Abstract
We used a novel atomic force microscopy (AFM)-based technique to compare the local viscoelastic properties of individual gram-negative (Escherichia coli) and gram-positive (Bacillus subtilis) bacterial cells. We found that the viscoelastic properties of the bacterial cells are well described by a three-component mechanical model that combines an instantaneous elastic response and a delayed elastic response. These experiments have allowed us to investigate the relationship between the viscoelastic properties and the structure and composition of the cell envelope. In addition, this is the first report in which the mechanical role of Lpp, the major peptidoglycan-associated lipoprotein and one of the most abundant outer membrane proteins in E. coli cells, has been quantified. We expect that our findings will be helpful in increasing the understanding of the structure-property relationships of bacterial cell envelopes.
The surface layers that isolate the interior of a bacterial cell from its external environment play a crucial mechanical role in the survival of the cell. They must be strong enough to maintain the cellular shape and resist turgor pressure yet, at the same time, be flexible enough to allow cell growth and division. Their elastic response is evident from their ability to recover from transient deformations, such as those induced by the incorporation of additional surface components (e.g., proteins) in response to changes in environmental conditions and the passage of small molecules across the cell boundary. It is therefore clear that understanding many aspects of cell physiology requires knowledge of the mechanical properties of cells.
The mechanical properties of the cell originate from the structural organization of the constituent lipids, sugar polymers, and proteins. Lipid molecules are brought together by their hydrophobic domains to form bilayers (membranes) that also incorporate different types of proteins. Polymeric strands of sugar molecules are typically cross-linked by flexible peptide molecules to form the peptidoglycan layer (27). Sometimes, an additional layer of proteins (S layer) is found on the outermost surface of the cell (7, 8, 40). Depending on the structural organization of the peptidoglycan and lipid bilayers, bacteria can generally be divided into gram-positive and gram-negative bacteria. In gram-positive cells, there is a relatively thick (20- to 35-nm) peptidoglycan layer that, together with the plasma membrane, sandwiches a viscous compartment called the periplasm (31, 32), whereas the envelope of gram-negative cells is made up of two lipid bilayers, the inner and outer membranes, separated by the periplasm, which contains a thin (3- to 8-nm) peptidoglycan layer (5, 33). In gram-negative bacteria, lipoproteins are associated with both the peptidoglycan layer and either the inner or outer membrane. Here, the “lipo” substituent is inserted into the hydrophobic domain of the membrane and the “protein” portion is linked to the peptidoglycan layer by either covalent or electrostatic bonds (18). Loss or altered expression of lipoproteins has been shown to affect cell shape generation and/or membrane integrity (10, 11, 13, 36, 43, 46), suggesting a possible mechanical role for these peptidoglycan-associated proteins.
Although the structure and chemistry of the gram-negative and gram-positive bacterial cell envelopes are well known, information about their mechanical properties has been difficult to elucidate. The simple stretching model used by Isaac and Ware (21) to describe the flexibility of bacterial cells indicated differences in the deformability of bacterial cells. Further advances in the characterization of the mechanical properties of bacterial cells were achieved by using bacterial threads, which are so-called macrofibers obtained from cultures of a cell-separation-suppressed mutant that were investigated by standard fiber-testing techniques (34, 48, 49). The requirement to use filament-forming mutants for this mechanical measurement has restricted the studies to date to the gram-positive bacterium Bacillus subtilis. In these studies, bacterial threads of B. subtilis were shown to be viscoelastic by performing creep experiments, a transient rheological technique in which a known force is applied to the material and the resulting extension (or deformation) is measured over time. The properties measured in these experiments were extrapolated to those of the individual cells, often with tenuous lines of inference. Recently, remarkable advances have been made in applying atomic force microscopy (AFM) to quantify the mechanical properties of individual microbial cells (15, 55). Typically, AFM force-indentation curves, which represent the relationship between a loading force and the depth of the indentation as the tip at the end of the AFM cantilever pushes onto the sample surface, are measured. Quantitative information on the elasticity of the sample is then obtained from the force required to achieve a certain depth of penetration (3, 16, 37, 51). It is only recently that direct creep measurements have become possible at the individual cell level by using AFM (50). In AFM creep experiments, the loading force is maintained at a constant value by controlling the cantilever deflection, while the displacement of the cantilever base generated by the sample response to the applied load is measured as a function of time. The sample creep response can be then analyzed with theoretical mechanical models to provide quantitative information on sample viscoelasticity.
In the present study, we used AFM creep experiments to probe and compare for the first time the local viscoelastic properties of individual gram-positive (B. subtilis) and gram-negative (Escherichia coli) bacterial cells. These experiments have allowed us to investigate the relationship between the viscoelastic properties and the structure and composition of the cell envelope. In addition, this is the first report in which the mechanical role of Lpp, the major peptidoglycan-associated lipoprotein and one of the most abundant outer membrane proteins in E. coli cells, has been quantified. We expect that our findings will be helpful in increasing the understanding of the structure-property relationships of bacterial cell envelopes.
MATERIALS AND METHODS
Bacterial strains, growth condition, and harvesting.
E. coli DLP79-22 (lpp+) (47) and E. coli DLP79-36 (a strain with a mutated lpp) (22) were kindly supplied by H. Tokuda, University of Tokyo, Tokyo, Japan. The E. coli strains, together with the B. subtilis 168 strain, were maintained on Trypticase soy agar (Becton, Dickinson and Company) and cultured in Trypticase soy broth (Becton, Dickinson and Company) at 37°C for 16 h on a rotary shaker (150 rpm) to late exponential growth phase. Bacteria were harvested by centrifugation (5 min at 1,150 × g), washed twice, and resuspended in deionized water.
