90%, and 95% for 20 min at each stage and then were subjected to 100% ethanol three times (for 20 min each time). This step and all subsequent steps were carried out on a rotator to aid infiltration through the spore coat. Ethanol was exchanged for propylene oxide (PO) twice, for 15 min each time, followed by a 1:1 mixture of PO and Epon resin for at least 1 h and undiluted Epon (with a few drops of PO) overnight. The spores were embedded in a flat molded tray with fresh resin and were cured in an oven at 65°C for 24 h. Sections (thickness, 40 nm) were cut on a Leica UCT ultramicrotome, contrasted with uranyl acetate and lead citrate, and imaged on a 120-kV FEI Spirit BioTWIN transmission electron microscope using an F415 Tietz charge-coupled device camera.
Infective dose determination for environmental spore contamination.
Spores from purified stock or from the feces of mice shedding high levels of C. difficile 630 were suspended in 10 ml of sterile PBS at levels of ∼5, 50, 500, 5,000, 50,000, and 500,000 spores per 10 ml. To enumerate spores from feces, fecal pellets were suspended in PBS to 100 mg/ml. These samples were mixed 1:1 with 100% ethanol for 1 h to kill the vegetative cells. Samples were then washed in sterile PBS and were then serially diluted and plated as described for pure spore preparations.
Within a sterile biosafety cabinet, each spore suspension was poured into a sterile polysulfone cage (without bedding) and spread evenly to cover the entire bottom surface (800 cm2). Contaminated cages were left to dry overnight, resulting in a level of environmental spore contamination of 0.006, 0.06, 0.6, 6, 60, or 600 spores per cm2. Naïve mice (n = 3) were aseptically (i.e., in a biosafety cabinet, while the technician was wearing a clean smock and gloves that had been disinfected with 2% Virkon) placed in each cage and left for 1 h. Mice were then aseptically removed and individually placed in sterile cages with standard bedding and food, and with water that contained 250 mg/ml of clindamycin. C. difficile-infected mice generally shed low levels of bacteria (∼500 CFU/g of feces). However, clindamycin treatment suppresses the intestinal microbiota and allows the clindamycin-resistant C. difficile 630 to overgrow the murine intestinal tract (T. D. Lawley et al., submitted for publication). After 4 days of clindamycin treatment, the feces of mice were cultured for C. difficile on selective Brazier's agar plates. Mice that had acquired C. difficile 630 from the contaminated cage would be shedding >108 CFU of C. difficile per g of feces, whereas mice that were not colonized would not be shedding C. difficile (detection limit, <101 CFU of C. difficile per g of feces). A fuller description of this murine infection model is given elsewhere (Lawley et al., submitted). All animal infections were performed in accordance with the United Kingdom Home Office Animals (Scientific Procedures) Act of 1986.
Spore protein solubilization and analysis.
Purified spore pellets were resuspended with NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) containing 10% 2-mercaptoethanol and were heated at 90°C for 10 min. The sample was centrifuged at 14,000 rpm for 15 min; the supernatant is referred to as the LDS-soluble fraction (see Fig. 4b), and the pellet corresponds to the insoluble fraction. The insoluble pellet (Fig. 4b) was further treated with 8 M urea-1 M Na2CO3, incubated for 10 min at room temperature, and centrifuged as before. The supernatant from this centrifugation is referred to as the urea-soluble fraction (Fig. 4). The urea-insoluble pellet (Fig. 4) was saved for further processing as described below.
FIG. 4.
Summary of scheme to determine the spore proteome. (a) SDS-PAGE gel of vegetative cells (veg) (left lane) and pure spores (right lane) after solubilization with LDS. (b) Flow diagram outlining the steps required to solubilize proteins from C. difficile spores and the number of extra proteins liberated at each stage. (c) Rarefaction curve showing the decrease in the number of extra proteins with each additional solubilization step.
The two soluble fractions were separated in a 4 to 12% NuPAGE Bis-Tris gel (Invitrogen). Gels were stained overnight with colloidal Coomassie blue (Sigma). Each lane was excised into 20 bands that were in-gel digested with trypsin (sequencing grade; Roche) overnight. Peptides were extracted from gel bands twice with 50% acetonitrile-0.5% formic acid (FA) and were dried in a SpeedVac system.
The “urea-insoluble pellet” (described above) was washed three times with high-performance liquid chromatography-grade water to remove the detergent and was then treated with 60% FA for 1 h at 25°C. The suspension was neutralized with ammonium hydroxide solution (Fluka). Digestion was performed overnight with trypsin (modified sequence grade; Promega). The sample was centrifuged to remove any insoluble components, and the supernatant was termed the “tryptic digest” (Fig. 4b).
MS.
The extracted peptides were redissolved in 0.5% FA and were analyzed with on-line nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS-MS) on an Ultimate 3000 Nano/Capillary LC system (Dionex) coupled to an LTQ FT Ultra MS (ThermoElectron) equipped with a nanospray ion source. Samples were separated on an RP analytical column (bridged ethyl hybrid C18 column; 75 μm [inner diameter] by 10 cm) (Waters) for 45 min (for weak gel bands) or 60 min (for intense gel bands) at a linear gradient of 4 to 32% CH3CN-0.1% FA, or for 120 min for the pellet digest.
