Abstract
Organisms coordinate biological activities into daily cycles using an internal circadian clock. The circadian oscillator proteins KaiA, KaiB, and KaiC are widely believed to underlie 24-h oscillations of gene expression in cyanobacteria. However, a group of very abundant cyanobacteria, namely, marine Prochlorococcus species, lost the third oscillator component, KaiA, during evolution. We demonstrate here that the remaining Kai proteins fulfill their known biochemical functions, although KaiC is hyperphosphorylated by default in this system. These data provide biochemical support for the observed evolutionary reduction of the clock locus in Prochlorococcus and are consistent with a model in which a mechanism that is less robust than the well-characterized KaiABC protein clock of Synechococcus is sufficient for biological timing in the very stable environment that Prochlorococcus inhabits.
Cyanobacteria are photosynthetic prokaryotes that are known to possess a true circadian clock. Gene expression and other biological activities follow rhythmic cycles with a circa 24-h period. Rhythmic behavior is maintained even in the absence of environmental stimuli such as light and temperature. The underlying core oscillator consisting of the clock proteins KaiA, KaiB, and KaiC is the only characterized prokaryotic circadian oscillator. It was previously demonstrated that these three proteins, together with ATP, can produce 24-h oscillations of KaiC phosphorylation in vitro (17). The essential roles of KaiA and KaiB in oppositely influencing KaiC phosphorylation are well documented for the oscillator of “Synechococcus elongatus” PCC 7942 (hereafter S. elongatus), the species for which most bacterial circadian research has been conducted. Thus, it is puzzling that marine cyanobacteria of the genus Prochlorococcus, probably the most abundant photosynthetic organisms on Earth (5, 28), contain homologs of only two of these clock proteins, KaiC and KaiB (3, 11, 20). Laboratory cultures (10) as well as natural Prochlorococcus populations (24) display a rhythmic cell cycle together with a daily periodicity of gene expression that can be explained by the functioning of a circadian clock. Alternatively, these rhythms could be controlled directly by the daylight (10). The functional role of the Kai proteins from Prochlorococcus has remained entirely unclear and has not been experimentally addressed thus far.
In the well-studied protein clock of S. elongatus, KaiC hexamers are at the center of the circadian oscillator, combining three intrinsic enzymatic activities: autokinase, autophosphatase, and ATPase. KaiA and KaiB modulate KaiC's activities in opposite manners. KaiA seems to be essential for the shift between autophosphatase and autokinase, and for generating KaiC phosphorylation rhythms, by stabilizing C-terminal residues of KaiC, the A-loops (12). Thus, the absence of KaiA should have consequences for the enzymatic activities of the remaining Kai proteins of Prochlorococcus. In this study, the previously unknown functions of the Prochlorococcus sp. strain MED4 protein KaiB (ProKaiB) and ProKaiC are examined. In our in vitro experiments, we analyzed the recombinant proteins ProKaiB and ProKaiC in direct comparison to the core oscillator of S. elongatus, which consists of S. elongatus KaiA (SynKaiA), SynKaiB, and SynKaiC. We show here that both clock proteins from Prochlorococcus sp. strain MED4 independently exhibit their known biochemical functions, although the influence of ProKaiB on ProKaiC dephosphorylation is different certainly due to the absence of KaiA, the third protein of the oscillator. For ProKaiC, we demonstrate ATPase activity as well as the phosphorylation of serine 427 (S427) and threonine 428 (T428) using mass spectrometry and high-resolution sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Moreover, we suggest that the deletion of kaiA is compensated by the enhanced autophosphorylation activity of ProKaiC. Our results might have further implications for the analysis of a possible timing mechanism in other bacterial species, such as purple bacteria that encode KaiB and KaiC homologs but that lack the KaiA component.
MATERIALS AND METHODS
Bacterial strains and culture.
Plasmids pMED4KaiC and pMED4KaiB were constructed by PCR amplification of the kaiC and kaiB sequences (CyanoBase accession numbers IPR013503 and IPR013474, respectively) from Prochlorococcus sp. strain MED4 using primers MED4_kaiC_fw (5′-GGATCCAAAGATAAAAAAATTAGTAAATC-3′), Med_kaiC_rev (5′-GCGGCCGCCTAATTTTTTTCAATTCCT-3′), MED4_kaiB_fw (5′-GGATCCGTGGCAAGAAAAAC-3′), and MED4_kaiB_rev (5′-GCGGCCGCTTAATTTTTTGTACCTC-3′). PCR fragments were subcloned into the pGEM-T vector (Promega), digested with BamHI and NotI, and cloned into the respective restriction endonuclease sites of vector pGEX-6P-1.