Isolation of MVs and biochemical characterization.
Membrane vesicles (MVs) from E. coli DLP79-22 and DLP79-36 were isolated following the protocol described previously (23). The protein was quantified with a micro-bicinchoninic acid protein assay kit (Pierce Bioassay) using bovine serum albumin as the standard. Samples were assayed in triplicate. The amount of 3-deoxy-d-manno-octolusonic acid (Kdo) present in the samples was determined by the periodic acid/thiobarbituric acid method described by Hancock and Poxton (17) and assayed in triplicate, and the results are given as means ± standard deviations. Samples were hydrolyzed for 30 min. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis assessment of protein composition was performed using the method described by Kadurugamuwa and Beveridge (23) with 40 μg of protein loaded per lane. Broad-range molecular weight markers were purchased from Bio-Rad.
Transmission electron microscopy (TEM).
Whole-mount preparations and embedding of cells for thin sections were performed as previously described (41). The sections were poststained with 2% (wt/vol) uranyl acetate and Reynold's lead citrate (52). All samples were examined using a Philips CM10 transmission electron microscope operating at an acceleration voltage of 80 kV under standard operating conditions. Images were archived using the iTEM program (version 5.0; Soft Imaging Systems, Münster, Germany).
AFM sample preparation.
An important requirement for AFM investigations is that the sample be immobilized on a surface. For this purpose, an aliquot of bacterial suspension of ∼105 cells per ml was allowed to adhere through electrostatic interactions to a very thin layer of poly-l-lysine on a glass substrate that was prepared as previously described (57). After 15 min, the bacterium-coated glass substrate was rinsed with deionized water to remove loosely attached bacteria and then transferred to the AFM for immediate measurement.
AFM imaging and force data acquisition.
All AFM measurements were conducted at room temperature under Milli-Q water (resistivity, 18.2 MΩ-cm) using an Asylum MFP-3D atomic force microscope (Asylum Research, Santa Barbara, CA). Unless otherwise stated, imaging of the cells was performed in contact mode at low applied force (∼1 nN) at a scan rate of 1 Hz using Si3N4 V-shaped cantilevers that have a pyramid-shaped tip with a typical radius of curvature of 20 nm (OTR4; Veeco). Force measurements were performed using Si3N4 V-shaped cantilevers with colloidal silicon oxide tips with a tip radius of curvature of 300 nm (Novascan Technologies, Inc.). Prior to use, the spring constant of each cantilever was determined using the thermal fluctuation method (29). Typically, the cantilever spring constant was 0.07 ± 0.01 N/m. To determine the elastic and viscous contributions to the mechanical properties of the cells, force-time curves were collected at the center of the tops of individual cells. Data were not collected near the edges of the bacterial cells, since slipping of the AFM tip can occur, which is known to give rise to inaccurate results (3). For force-time curve acquisition, the AFM tip was lowered at a constant rate of 72 nN/s toward the cell surface until a preset value of the loading force, F0, was reached. The loading force F0 was then held constant for a 10-s period by controlling the cantilever deflection, d, where F0 is equal to k × d and k is the cantilever spring constant, and the cantilever base displacement was measured by monitoring the vertical movement (Z) of the z-piezoelectric transducer. For samples such as bacterial cells that are less stiff than the cantilever, the deflection of the cantilever during the approach of the AFM tip to the sample can be assumed to result exclusively from the mechanical indentation of the cell (50). Because of this, the approach part of the force-time curves was used to generate force-indentation curves according to the method described previously (3, 38). The data collected during the time of contact between the AFM tip and the bacterial cell represents the time-dependent deformation of the bacterial cell in the presence of a constant loading force, i.e., the cell creep response, which is a distinct feature of viscoelastic materials. A more detailed description of force-time curve acquisition and interpretation can be found in A-Hassan et al. and Vadillo-Rodriguez et al. (1, 50). To investigate the dependence of the cell mechanical response on the magnitude of the loading force F0, the force was varied between 2 and 10 nN. Three force-time curves were collected per cell for each value of F0, and five cells from different cultures were studied. Thus, each set of the cell viscoelastic parameters reported in this study was calculated as the average of the values obtained from the analysis of 15 force-time curves for each experimental condition. The reproducibility of consecutive force-time curves obtained under the same experimental conditions indicated that there was no plastic deformation (i.e., irreversible deformation) taking place on the time scale of the measurements. As a control, a set of force-time curves were recorded on clean glass substrates for each experimental condition investigated.
Evaluation of the creep response: viscoelastic parameters.