The MS was operated in the standard data-dependent acquisition mode controlled by Xcalibur, version 2.0. The instrument was operated with a cycle of 1 MS (in the Fourier transform ion cyclotron resonance cell) acquired at a resolution of 100,000 at m/z 400. The top three (for gel band extracts) or top five (for the pellet digest) most abundant multiply charged ions were subjected to MS-MS in the linear ion trap.
The raw data files were processed by BioWorks, version 3.3 (ThermoElectron), and were then submitted to a database search in the Mascot server, version 2.2 (Matrix Science, Ltd.), against an in-house-built C. difficile database by using the following search parameters: trypsin/P with three miscleavages, 20 ppm for MS, 0.5 Da for MS-MS, and nine variable modifications: acetyl (N-term), carbamidomethyl (C), deamidated (NQ), dioxidation (M), formyl (N-term), Gln→pyro-Glu (N-term Q), Glu→pyro-Glu (N-term E), methyl (E), and oxidation (M). Protein identifications were filtered using a 5% false discovery rate per peptide from a reverse database search in Mascot, combined with manual checking for single-peptide match-based protein identifications.
Bioinformatic analysis.
Whole-genome and spore proteome contents were analyzed using Artemis (8). Reciprocal FASTA analysis was performed as previously described (33) using the genomes of Clostridium acetobutylicum (AE001437), C. bartlettii (B0A9S8), C. beijerinckii (CP000721), C. botulinum (AM412317), C. cellulolyticum (CP001348), C. kluyveri (CP000673), C. novyi (CP000382), C. perfringens (CP000246), C. phytofermentans (CP000885), C. tetani (AE015927), C. thermocellum (CP000568), and B. subtilis (AL09126).
RESULTS
Purification of spores from C. difficile 630.
Since little is known about the structure and composition of C. difficile spores, we developed a protocol for their purification. C. difficile 630 was selected for the initial analysis, because the complete genome sequence is available (34). Routine culture experiments using C. difficile 630 grown statically for 5 to 7 days in Wilson's broth (45) showed that the strain reproducibly produced ∼106 to 107 spores/ml, which was 10- to 100-fold higher than the number produced by the same strain grown in BHI broth (data not shown). We therefore grew C. difficile 630 for 7 days in Wilson's broth prior to spore purification.
Initial attempts to purify C. difficile spores were guided by an established B. subtilis spore purification protocol (9). This protocol exploits serial washing of bacterial cells in cold water to lyse vegetative cells, followed by density gradient centrifugation to separate spores from the remaining vegetative cells and debris. Although we found that this protocol did enrich for C. difficile spores, there was significant contamination with dead vegetative cells and an unidentified extracellular matrix that appeared to facilitate the clumping of bacteria (Fig. 1a). Therefore, to isolate highly purified spores, we developed a protocol based on the sequential treatment of C. difficile cultures with detergent, lysozyme, sonication, and proteinase K, followed by sucrose layer centrifugation (see Materials and Methods). This protocol effectively and reproducibly yielded spores of high purity (>99.99% free from vegetative cells) (Fig. 1a and b).
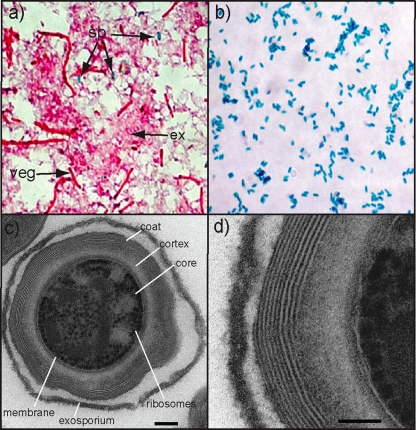
FIG. 1.
Visualization of pure C. difficile spores. (a) Endospore stain of C. difficile culture showing the pink vegetative cells (veg) and pink extracellular matrix (ex), with a few interspersed green spores (sp). (b) Purified C. difficile spores are stained green. (c) Transmission electron microscopy of sectioned C. difficile spores, demonstrating the spore ultrastructure including the exosporium, coat, cortex, core, membrane, and ribosomes. Bar, 100 nm. (d) Magnified section of outer surface of spore. Bar, 50 nm.
Transmission electron microscopic examination of spores showed that they were composed of an electron-dense outer surface with a spore coat structure composed of concentric inner rings (Fig. 1c and d). The outer surface of the spore coat often contained an exosporium (Fig. 1c). Within the spore coat was a clearer cortex region demarcated by a putative inner membrane that defines the core. Within the core were globular structures that appeared to be densely packed ribosomes, with intervening regions that likely contain condensed nucleoprotein complexes. A thorough examination of the microscope grids failed to identify any evidence of vegetative cells or obvious cell debris. Thus, using the novel protocol, we were able to isolate intact and well-defined C. difficile spores at a high level of purity.
Pure C. difficile spores are viable and maintain their resistant nature.