The recombinant Prochlorococcus glutathione S-transferase (GST)-KaiB and GST-KaiC fusion proteins were produced in Escherichia coli BL21 cells. For the production of the recombinant ProKaiC and ProKaiB proteins, cells were cultured overnight at 37°C in 100 ml of LB medium (1) containing 75 μg ml−1 of ampicillin. The ProKaiC expression culture was diluted in 1 liter of LB medium and incubated for 5 h at 18°C. After the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), cells were incubated at 18°C for 60 h before harvest. The expression of ProKaiB was carried out for 21 h at 37°C. IPTG (1 mM) was added after the first 3 h. Cell pellets were resuspended in 20 ml of cold extraction buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1 mM ATP, 5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM EDTA) and lysed by the addition of 17,000 U lysozyme (BioChemika) and 125 U of Benzonase nuclease (Novagen) and sonication. The recombinant circadian clock proteins from Synechococcus elongatus PCC 7942 were produced in E. coli BL21 strains kindly provided by T. Kondo (Nagoya University, Japan). The recombinant GST fusion proteins ProKaiB, ProKaiC, SynKaiA, SynKaiB, and SynKaiC were purified using glutathione-Sepharose 4B and PreScission protease (GE Healthcare, Uppsala, Sweden) as described previously (9, 19). Protein concentrations were estimated according to methods described previously by Lowry et al. (15) as well as by a visual comparison of the different protein amounts on Coomassie-stained SDS gels.
Phosphosite mapping of KaiC from Prochlorococcus sp. strain MED4.
In order to generate nonphosphorylated forms of ProKaiC, samples of 10 μg ProKaiC from two different preparations (prepared in the presence of 0.2 mM ATP or 0.5 mM ATP) were treated with 2 U of λ protein phosphatase (New England Biolabs) for 6 h. As a control, samples were incubated without phosphatase for 6 h. After phosphatase treatment, phosphorylated and nonphosphorylated forms of ProKaiC were separated by SDS-PAGE (11% T with 0.67% C) according to methods described previously (18).
Mass spectrometric analysis of ProKaiC phosphorylation sites was performed essentially as described previously (22, 23). Briefly, Coomassie-stained protein bands were excised from SDS-PAGE gels, minced, reduced with Tris(2-carboxyethyl)phosphine, and alkylated with iodoacetamide prior to proteolytic digestion. In four parallel in-gel reactions, KaiC was digested with the proteases proteinase K, thermolysin, elastase, and trypsin. The resulting peptides were extracted from the gel, and phosphopeptides were enriched from the complex peptide mixture on a TiO2 column. Mass spectrometry was performed using a reversed-phase nanoflow liquid chromatograph coupled to a tandem mass spectrometer (CapLC and Q-ToF Micro; Micromass, Manchester, United Kingdom). Peak lists from nanoflow liquid chromatography (NanoLC)-tandem mass spectrometry (MS/MS) raw data were generated by Mascot Distiller 2.0 (Matrix Science, London, United Kingdom). Mascot Server 2.1 (Matrix Science) was used for database searching against the SwissProt database. ProKaiC phosphorylation sites were mapped using Phosm software (22).
In vitro assay of KaiC phosphorylation and dephosphorylation.
Phosphorylation and dephosphorylation assays were performed as described previously (17), with [γ-32P]ATP, but were carried out at 18°C when ProKaiC was used. Images were analyzed by Personal Molecular Imager FX (Bio-Rad) and Quantity One software (Bio-Rad).
ATPase assay.
The analysis of ATPase activity is based on measuring the concentration of the released orthophosphate colorimetrically (14, 27). The assay was carried out with 0.3 μM KaiC (and 0.3 μM KaiB) of Prochlorococcus sp. strain MED4 (at 18°C) or KaiC of Synechococcus (at 30°C) as a control. Aliquots were sampled every 2 h. Assay buffer served as a negative control. The final buffer composition in the incubation mixtures was 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM ATP, 1 mM dithiothreitol, and 1 mM EDTA. Data were corrected for negative control and zero-time values.
Genome data and software tools.
Cyanobacterial genomes were downloaded from the NCBI GenBank server (ftp://ftp.ncbi.nih.gov/GenBank/genomes/Bacteria/). For visualization of genomes and gene arrangements as well as for extraction of gene and intergenic sequences, Artemis software (22) was used. Sequence similarities and E values were calculated using NCBI BLASTp (23, 24). Multiple alignments were constructed with CLUSTALW 1.83 (25), and the phylogenetic 16S rRNA tree was constructed using the neighbor-joining method (26).
RESULTS
kaiA gene deletion during evolution of the kai operon.