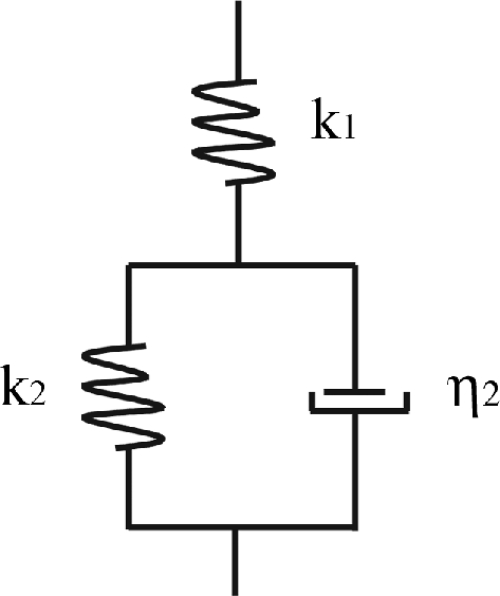
The viscoelastic behavior of materials can be modeled as combinations of elastic elements (springs) and viscous elements (dashpots) (53). These models are used to derive equations that describe the deformation of the material under investigation. One of the simplest models that predicts creep behavior is called the standard solid (14), which is shown schematically in Fig. 1. It consists of an elastic spring, which describes an instantaneous elastic deformation, placed in series with a parallel combination of a spring and a dashpot (Kelvin-Voigt element), which describes a delayed elastic deformation. We have used the standard solid model to interpret the creep data obtained for bacterial cells in the present study, since we obtained evidence for both an elastic and a delayed elastic response. We have not allowed for the possibility of viscous flow because the deformations observed in the present experiments are reversible to within the precision of the experiment. Based on the standard solid model, we have derived the following equation, which we have used to describe the experimentally observed creep response:
 |
(1) |
where Z(t) is the position of the z-piezoelectric transducer as a function of time (t), F0 is the magnitude of the loading force, k1 and k2 are the spring constants, and η2 is the viscosity characterizing the cell surface. The ratio η2/k2 is the so-called characteristic response time, τ, corresponding to the time during which the sample deforms by 1 − exp(−1) (or 63.2%) of the total creep deformation. The fits of the creep deformation data to this model were found to be very good, with linear correlation coefficient values that were close to 1 (R2 ≥ 0.96).
FIG. 1.
Schematic diagram of the standard solid model used to obtain the cell viscoelastic constants. The model consists of an elastic spring with stiffness k1, which describes the instantaneous elastic deformation, in series with a parallel combination of a spring with stiffness k2 and a dashpot with viscosity η2, which describe the delayed elastic deformation.
RESULTS AND DISCUSSION
TEM and AFM image analysis.
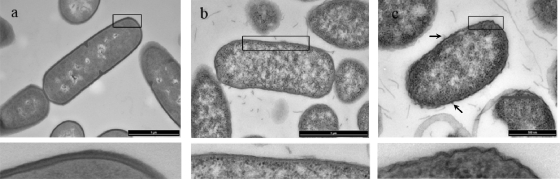
TEM micrographs of thin sections of a gram-positive bacterium, B. subtilis 168, and two gram-negative cells, E. coli (lpp+) and its Lpp mutant E. coli (lpp), are shown in Fig. 2a, b, and c, respectively, together with enlarged views of cross sections through their cell envelopes. The profound structural differences between gram-positive and gram-negative cells can be clearly observed. The peptidoglycan layer of B. subtilis 168 was ∼26 nm thick and amorphous in appearance. The outermost surface of the gram-negative cells presented the typical bilayered profile expected of membranes, with the peptidoglycan layer appearing as a thin electron-dense layer lying in close apposition to the inner surface of the outer membrane. In addition, while the outer surface of the gram-negative wild-type cell appeared smooth, that of its mutant showed uneven, small, irregular protuberances, some of which were observed to pinch off (Fig. 2c), forming MVs of various sizes. Whole-mount observations of the two strains indicated that the wild-type strain produced few MVs, whereas the mutant strain produced substantial amounts of MVs. The wild-type strain was shown to produce 118 ± 4 μg MV protein/liter of cell culture, whereas, in agreement with the visual observations made by TEM, the mutant strain produced a much larger concentration: 2,711 ± 109 μg MV protein/liter of cell culture, ∼23 times that of the wild-type strain. Previous observations of the relative quantities of MVs produced by other E. coli wild-type and lpp strains have been described (5, 11, 12, 42), with similar results reported. The morphological alteration of the lpp mutant strain indicated that the peptidoglycan-lipoprotein complex was involved in maintaining the integrity of the cell envelope structure (10). The total peptidoglycan-lipoprotein complex is a covalent structure in which the lipid is inserted into the hydrophobic domain of the outer membrane and the protein is covalently linked to the peptidoglycan. The link between the lipoprotein and the peptidoglycan is lysine, and trypsin digestion of this bond was shown by Braun and Rehn (10) to cause structural distortion of the E. coli cell envelope. From this point of view, the peptidoglycan-associated Lpp can be considered to be a true structural protein. Examination of Fig. 2b also indicates that the peptidoglycan-lipoprotein complex is evenly distributed over the entire cell surface layer.
FIG. 2.
Transmission electron micrographs of thin sections of B. subtilis 168 (a), E. coli (lpp+) (b), and E. coli (lpp) (c) cells. The bar shown in each image corresponds to 1 μm (a and b) and 500 nm (c). The arrows in panel c indicate membrane vesicles blebbing from the surface of the cell. The area enclosed by the rectangle shown in each image is shown in an enlarged view below each image.