The number of spores yielded in individual preparations was routinely estimated by counting samples visually using a light microscope, because spores are easily recognized by their characteristic phase-bright appearance (23). Next, we sought to determine if the spores remained viable after exposure to the relatively harsh purification conditions. Compared to the number of spores estimated by visual counting, only ∼0.1 to 1% of the spores routinely germinated and grew out as colonies on BHI agar plates (Fig. 2a). However, by supplementing BHI agar with 0.1% of the germinant cholate, taurocholate, or glycocholate (40), we observed 100- to 1,000-fold increases in the germination rate and outgrowth efficiency. Hence, the purified spores remain viable and retain their sensitivities to cholate-based germinants (40, 45).
FIG. 2.

Pure C. difficile spores are viable and maintain their resistant nature. (a) Germination of pure C. difficile spores in response to cholate, taurocholate, or glycocholate. Spores were serially diluted and plated onto BHI agar plates containing 0.1% of the indicated germinant. Spores were also enumerated visually with a light microscope. (b) Heat resistance properties of spores. Spores were incubated in distilled water heated to 60°C or 70°C for the indicated times. (c) Chemical resistance properties of spores. Spores were incubated in water, 70% ethanol (EtOH), or 1% Virkon for 20 min at room temperature. After incubation, spores were serially diluted and plated onto BHI agar plates containing 0.5% taurocholate. Germinated spores were counted after 48 h of growth at 37°C under anaerobic conditions. The detection limit of 5 × 101 C. difficile CFU/ml is indicated by a dashed horizontal line.
The ability to prepare pure spores allowed us to quantitatively evaluate the sensitivities of spores to different chemical or physical treatments. Heating cultures of Clostridium or Bacillus spp. to 60°C for 10 min is a routine method of inactivating vegetative cells while retaining the spores in a viable state (23, 40). To test if purified C. difficile spores remain resistant to high temperatures, they were incubated in water heated to 60°C or 70°C. Significantly, C. difficile 630 spores remained completely viable when incubated at 60°C for as long as 24 h (Fig. 2b). Even at 70°C, high levels of C. difficile were recovered after 3 h of incubation, but these levels dropped to <1% of the input after 24 h (Fig. 2b). Further, we found that 70% ethanol had no obvious or reproducible effect on the viability of the spores, whereas the sporicidal agent Virkon (1%) efficiently inactivated the spores (Fig. 2c). Therefore, the purified spores are resistant to prolonged exposure to high temperatures and 70% ethanol but are effectively inactivated by sporicidal agents.
Environmental C. difficile spores are highly infective.
We next evaluated the host infectivity of purified spores by calculating the dose required to infect laboratory mice. To do so, we contaminated individual cages, without bedding, with differing concentrations of spores. Naïve recipient mice were aseptically placed in contaminated cages for 1 h; then these mice were placed individually in sterile cages for 4 days and were provided with water containing clindamycin. C. difficile 630 is resistant to clindamycin, so clindamycin treatment of mice that acquired spores from the contaminated cages triggers C. difficile overgrowth and high-level fecal shedding (>108 CFU of C. difficile/g of feces) (Lawley et al., submitted). In contrast, mice that did not acquire spores from contaminated cages would not shed any C. difficile organisms after clindamycin treatment.
Based on these experiments, we found that environmental spores infected mice in a dose-dependent manner (Fig. 3a). Using the Reed-Muench formulation (25), we determined that the dose required to infect 50% of the mice (ID50) under these conditions is ∼7 spores/cm2 (Fig. 3a). This ID50 is comparable to that determined from C. difficile 630 spores derived from the feces of C. difficile-infected mice (4 spores/cm2) (Fig. 3a). Importantly, infection of mice with pure spores resulted in asymptomatic intestinal carriage of C. difficile (∼500 CFU of C. difficile/g of feces), which lasted several weeks after the cessation of clindamycin treatment (Fig. 3b). These results demonstrate that purified spores are highly infective to mice.
FIG. 3.
Environmental C. difficile 630 spores are highly infective. (a) Infective dose curves of pure and feces-derived C. difficile 630 spores in mice (n = 3) after exposure to environmental spore contamination. Based on the Reed-Muench formulation, the ID50 of environmental spores with a 1-h exposure is 4 spores/cm2 for fecal spores (gray lines) and 7 spores/cm2 for pure spores (black lines). Data are representative of two independent experiments. (b) Fecal shedding of C. difficile 630 from infected mice (n = 6) demonstrates that spores establish intestinal carriage in infected mice. The broken gray line represents the expected shedding pattern during the induction of high-level C. difficile shedding. The detection limit of 5 × 101 C. difficile CFU/g of feces is indicated by a dashed horizontal line.
Determination of the C. difficile spore proteome.
The proteome profiles resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of vegetative cells and purified spore preparations are shown in Fig. 4a. The proteome from vegetative cells shows a very dense banding pattern, because the protein composition is more complex than that for pure spores. Moreover, prominent polypeptide signatures do not overlap between the two samples, indicating that the profiles are distinct and therefore have very different compositions. To further define the structure and composition of purified C. difficile spores, an exhaustive proteomic analysis was performed by LC-MS-MS, as outlined in Fig. 4b. Spore proteins were extracted by two solublization steps, the first using standard NuPAGE LDS sample buffer (with 10% 2-mercaptoethanol) and the second using 8 M urea in 1 M Na2CO3. Polypeptides in these fractions were separated by SDS-PAGE and analyzed by LC-MS-MS. The first solubilization step produced ∼233 polypeptides, and the second step produced an additional 71 polypeptides (Fig. 4b). Direct tryptic digestion was performed on the remaining insoluble pellet, and analysis of this sample led to assignment of a further 32 polypeptides. Multiple treatment steps were used to ensure thorough analysis of the spore proteome (Fig. 4c). There are no obvious trends between spore protein solubility and functionality (see the color coding in Table S1 in the supplemental material). Overall, ∼336 proteins were identified as components of purified C. difficile 630 spores.
Genome mapping of the C. difficile spore proteome.
In order to analyze the spore proteome further, the spore-associated polypeptides were mapped onto the annotated C. difficile 630 genome (Fig. 5) (34). The genes encoding the spore-associated polypeptides are dispersed throughout the C. difficile 630 genome, with a notable cluster of ribosomal genes near the origin of replication (Fig. 5a). The proteomics data revealed three novel small proteins that were not previously identified in the annotated genome (34). The coding sequences (CDS) encoding these proteins were named CD1595A, CD2687A, and CD3439A.
FIG. 5.
Representation of the C. difficile strain 630 genome, highlighting genes encoding the spore proteome. (a) Circular representation. The concentric circles represent the following (from outside in): first and second circles, all CDS (transcribed clockwise and counterclockwise); third circle, locations of prophage (red), conjugative transposons (blue), and carbohydrate utilization (green) and cell wall (orange) loci; fourth and fifth circles, CDS that code for spore proteins (transcribed clockwise and counterclockwise); sixth circle, G+C content (plotted using a 10-kb window). (b) Linear representation of a 27.8-kb region of the C. difficile strain 630 genome that contains the genes encoding several spore proteins, including a hypothetical cysteine-rich protein (CD1067) that is abundant in C. difficile spores. Red horizontal lines above CDS indicate those that encode proteins found in mature C. difficile spores. The genes are color coded as follows: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface association; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical protein; brown, pseudogenes; pink, phage and insertion elements; gray, miscellaneous.
Classification of the spore-associated polypeptides according to the functional scheme used in the annotated genome revealed a number of trends (Fig. 6; see also Table S1 in the supplemental material). Of the 60 predicted proteins in the sporulation/germination class that are present in the entire genome (34), 14 (∼25%) were detected as spore-associated peptides (Fig. 6; see also Table S1 in the supplemental material). Many of the in silico-predicted sporulation proteins apparently absent from the purified spores are likely associated with the earlier stages of spore formation in other bacteria (42); hence, these would not necessarily be expected to be present in the mature C. difficile spore structure.
FIG. 6.
Representation of the distribution and abundance, by functional class, of C. difficile 630 spore proteins. Shown are the absolute number of proteins (filled bars, left axis) and the percentage of each functional class in spores relative to the total genome (shaded bars, right axis). Functional classes are described in reference 34.
The most common functional class of the spore proteome, constituting 25% of all the detected polypeptides, was translation (Fig. 6; see also Table S1 in the supplemental material). This is in agreement with the electron microscopic analysis, which revealed a high density of ribosomes in the core cytoplasm of the C. difficile spores. Interestingly, the majority (15 out of 16) of the total predicted chaperone complement of C. difficile was found to be present in the spore proteome (Fig. 6), suggesting the importance of maintaining the folded structure of proteins in these dormant bodies under harsh conditions and within the densely packed spore core. Proteases were also relatively abundant, with 16 of 40 annotated in the genome present (Fig. 6) (>2.5-fold overrepresentation). Stress response proteins, including rubrerythrins, tellurium resistance proteins, bacterioferritin, catalase, and superoxide dismutase, are present within the spore and may contribute to the protection of spores from oxidative stress during germination (see Table S1 in the supplemental material).
Proteins involved in regulation are underrepresented in the spore proteome (Fig. 6). The late-sporulation-specific sigma factor σG (SigG) was the only putative sigma factor detected, and peptides for only two predicted transcription factors were identified. These were CcpA (Fig. 5b), a homologue of the carbon catabolite repressor important for sporulation in C. perfringens (43), and CodY, the GTP- and branched-chain amino acid-activated pleiotropic transcriptional repressor of early-stationary-phase genes (10). The presence of CodY in C. difficile spores suggests that during germination the spore represses genes associated with sporulation and stationary-phase growth.