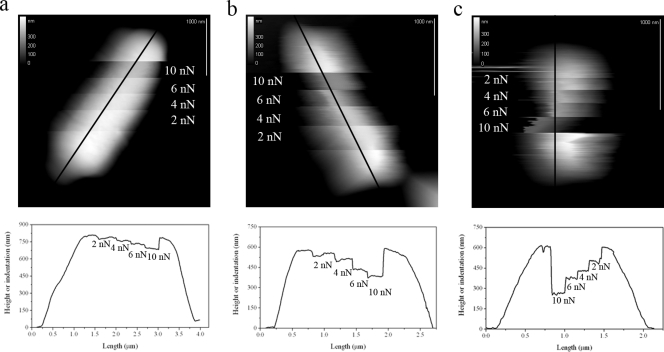
Individual cells of B. subtilis 168, E. coli (lpp+), and E. coli (lpp) strains were also imaged with AFM using pyramidal tips while increasing the loading force during the collection of the images (Fig. 3). These images are intended only to show the relative deformability of each bacterial strain, and they were not used to obtain quantitative mechanical deformation values; only measurements performed at the centers of the bacterial cells were used for quantitative analysis, as noted above. Nonetheless, these measurements revealed that, for the same loading force, the cell surface of the lpp mutant strain (see cross section in Fig. 3c) was more easily deformed than that of either its parent strain (Fig. 3b) or the gram-positive cell (Fig. 3a), with the latter being the least deformable of the three strains. Subsequent imaging of the same cells at low values of the loading force showed that, for all three cell types, the deformation previously induced was completely reversible, indicating that both gram-positive and gram-negative cells are capable of experiencing reversible, nondestructive deformations. The extent of the reversible deformation depends on the composition and interaction of the various structural components of the cell envelope. For instance, the peptidoglycan layer is a single, covalently linked macromolecular structure that has been shown to be quite elastic (25, 30, 56), a property that is mainly attributed to the flexibility of the tetrapeptide molecules that cross-link the rather rigid glycan strands. It is reasonable to assume that the elastic strength of the cells will likely depend on the thickness of the peptidoglycan layer, and thus, for the same values of the loading force, B. subtilis 168 was expected to be less deformable than the E. coli strains. The differences found in the levels of deformability between the E. coli strains further suggest that it is not only the thickness of the peptidoglycan layer, but also the bound form of the peptidoglycan-lipoprotein complex, which contributes to the rigidity of the cell envelope. Lipid bilayers, on the other hand, can deform only slightly (between 2 and 5%) before rupturing or buckling upon compression (24), and therefore, it is perhaps surprising that the cells can withstand the large deformations induced by the AFM tip without apparent disruption of their envelopes. It is important to note that large curvatures of the outer and inner membranes are known to occur naturally for bacterial cells, leading to cell division or to the formation of MVs (6, 24). Recent studies have proposed that membrane curvature in living cells is generated by specialized proteins that are bound to or inserted into the lipid bilayer (26, 39). The outer membrane of gram-negative cells and the inner membrane of gram-negative and gram-positive cells are known to contain numerous proteins, some of which have yet to be identified (4, 45). It is possible that some of these proteins provide bacterial cell membranes with the elasticity that is required for a variety of physiological processes to take place, such as growth and division. The values of the ratio of Kdo, a molecule found solely in lipopolysaccharide and therefore in the outer membranes, to protein indicated similarity between the wild-type and mutant E. coli cells (16 ± <1 μg Kdo/mg cell protein and 17 ± <1 μg Kdo/mg cell protein, respectively). In contrast, the MVs from the two strains were found to differ substantially—the parent strain MV contained 57 ± 2 μg Kdo/mg MV protein, and the Lpp mutant contained 669 ± 36 μg Kdo/mg MV protein and was indicative of alterations to the cell surface, as indicated by TEM and AFM images. More detailed analysis of the outer membrane chemistry would be beneficial to understanding whether the loss of Lpp has knock-on effects, which results in significantly altered chemistry, or whether the loss of this single protein simply causes these drastic alterations. This is even more important when one considers the impact of environmental conditions upon cell physiology (19). For example, under certain conditions, Lpp may be downregulated and the chemistry of peptidoglycan altered (2, 20), with possible implications for the regulation of MV production during pathogenesis.
FIG. 3.
Representative examples of AFM topographic images of individual cells of B. subtilis 168 (a), E. coli (lpp+) (b), and E. coli (lpp) (c) obtained in contact mode with an AFM pyramid-shaped tip by increasing the loading force F0 in a stepwise fashion. The vertical white bars in each image correspond to 1 μm. The cross sections shown below each topographic image correspond to the black lines shown in the images along the length of the cell. The images and cross sections are intended to show the relative deformability of each bacterial strain, and no quantitative data were obtained from these images.
EM and AFM imaging also showed that the E. coli (lpp) cells, lacking the structural link between the outer membrane and the peptidoglycan layer, are shorter than the wild-type E. coli (lpp+) cells. EM image analysis provided an average size of 1.3 ± 0.2 μm by 0.8 ± 0.1 μm for the E. coli mutant cells, whereas the average size was 2.1 ± 0.4 μm by 0.8 ± 0.1 μm for the E. coli wild-type cell. Nevertheless, the growth and division of the mutant strain was not inhibited or impaired, indicating that the bound form of this particular peptidoglycan-lipoprotein complex is dispensable for the vital process of cell growth and division.
Force-indentation curves: cell surface elasticity.
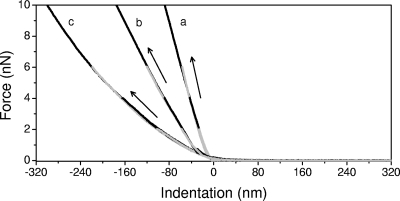
Elastic properties, i.e., reversible deformation of cells, can be quantified by measuring AFM force-indentation curves, which represent the relationship between the loading force and the depth of indentation as the AFM tip is pushed against the cell surface. An example of force-indentation curves for B. subtilis 168, E. coli (lpp+), and E. coli (lpp) is shown in Fig. 4 for applied force values of 2, 4, 6, and 10 nN. These curves were collected using a 600-nm-diameter sphere as the probe tip to minimize local strains at the point of contact and the possibility of damage to the cells. As expected for elastic materials, the relationship between the loading force and the indentation was linear for the bacterial cells, which allowed an estimation of the effective spring constant by calculating the ratio of the loading force to the depth of indentation (42). For the E. coli (lpp) cells, we observed a slight deviation from linearity in the loading-force-indentation behavior, but the deviation was small enough that we were able to reliably calculate the effective spring constant of the cells. The values of the effective spring constants were 0.103 ± 0.015 N/m, 0.045 ± 0.010 N/m, and 0.026 ± 0.006 N/m for B. subtilis 168, E. coli (lpp+), and E. coli (lpp), respectively, and these values were independent of the value of the loading force, F0. We note that the values reported in the literature for the stiffness of the surface of bacterial cells range between 0.016 and 0.053 N/m for gram-negative and gram-positive cells (35). The gram-positive B. subtilis 168 is ∼2.3 times stiffer than the E. coli wild type and ∼4.0 times stiffer than the E. coli mutant. Curiously, the ratio of the effective cell spring constants for B. subtilis 168 and the E. coli mutant closely corresponds to the ratio of the thicknesses of their respective peptidoglycan layers (∼3.9) (32, 33), suggesting that the elastic behavior of the cells might be dominated by the elastic nature of the peptidoglycan layer. However, the differences between the elastic constant values for the E. coli strains indicated that the link between the outer membrane and the peptidoglycan layer also contributes to the elasticity of the cells. The presence of the bound form of the peptidoglycan-lipoprotein complex increased the rigidity of E. coli cells by a factor of 1.7.
FIG. 4.
Approach force-indentation curves for B. subtilis 168 (a), E. coli (lpp+) (b), and E. coli (lpp) (c) cells for loading-force values F0 of 2, 4, 6, and 10 nN obtained by using colloidal AFM tips. The black arrows indicate the approach directions.
Although the effective elastic spring constant is the most commonly reported parameter to characterize the mechanical properties of cells (16, 33, 51, 56), it does not provide a complete description. As we will see below, bacterial cells are more generally described as viscoelastic materials.
Creep-deformation curves: cell surface viscoelasticity.
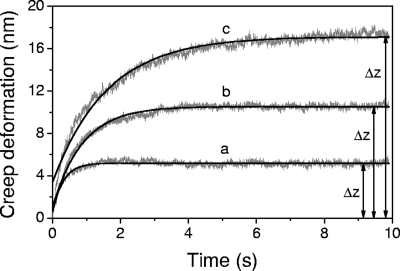
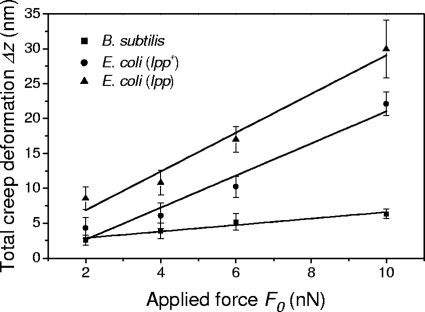
The data collected during the time of contact between the AFM tip and the bacterial cells in force-time curves demonstrate that the cells undergo a time-dependent deformation in response to a constant applied force, i.e., they creep. Because creep deformation is a distinct characteristic of viscoelastic materials, bacterial cells are more properly described as viscoelastic. An example of creep deformation for B. subtilis 168, E. coli (lpp+), and E. coli (lpp) is presented in Fig. 5 for an applied force of 6 nN. It can be seen that, for all three cells, the creep deformation increased monotonically with time, asymptotically approaching an equilibrium value, Δz. This behavior was observed for all of the values of the loading force F0 used in this study. We also found that the overall deformation, Δz, generated during the 10-s period of contact between the AFM tip and the cell surface was directly proportional to the loading force for each cell type investigated, as shown in Fig. 6, indicating that the experiments were performed in the linear viscoelastic regime.
FIG. 5.
Creep deformation as a function of time for B. subtilis 168 (a), E. coli (lpp+) (b), and E. coli (lpp) (c) cells obtained for an applied force F0 value of 6 nN using colloidal AFM tips. Note that the creep curves have been offset to begin at 0 nm, whereas for the mathematical analysis of these curves using equation 1, the first data point was taken as the indentation measured from the corresponding force-indentation curve after the desired load F0 was reached. The total creep deformation experienced by the cells is indicated in the plot as Δz.
FIG. 6.
Total creep deformation Δz as a function of the loading force F0 for B. subtilis 168, E. coli (lpp), and E. coli (lpp+) cells measured using a colloidal AFM tip. The straight lines were calculated using the best-fit parameter values for each data set. The error bars correspond to the standard deviations of the average values obtained from the analysis of 15 creep curves for each loading force and cell type investigated.