A total of 41 hypothetical proteins were detected in the proteome (Fig. 6), validating the annotation of these CDS (34). One of the more abundant spore proteins was the hypothetical protein CD1067, a 405-amino-acid cysteine-rich protein (10% Cys) encoded within a distinct genomic region containing several spore genes (Fig. 5b). Despite its presence at high levels in the spore preparations, coverage across the length of the CD1067 protein was distinctly nonuniform; peptides rich in cysteine residues were infrequently detected, regardless of the presence of 2-mercaptoethanol in the solubilization buffer. This suggests that these sulfur-containing residues may not be accessible to trypsin digestion and might be involved in extensive disulfide bridge formation or might be otherwise modified in the spore. CD1067 has an orthologue in C. bartlettii but in no other sequenced bacterium, including Clostridium or Bacillus spp. Interestingly, the N-terminal region of the protein encoded by CD1067 also shows limited similarity (52%) to a small cysteine-rich outer membrane protein, Chlamydia trachomatis OmcA, which provides structural integrity to the outer envelope of the infectious elementary body (1).
Metabolic proteins are well represented, with 34 classified in energy and 20 in central/intermediary metabolism (Fig. 6; see also Table S1 in the supplemental material). Anabolic pathways, such as purine, pyrimidine, and amino acid biosynthesis, are poorly represented, while many of the components of the catabolic glycolytic pathway were detected. Also present are enzymes required for the fermentation of amino acids via the Stickland reactions (16, 17, 19), including coenzyme A dehydrogenase enzymes and electron transfer proteins (CD1054 to CD1059) (18) (Fig. 5b). KEGG pathway analysis (http://www.genome.ad.jp/kegg) (see Table S1 in the supplemental material) demonstrated that nearly complete pathways for gluconeogenesis (production of glucose from pyruvate) and carbon fixation, as well as enzymes that convert d-lactate, phosphoenolpyruvate, and formate into pyruvate, are present within spores. Further, the nearly complete pentose phosphate pathway (production of ribose and NADPH from glucose) is also present (see Table S1 in the supplemental material).
In total, 39 of the spore-associated proteins are predicted to be secreted or surface associated (Fig. 6). In accordance with this observation, SecA, involved in the translocation of proteins across the cell membrane, and a foldase (PrsA) were also represented in the data set. Almost half of these (21 CDS) have no predicted function in the genome annotation. Of those that do, six are predicted substrate-binding proteins for ABC transporters. Only one transporter (CD2465), predicted to facilitate the import of amino acids, is found in the spore proteome. Interestingly, a subset of the surface-associated proteins contain functional domains that are generally associated with adherence to host cells, such as the collagen-like glycoprotein BclA (CD0332), two surface layer proteins (CD2791 and CD2793 [SlpA]) that contain cell wall binding domains, and a putative hemagglutinin/adhesin (CD0514) (see Table S1 in the supplemental material).
Comparison of C. difficile spore genes to other clostridial genomes.
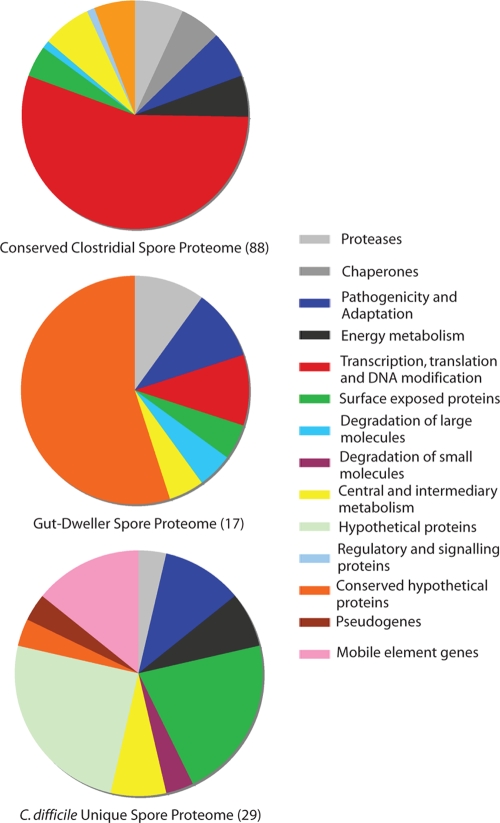
We performed reciprocal FASTA analysis to compare the C. difficile genes that encode the spore proteome to the sequenced genomes of 11 clostridial species, including C. acetobutylicum, C. bartlettii, C. beijerinckii, C. botulinum, C. cellulolyticum, C. kluyveri, C. novyi, C. perfringens, C. phytofermentans, C. tetani, and C. thermocellum. We found that 51 to 70% of the C. difficile spore proteome genes have orthologues in individual clostridia (see Fig. S1 in the supplemental material). However, only 26% (88 of 336) of the C. difficile spore proteome genes are well conserved among all the tested clostridial genomes (see Table S1 in the supplemental material). The majority of the C. difficile spore genes that are conserved in the 11 clostridial genomes encode proteins associated with translation, protein folding, and degradation and glycolysis (Fig. 7, top). The C. bartlettii genome contains the highest proportion of orthologues (70%) shared with the C. difficile spore proteome (see Fig. S1 in the supplemental material). C. bartlettii is a component of the human intestinal microbiota and is phylogenetically the closest available clostridial genome to C. difficile (16S rRNA genes place these two species in cluster XI [38]). Interestingly, among the 336 genes of the C. difficile spore proteins, 17 (5% of the C. difficile spore proteome) have orthologues only in C. bartlettii (Fig. 7, center). The majority of these genes encode proteins with no known function, including the C. difficile gene CD1067, encoding the cysteine-rich protein discussed above. However, many of the genes encode proteins associated with macromolecular degradation, such as putative chitinase/thioredoxin (CD1433), polysaccharide deacetylase (CD1319), peptidases (CD0684, CD3221, CD2246), and an amidohydrolase (CD2841) (see Table S1 in the supplemental material).