Since we obtained evidence for both an elastic and a delayed elastic response for the B. subtilis 168 and the two E. coli strains, we fitted the creep deformation data to the standard solid model to extract the cell viscoelastic parameters, i.e., k1, k2, and η2 as defined in equation 1 and Fig. 1. An example of a fit of typical creep deformation data to the standard solid model for each cell investigated is shown in Fig. 5. The average values of the best-fit parameters obtained for the different values of the loading force F0 are listed in Table 1 for the different cells, since the values do not vary significantly within the range of F0 values studied. This finding verifies that our experiments were carried out in the linear regime of viscoelasticity. The parameters of the standard solid model used to describe the creep response of the cells have well-defined physical meanings. The spring constant k1 is a measure of the effective spring constant of the cells and therefore determines the instantaneous elastic deformation that the cells undergo after the sudden application of a force, F0. It is for this reason that the values obtained for k1 for each of the cells studied is the same as the sample stiffness determined from the analysis of the force-indentation curves. The spring constant k2 and the dashpot viscosity, η2 characterize the delayed elastic deformation. The ratio η2/k2 is the response time τ, and it characterizes the time during which the delayed elastic deformation occurs. We found that the values of τ are 2.55, 1.12, and 0.91 s for B. subtilis 168, E. coli (lpp+), and E. coli (lpp), respectively, indicating that B. subtilis 168 cells reach the asymptotic creep deformation value Δz faster than the E. coli strains, with the lpp mutant taking the longest time to reach Δz. The value of the spring constant k2 also determines the magnitude of Δz and thus is a measure of the stiffness of the cells for the delayed elastic response. In this regard, B. subtilis 168 is stiffer in its delayed elastic response than the E. coli strains (see the values of k2 in Table 1, as well as the extent of the total deformation experienced by the cells during creep in the example shown in Fig. 5), and the lpp mutant is again the softer of the two E. coli strains. Note that the applied force in force-time curves was ramped at a constant rate of 72 nN/s. For the range of loading forces investigated (2, 4, 6, and 10 nN), this corresponds to a total loading time that varies between 0.03 and 0.14 s to achieve the desired load. This loading time is substantially shorter than the characteristic response time τ of the cells, and it is for this reason that the total deformation experienced by the cells in the AFM creep experiments can be separated into two contributions: an immediate elastic deformation defined by k1, followed by a delayed elastic deformation defined by the viscoelastic constants k2 and η2.
TABLE 1.
Summary of the best-fit viscoelastic constants of bacterial cellsa
| Parameter | Value
|
||
|---|---|---|---|
| B. subtilis 168 | E. coli (lpp+) | E. coli (lpp) | |
| k1 (N/m) | 0.10 ± 0.02 | 0.045 ± 0.010 | 0.026 ± 0.006 |
| k2 (N/m) | 1.2 ± 0.3 | 0.54 ± 0.10 | 0.33 ± 0.06 |
| η2 (N·s/m) | 3.0 ± 0.6 | 0.61 ± 0.28 | 0.30 ± 0.07 |
| τ (s) | 2.6 ± 1.1 | 1.1 ± 0.2 | 0.91 ± 0.30 |
The values were obtained from a least-squares fit of the experimental creep response of the cells to equation 1 derived from the standard solid model (Fig. 1). The parameters k1 and k2 are the stiffness values of the elastic springs, η2 is the viscosity of the dashpot, and τ is the characteristic response time defined as the ratio η2/k2. The average values of the best-fit parameters obtained for the different values of the loading force F0 (2, 4, 6, and 10 nN) have been listed, since the values do not vary significantly with the value of F0.
The contributions of the different cell surface layers to the mechanical properties of the cells can also be separated based on differences in their physical natures. For instance, cell stiffness, which is required to maintain shapes other than spherical, can only be a property of the cell wall. This was shown in a study of the disruption of rod-shaped bacteria with chemical treatments, which resulted in the formation of round, wall-less cells (i.e., spheroplasts), with the isolated peptidoglycan sacculi retaining the rod shape of intact cells (28, 54). Moreover, while spheroplasts are osmotically sensitive, isolated E. coli sacculi have been shown to reversibly expand or contract up to 300% from their relaxed state in response to electrostatic factors (25). Mechanical perturbation of isolated sacculi studied using AFM later confirmed that this surface layer is flexible and elastic, springing back readily to its original position after the external pressure applied by the AFM tip was removed. Spheroplasts were found to be so easily deformed by the AFM tip that it has not been possible to estimate an effective spring constant for the wall-less cells, i.e., they appeared softer than the soft AFM cantilevers typically used for the study of cells (44). Therefore, the results of the present study, as well as the previous studies mentioned above, led us to suggest that the elastic response of the cells is largely dominated by the physical nature of the peptidoglycan network. Moreover, it has been frequently suggested that the peptidoglycan-associated proteins found in E. coli might also contribute to the rigidity of the cells (9, 10, 46), as discussed above. In the present study, we found that, in the absence of the bound form of the lipoprotein, the envelope of the E. coli cells was 42% less rigid (see the values of the effective cell spring constant k1 in Table 1), and thus, for the first time, we have demonstrated quantitatively the in vivo mechanical role of the peptidoglycan-lipoprotein complex.
Although the elastic nature of the peptidoglycan layer and, to a lesser degree, the nature of the bonding between the peptidoglycan layer and the cell membranes likely dominate the elastic response of the cells, it is also necessary to assess the contribution of the membranes to the viscoelastic response of the cells. On the nanoscale, components such as lipids and proteins are in continual motion. It is this motion that provides membrane bilayers with their liquid-like character. Because of the inertial and viscous properties of the bilayer, it will provide resistance to deformation upon the application of an external force. This resistance to deformation likely introduces a temporal delay in the response of the cell to external forces, leading to the measured delayed elastic deformations. A number of properties affect membrane fluidity, including the bilayer thickness, the hydration state of the lipid head groups, interfacial polarity, and charge. Quantification of the delayed elastic response for the cells investigated in the present study showed that the gram-negative E. coli lpp mutant is less viscous than the wild type and thus offers less resistance to deformation (see the values of the viscous parameter η2 in Table 1). It has been estimated that about 250,000 lipoprotein molecules per E. coli cell are distributed 10.3 nm apart along the glycan strands of the peptidoglycan (9). Whereas the protein portion of these lipoproteins is covalently linked to the peptidoglycan, the lipid portion is deeply embedded in the hydrophobic domain of the outer membrane, strongly interacting with other membrane components. It is conceivable that this strong interaction could reduce the fluidity of the membrane, which is consistent with the higher viscosity values measured for the E. coli wild type than for the E. coli mutant. The gram-positive B. subtilis 168 cells were found to be 80 to 90% more viscous than the E. coli cells (see the values of η2 in Table 1). This is not an unexpected result, considering that the envelopes of gram-positive bacteria consist of only a single membrane, i.e., the plasma membrane, buried below a relatively thick peptidoglycan layer. The magnitude of the instantaneous elastic deformations due to the initial loading of the applied force F0 ranged between 25 and 84 nm for B. subtilis 168 cells for the range of loading forces investigated (see the indentation values in Fig. 4). This means that the deformations initially produced by the AFM tip are equal to or larger than the thickness of the peptidoglycan layer of this gram-positive cell, and hence, we are also probing the properties of its plasma membrane during the measurement.