FIG. 7.
Comparative analysis of the C. difficile 630 spore proteome to 11 clostridial genomes. Diagrams summarize the C. difficile spore protein functional classes shared among clostridia (top), shared between C. difficile and C. bartlettii (center), and unique to C. difficile (bottom).
We found that 29 spore genes, representing 9% of the spore proteome, are currently unique to C. difficile relative to the representative clostridial genomes (Fig. 7, bottom; see also Table S1 in the supplemental material). Many of these genes encode hypothetical proteins. Interestingly, five genes unique to C. difficile are encoded within various mobile elements, including those from prophage 1 (CD0939 and CD0940), a genomic island (CD1898; putative phage endolysin), Tn5398 (CD2010; erythromycin resistance factor; note that CD2010 and CD2007 are identical, but the latter is likely not expressed due to a potential promoter deletion [12]), and prophage 2 (CD2926) (see Table S1 in the supplemental material), although no spore-associated genes mapped to any of the seven conjugative transposons (Fig. 5). These observations imply that the acquisition of some classes of mobile elements by C. difficile can contribute directly to the mature spore composition. Importantly, predicted surface-exposed proteins represent 17% (5 of 29) of the unique C. difficile spore proteome, including a putative exosporium glycoprotein, BclA1 (CD0332), the S-layer protein SlpA (CD2793) (11), and its paralogue CD2791.
DISCUSSION
Here we describe a novel protocol for the isolation of highly purified spores from cultures of a human-virulent C. difficile strain. Purified spores remain viable for extended periods, respond to cholate germinants, and retain their resistance to heat and ethanol. The availability of purified spores facilitated, for the first time, an estimate of the infectious dose of this pathogen, as assessed by an assay that mimics environmental spore contamination. Hence, the C. difficile spores were not irreversibly damaged or significantly altered by the purification process. This spore purification protocol will be valuable for purifying spores from other C. difficile strains, particularly those with genomes being sequenced (http://www.sanger.ac.uk/Projects/C_difficile), as well as for purifying spores from other clostridial species. The availability of pure C. difficile spores also facilitated the first detailed proteomic and bioinformatic analysis of spores from any Clostridium species. The spore proteome of ∼336 polypeptides defined here likely harbors proteins that contribute to the high level of resistance displayed by spores during host-to-host transmission and survival in the hospital environment, as well as those that stimulate germination and tissue colonization after entry into the mammalian intestinal tract.
C. difficile does not encode the classical germination receptors present in bacilli and many Clostridium species (33, 34). Perhaps this implies a novel mechanism of germination by C. difficile spores. C. difficile spores germinate in response to bile-associated cholate derivatives in vitro (Fig. 2) (40), suggesting that C. difficile spores germinate within the intestinal tract. Thus, C. difficile spores are likely to possess a novel receptor(s) for cholate-based compounds. There are 29 predicted cell surface proteins and 12 predicted transport/binding proteins within the spore-associated proteome that could directly engage with the extracellular environment and are therefore considered candidate germination receptors. The abundance of ribosome-associated proteins, translation factors, and RNA polymerases indicates that high levels of translation are required to drive primary biological processes upon germination. The presence of apparently intact ribosomes within the spore indicates that these molecular machines are robust enough to survive the purification process that destroys the vegetative cells. The profusion of chaperones in the C. difficile spore proteome probably plays a crucial role in preventing the unfolding of proteins during germination.
Analysis of the spore proteome suggests that amino acid fermentation (Stickland reaction), to produce energy, and pyruvate metabolism, to produce carbohydrates, are important early requirements for a germinating C. difficile spore. To supply the translation machinery and Stickland reactions (16, 17) with substrates, spores may rely on stored reserves of amino acids within intact proteins. This hypothesis is supported by the presence of proteases/peptidases within the spore proteome that could degrade spore proteins during the regeneration of vegetative cells. Indeed, Bacillus and Clostridium spores are known to degrade specific proteins during germination (31, 32, 37). Surprisingly, very few proteins within the spore are dedicated to DNA replication, cell division, and transcriptional regulation (Fig. 5). The lack of proteins involved in transcriptional regulation suggests that during germination the initial transcriptional patterns are limited and likely predetermined. Therefore, upon germination, C. difficile spores likely undergo a period of macromolecular degradation, amino acid fermentation, pyruvate metabolism, and translation prior to engaging in transcription, replication, and cell division. Interestingly, DNA replication initiates after a lag period upon germination of Bacillus megaterium spores (35).