In summary, we have demonstrated that gram-positive and gram-negative bacteria have a viscoelastic response to an external compressive force applied by an AFM tip. In particular, the cells exhibit a viscoelastic solid-like behavior that is characterized by an instantaneous elastic deformation followed by a delayed elastic deformation. Comparison of the results obtained for the different strains suggests that the instantaneous elastic response might be dominated by the properties of the peptidoglycan layer and the nature of its association with the membranes, whereas the delayed elastic response is more likely to arise from the liquid-like character of the cell membranes. It is remarkable that the influence of the peptidoglycan-lipoprotein Lpp was easily discerned in the viscoelastic response of E. coli cells, quantitatively corroborating for the first time the mechanical role of the peptidoglycan-lipoprotein complex. We envision that this type of measurement will play an important role in helping to elucidate structure-property relationships for bacterial cells. Ultimately, estimation of cell mechanical properties could become a valuable technique for testing the effects of drugs that target different components of the cell envelope.
Acknowledgments
We gratefully acknowledge financial support from the Advanced Foods and Materials Network (AFMnet), the Canadian Foundation for Innovation, and the Natural Sciences and Engineering Research Council of Canada. J.R.D. acknowledges support from the Canada Research Chairs (CRC) program. The TEM measurements were performed in the NSERC Guelph Regional Integrated Imaging Facility (GRIIF), which has been partially funded by an NSERC-MFA grant.
We thank Dianne Moyles for performing the TEM measurements.
This work is dedicated to the late Terry Beveridge, our good friend and superb colleague, who provided the motivation for this work.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.A-Hassan, E., W. F. Heinz, M. D. Antonik, N. P. D'Costa, S. Nageswaran, C. A. Schoenenbenger, and J. H. Hoh. 1998. Relative microelastic mapping of living cells by atomic force microscopy. Biophys. J. 741564-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anfora, A. T., B. J. Haugen, P. Roesch, P. Redford, and R. A. Welch. 2007. Roles of serine accumulation and metabolism in the colonization of the murine urinary tract by Escherichia coli CFT073. Infect. Immun. 755298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnoldi, M., C. M. Kacher, E. Bauerlein, M. Radmacher, and M. Fritz. 1998. Elastic properties of the cell wall of Manetospirillum gryphyswaldense investigated with atomic force microscopy. Appl. Phys. 66S613-S617. [Google Scholar]
- 4.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubès. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 1804872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1814725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beveridge, T. J. 1989. The structure of bacteria, p. 1-65. In J. S. Poindexter and E. R. Leadbetter (ed.), Bacteria in nature, vol. 3. Plenum Press, New York, NY. [Google Scholar]
- 8.Beveridge, T. J. 1981. Ultrastructure, chemistry, and function of the bacterial wall. Int. Rev. Cytol. 72229-317. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V. 1973. Molecular organization of the rigid layer and the cell wall of Escherichia coli. J. Infect. Dis. 128S9-S15. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., and K. Rehn. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10426-438. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., A. Bernadac, M. Gavioloi, J. C. Lazaron, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubes. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38904-915. [DOI] [PubMed] [Google Scholar]
- 13.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29359-367. [DOI] [PubMed] [Google Scholar]
- 14.Findley, W. N., J. S. Lai, and K. Onaran. 1989. Creep and relaxation of nonlinear viscoelastic materials with an introduction to linear viscoelasticity. Dover Publications, Inc., New York, NY.
- 15.Gaboriaud, F., and Y. Dufrene. 2007. Atomic force microscopy of microbial cells: application to nanomechanical properties, surface forces and molecular recognition. Colloids Surf. B 5410-19. [DOI] [PubMed] [Google Scholar]
- 16.Gaboriaud, F., S. Bailet, E. Dague, and F. Jorand. 2005. Surface structure and nanomechanical properties of Shewanella putrefaciens bacteria at two pH values (4 and 10) determined by atomic force microscopy. J. Bacteriol. 1873864-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, I., and I. Poxton. 1988. Bacterial cell surface techniques. John Wiley and Sons, Chichester, United Kingdom.
- 18.Hancock, R. E., W. R. Siehnel, and N. Martin. 1990. Outer membrane proteins of Pseudomonas. Mol. Microbiol. 41069-1075. [DOI] [PubMed] [Google Scholar]
- 19.Haugen, B. J. 2007. Characterization of the response of uropathogenic Escherichia coli strain CFT073 to d-serine. Ph.D. thesis. University of Wisconsin, Madison.