One of the more abundant spore proteins was the cysteine-rich hypothetical protein CD1067. C. bartlettii is the only other bacterium that possesses an orthologue (42% identity), although we did note limited homology to the surface protein OmcA from the transmissible form of Chlamydia trachomatis (1). Interestingly, B. subtilis spores contain the cysteine-rich proteins CotY and CotZ (these have no detectable homology with CD1067) in their outer spore coat, which influence the accessibility of germinants to their receptors (46). Thus, it may be envisaged that a highly abundant and cysteine-rich surface protein could form a rigid, disulfide-linked mesh structure around the C. difficile spore under oxidative conditions (i.e., in the environment) but could become weakened under reduced conditions (i.e., in the intestinal tract) to facilitate germination. If so, CD1067 is a candidate marker for C. difficile spore detection. Further, unique surface-exposed proteins on spores are also excellent candidates for reverse vaccinology (28) to develop a protective vaccine against C. difficile. A prime antigen candidate for the C. difficile spore is the collagen-like protein BclA1, which has an orthologue in B. anthracis spores. Immunization with BclA protects animals from B. anthracis spore colonization by increasing opsonophagocytic uptake of spores and inhibiting intramacrophage germination (6, 14). Another candidate is the unique C. difficile surface layer protein SlpA, since it is present on the surfaces of vegetative cells (11) and also in spores of C. difficile.
Our previous bioinformatic analysis of the genome of C. difficile 630 has shown that many of the genes encoding classical spore proteins found in other bacteria are not obviously present in the C. difficile 630 genome (33, 34). For example, orthologues of only 18 of 70 B. subtilis spore coat proteins could be identified in the genome of C. difficile 630 (15). Our comparative genome analysis identified only 88 C. difficile spore genes for which orthologues are present in the genomes of 11 Clostridium species. These genes should represent the core spore genes that were inherited from the common ancestor of the clostridia. The majority of these genes encode proteins that function in the production, stability, and degradation of spore proteins, likely reflecting the importance of protein turnover during clostridial sporulation and germination.
The C. difficile 630 spore proteome contains several unique features that could be exploited for the detection and discrimination of C. difficile spores, providing candidate targets for future diagnostic tools. Current diagnostics is based on the detection of C. difficile toxins A and B in patient stool specimens with antibodies, generally to confirm C. difficile as the causative agent of diarrhea (26). Accurate detection of C. difficile spores from hospital patients, especially those excreting high levels of spores, and from the environment should improve infection control measures targeted at preventing C. difficile disease in hospitals.
Supplementary Material
Acknowledgments
We are grateful to the Washington University Genome Sequencing Center for permission to use the C. bartlettii genome data. We thank Neil Fairweather and Stephen Bentley for comments on the manuscript and Cordellia Brandt, Nicola Goodwin, and Claire Raisen for assistance with the animal experiments.
This work was funded by the Wellcome Trust and a Royal Society of London fellowship to T.D.L.
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, J. E., M. C. Cerrone, P. R. Beatty, and R. S. Stephens. 1990. Cysteine-rich outer membrane proteins of Chlamydia trachomatis display compensatory sequence changes between biovariants. Mol. Microbiol. 41543-1550. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. G. 2006. The new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 145758-764. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. G., T. Chang, N. S. Taylor, and A. B. Onderdonk. 1979. Colitis induced by Clostridium difficile. Rev. Infect. Dis. 1370-378. [DOI] [PubMed] [Google Scholar]
- 4.Borriello, S. P. 1998. Pathogenesis of Clostridium difficile infection. J. Antimicrob. Chemother. 41(Suppl. C)13-19. [DOI] [PubMed] [Google Scholar]
- 5.Borriello, S. P., and P. Honour. 1981. Simplified procedure for the routine isolation of Clostridium difficile from faeces. J. Clin. Pathol. 341124-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahmbhatt, T. N., S. C. Darnell, H. M. Carvalho, P. Sanz, T. J. Kang, R. L. Bull, S. B. Rasmussen, A. S. Cross, and A. D. O'Brien. 2007. Recombinant exosporium protein BclA of Bacillus anthracis is effective as a booster for mice primed with suboptimal amounts of protective antigen. Infect. Immun. 755240-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazier, J. S. 2008. Clostridium difficile: from obscurity to superbug. Br. J. Biomed. Sci. 6539-44. [DOI] [PubMed] [Google Scholar]
- 8.Carver, T., M. Berriman, A. Tivey, C. Patel, U. Bohme, B. G. Barrell, J. Parkhill, and M. A. Rajandream. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 242672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, W. H., D. Chen, Y. Q. Li, A. E. Cowan, and P. Setlow. 2007. How moist heat kills spores of Bacillus subtilis. J. Bacteriol. 1898458-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 11.Fagan, R. P., D. Albesa-Jove, O. Qazi, D. I. Svergun, K. A. Brown, and N. F. Fairweather. 2009. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 711308-1322. [DOI] [PubMed] [Google Scholar]