- 20.Haugen, B. J., S. Pellett, P. Redford, H. L. Hamilton, P. L. Roesch, and R. A. Welch. 2007. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect. Immun. 75278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaac, L., and G. C. Ware. 1974. The flexibility of bacterial cell walls. J. Appl. Bacteriol. 37335-339. [DOI] [PubMed] [Google Scholar]
- 22.Ito, H., A. Ura, Y. Oyamada, H. Yoshida, J. I. Yamagishi, S. I. Narita, S. I. Matsuyama, and H. Tokuda. 2007. A new screening method to identify inhibitors of the Lol (localization of lipoproteins) system, a novel antibacterial target. Microbiol. Immnol. 512663-2670. [DOI] [PubMed] [Google Scholar]
- 23.Kadurugamuwa, J., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 1773998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch, A. L. 1998. The biophysics of the gram-negative periplasmic space. Crit. Rev. Microbiol. 2423-59. [DOI] [PubMed] [Google Scholar]
- 25.Koch, A. L., and S. Woeste. 1992. Elasticity of the sacculus of Escherichia coli. J. Bacteriol. 1744811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlov, M. M. 2007. Bending over to attract. Nature 447387-388. [DOI] [PubMed] [Google Scholar]
- 27.Labischinski, H., G. Barnickel, H. Bradaczek, and P. Giesbrecht. 1979. On the secondary and tertiary structure of murein. Low- and medium-angle X-ray evidence against chitin-based conformations of bacterial peptidoglycan. Eur. J. Biochem. 95147-155. [DOI] [PubMed] [Google Scholar]
- 28.Lederberg, J. 1956. Bacterial protoplasts induced by penicillin. Proc. Natl. Acad. Sci. USA 42574-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, R., and M. Maaloum. 2002. Measuring the spring constant of atomic force microscope cantilevers: thermal fluctuations and other methods. Nanotechnology 1333-37. [Google Scholar]
- 30.Marquis, R. E. 1968. Salt-induced contraction of bacterial cell walls. J. Bacteriol. 95775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matias, V. R. F., and T. J. Beveridge. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 1881011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matias, V. R. F., and T. J. Beveridge. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56240-251. [DOI] [PubMed] [Google Scholar]
- 33.Matías, V. R. F., A. Al-Amoudi, J. Dubochet, and T. J. Beveridge. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 1856112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendelson, N. H., and J. J. Thwaites. 1989. Cell wall mechanical properties as measured with bacterial thread made from Bacillus subtilis. J. Bacteriol. 1711055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendez-Vilas, A., A. M. Gallardo-Moreno, and M. L. Gonzales-Martin. 2007. Atomic force microscopy of mechanically trapped bacterial cells. Microsc. Microanal. 1355-64. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F., C. Kirkham, and A. J. Lesse. 2006. Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect. Immun. 745169-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penegar, I., C. Toque, S. D. A. Cornell, J. R. Smith, and S. A. Campbell. 2001. Nano-indentation measurements of the marine bacteria Sphingomonas paucimobilis using the atomic force microscope, p. 5-15. In J. A. Lewis (ed.), 10th International Congress on Marine Corrosion and Fouling, University of Melbourne, February 1999: additional papers. Aeronautical and Maritime Research Laboratory, Melbourne, Australia.
- 38.Radmacher, M., M. Fritz, and P. K. Hansma. 1995. Imaging soft samples with the atomic force microscope: gelatin in water and propanol. Biophys. J. 69264-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynwar, B. J., G. Illya, V. A. Harmandaris, M. M. Muller, K. Kremer, and M. Deserno. 2007. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447461-464. [DOI] [PubMed] [Google Scholar]
- 40.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 1885945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shadwick, R. E. 1999. Mechanical design in arteries. J. Exp. Biol. 2023305-3313. [DOI] [PubMed] [Google Scholar]
- 43.Sonntag, I., H. Schwarz, Y. Hirota, and U. Henning. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan, C. J., S. Venkataraman, S. T. Retterer, D. P. Allison, and M. J. Doktycz. 2007. Comparison of the indentation and elasticity of E. coli and its spheroplasts by AFM. Ultramicroscopy 107934-942. [DOI] [PubMed] [Google Scholar]
- 45.Sundararaj, S., A. Guo, B. Habibi-Nazhad, M. Rouani, P. Stothard, M. Ellison, and D. S. Wishart. 2004. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 32D293-D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 1671-9. [DOI] [PubMed] [Google Scholar]
- 47.Terada, M., T. Kuroda, S. I. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 27647690-47694. [DOI] [PubMed] [Google Scholar]
- 48.Thwaites, J. J., and N. H. Mandelson. 1985. Biomechanics of bacterial walls: studies of bacterial thread made from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 822163-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thwaites, J. J., and N. H. Mendelson. 1991. Mechanical behaviour of bacterial cell walls. Adv. Microb. Physiol. 32173-222. [DOI] [PubMed] [Google Scholar]
- 50.Vadillo-Rodriguez, V., T. J. Beveridge, and J. R. Dutcher. 2008. Surface viscoelasticity of individual gram-negative bacterial cells studied using atomic force microscopy. J. Bacteriol. 1904225-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velegol, S. B., and B. E. Logan. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir 185256-5262. [Google Scholar]
- 52.Venable, J. H., and R. Coggeshall. 1965. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 25407-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, I. M., and D. W. Hadley. 1993. An introduction to the mechanical properties of solid polymers. John Wiley & Sons, Hoboken, NJ.
- 54.Weibull, C. 1953. The isolation of protoplasts from Bacillus megaterium by controlled treatment with lysozyme. J. Bacteriol. 66688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, C. J., and I. Amstrong. 2006. The application of atomic force microscopy force measurements to the characterization of microbial surfaces. Surf. Interface Sci. 381419-1428. [Google Scholar]
- 56.Yao, X., M. Jericho, D. Pink, and T. Beveridge. 1999. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 1816865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, L., D. Schaefer, and M. R. Marten. 2005. Assessment of elasticity and topography of Aspergillus nidulans spores via atomic force microscopy. Appl. Environ. Microbiol. 71955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]