- 12.Farrow, K. A., D. Lyras, and J. I. Rood. 2000. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm(B) genes. Antimicrob. Agents Chemother. 44411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerding, D. N., C. A. Muto, and R. C. Owens, Jr. 2008. Measures to control and prevent Clostridium difficile infection. Clin. Infect. Dis. 46(Suppl. 1)S43-S49. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, U. K., R. Boehm, and W. Beyer. 2006. DNA vaccination against anthrax in mice—combination of anti-spore and anti-toxin components. Vaccine 244569-4571. [DOI] [PubMed] [Google Scholar]
- 15.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61555-588. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, S., M. Calos, A. Myers, and W. T. Self. 2006. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 1888487-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson-Rosario, S., D. Cowart, A. Myers, R. Tarrien, R. L. Levine, R. A. Scott, and W. T. Self. 2009. Auranofin disrupts selenium metabolism in Clostridium difficile by forming a stable Au-Se adduct. J. Biol. Inorg. Chem. 14507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson, S., L. G. Burman, and T. Akerlund. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 1543430-3436. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J., D. J. Darley, W. Buckel, and A. J. Pierik. 2008. An allylic ketyl radical intermediate in clostridial amino-acid fermentation. Nature 452239-242. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., and B. A. McClane. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 4e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 23.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 1461106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moir, A. 2006. How do spores germinate? J. Appl. Microbiol. 101526-530. [DOI] [PubMed] [Google Scholar]
- 25.Ozanne, G. 1984. Estimation of endpoints in biological systems. Comput. Biol. Med. 14377-384. [DOI] [PubMed] [Google Scholar]
- 26.Planche, T., A. Aghaizu, R. Holliman, P. Riley, J. Poloniecki, A. Breathnach, and S. Krishna. 2008. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect. Dis. 8777-784. [DOI] [PubMed] [Google Scholar]
- 27.Plomp, M., T. J. Leighton, K. E. Wheeler, H. D. Hill, and A. J. Malkin. 2007. In vitro high-resolution structural dynamics of single germinating bacterial spores. Proc. Natl. Acad. Sci. USA 1049644-9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappuoli, R. 2000. Reverse vaccinology. Curr. Opin. Microbiol. 3445-450. [DOI] [PubMed] [Google Scholar]
- 29.Riggs, M. M., A. K. Sethi, T. F. Zabarsky, E. C. Eckstein, R. L. Jump, and C. J. Donskey. 2007. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin. Infect. Dis. 45992-998. [DOI] [PubMed] [Google Scholar]
- 30.Rupnik, M. 2007. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin. Microbiol. Infect. 13457-459. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez-Salas, J. L., M. L. Santiago-Lara, B. Setlow, M. D. Sussman, and P. Setlow. 1992. Properties of Bacillus megaterium and Bacillus subtilis mutants which lack the protease that degrades small, acid-soluble proteins during spore germination. J. Bacteriol. 174807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Salas, J. L., and P. Setlow. 1993. Proteolytic processing of the protease which initiates degradation of small, acid-soluble proteins during germination of Bacillus subtilis spores. J. Bacteriol. 1752568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebaihia, M., M. W. Peck, N. P. Minton, N. R. Thomson, M. T. Holden, W. J. Mitchell, A. T. Carter, S. D. Bentley, D. R. Mason, L. Crossman, C. J. Paul, A. Ivens, M. H. Wells-Bennik, I. J. Davis, A. M. Cerdeno-Tarraga, C. Churcher, M. A. Quail, T. Chillingworth, T. Feltwell, A. Fraser, I. Goodhead, Z. Hance, K. Jagels, N. Larke, M. Maddison, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, B. White, S. Whithead, and J. Parkhill. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 171082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 35.Setlow, P. 1973. Deoxyribonucleic acid synthesis and deoxynucleotide metabolism during bacterial spore germination. J. Bacteriol. 1141099-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6550-556. [DOI] [PubMed] [Google Scholar]
- 37.Setlow, P., and W. M. Waites. 1976. Identification of several unique, low-molecular-weight basic proteins in dormant spores of Clostridium bifermentans and their degradation during spore germination. J. Bacteriol. 1271015-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, Y. L., C. X. Liu, M. McTeague, P. Summanen, and S. M. Finegold. 2004. Clostridium bartlettii sp. nov., isolated from human faeces. Anaerobe 10179-184. [DOI] [PubMed] [Google Scholar]
- 39.Songer, J. G., and M. A. Anderson. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 121-4. [DOI] [PubMed] [Google Scholar]
- 40.Sorg, J. A., and A. L. Sonenshein. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 1902505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorg, J. A., and A. L. Sonenshein. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 1911115-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30297-341. [DOI] [PubMed] [Google Scholar]
- 43.Varga, J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 1865221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vonberg, R. P., E. J. Kuijper, M. H. Wilcox, F. Barbut, P. Tull, P. Gastmeier, P. J. van den Broek, A. Colville, B. Coignard, T. Daha, S. Debast, B. I. Duerden, S. van den Hof, T. van der Kooi, H. J. Maarleveld, E. Nagy, D. W. Notermans, J. O'Driscoll, B. Patel, S. Stone, and C. Wiuff. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin. Microbiol. Infect. 14(Suppl 5)2-20. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, K. H. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 181017-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 1753757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.