Most of the research on the cyanobacterial clock has been done on the model freshwater unicellular cyanobacterium S. elongatus. The genome of this organism contains single copies of the three kai genes, which are located together in a single gene cluster. However, the set of core clock genes differs among the various groups of cyanobacteria. Many cyanobacterial genomes contain more than just one copy per kai gene, possibly indicating additional layers of complexity within the clock. In contrast, cyanobacteria belonging to the Prochlorococcus group do not even harbor one complete set of genes for the three-protein oscillator. Comparative analyses revealed that all Prochlorococcus genomes sequenced so far contain the genes for the KaiB and KaiC proteins but lack a kaiA gene. Moreover, it can be demonstrated at the sequence level that the kaiA clock component of the kai operon was deleted by genome reduction within the genus Prochlorococcus (for further details, see the supplemental material) (6). From this fact, the question arose of whether the remaining genes still encode functional ProKaiC and ProKaiB proteins and, therefore, could be assumed to be involved in a timing mechanism of the living Prochlorococcus cell.
Conservation of KaiC phosphorylation sites S427 and T428 in Prochlorococcus sp. strain MED4.
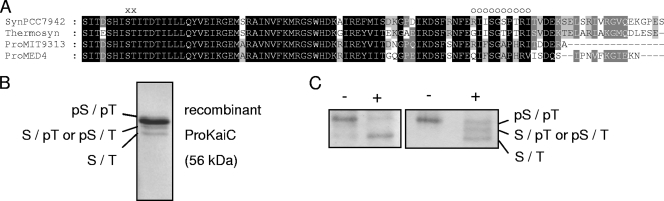
To drive the well-studied KaiC phosphorylation cycle, sequential phosphorylation reactions of S431 and T432 are important (18, 21). The phosphorylation state of these two amino acid residues regulates autokinase and autophosphatase activities of KaiC from S. elongatus. Indeed, within the amino acid sequence of KaiC from Prochlorococcus sp. strain MED4, we found a serine (S427) next to a threonine (T428) residue aligning to the known phosphorylation sites S431 and T432 in KaiC of S. elongatus (Fig. 1A).
FIG. 1.
(A) Sequence alignment of KaiC C-terminal regions from Prochlorococcus sp. strain MED4 (ProMED4), Prochlorococcus sp. strain MIT9313 (ProMIT9313), Synechococcus elongatus PCC 7942 (SynPCC7942), and “Thermosynechococcus elongatus” BP-1 (Thermosyn), where S431 and T432 (indicated by “x”), the sites of phosphorylation in S. elongatus, are conserved in Prochlorococcus sp. strain MED4 as S427 and T428, respectively. Several residues differ within the A-loop (indicated by “o”) and the C-tail between Prochlorococcus and Synechococcus strains. (B) NanoLC-MS/MS analysis of ProKaiC (purified in the presence of 0.2 mM ATP) revealed the presence of the doubly phosphorylated form pS427/pT428 in the top band and of the singly phosphorylated form in the middle band, respectively. No phosphopeptides could be detected in the faster-migrating band. (C) Dephosphorylation of ProKaiC with λ protein phosphatase (+) of two different preparations (in the presence of 0.2 mM ATP [left] and 0.5 mM ATP [right] during protein purification). After staining, three bands of different mobilities were detected using a high-resolution gel system.
Using high-resolution SDS-PAGE for the optimal separation of different KaiC phosphorylation forms, we obtained mainly one band for recombinant ProKaiC after purification (Fig. 1B). After incubation of ProKaiC with λ protein phosphatase, faster-migrating protein bands appeared (Fig. 1C), possibly representing dephosphorylated ProKaiC forms. In order to identify the phosphorylation state of ProKaiC in the three different migrating bands, we mapped phosphorylation sites by NanoLC-MS/MS (see Fig. S4 in the supplemental material for MS/MS spectra). In the mass spectrometric analysis, we detected peptides representing three phosphorylation states in ProKaiC (see Table S1 in the supplemental material). The main fraction (Fig. 1B, top band) consisted of the double-phosphorylated (pS427 and pT428) form. Only relatively long periods of incubation (4 to 6 h) with a phosphatase resulted in significant amounts of singly phosphorylated (pS427 or pT428) and nonphosphorylated KaiC forms as found in the middle and bottom bands, respectively, in Fig. 1B. Thus, high-resolution SDS-PAGE and mass spectrometric analyses of Prochlorococcus sp. strain MED4 KaiC revealed that the two amino acid residues S427 and T428 can be phosphorylated in this species (Fig. 1A), suggesting that sequential phosphorylation reactions might also underlie the daily timing mechanism in Prochlorococcus sp. strain MED4.
KaiC from Prochlorococcus sp. strain MED4 shows autophosphorylation without KaiA.
Although KaiC sequences of S. elongatus and Prochlorococcus sp. strain MED4 are very similar (identities of 373/497 [75%]) (see Fig. S3 in the supplemental material), several residues differ within the C tail and the adjacent region, termed the A-loop (12) (Fig. 1A). These A-loops were previously proposed to possess an important function, as they likely determine the steady-state level of phosphorylation of KaiC (12). In that model for S. elongatus, KaiA directly binds to the A-loops and the tail segment of KaiC, thereby increasing phosphorylation levels.
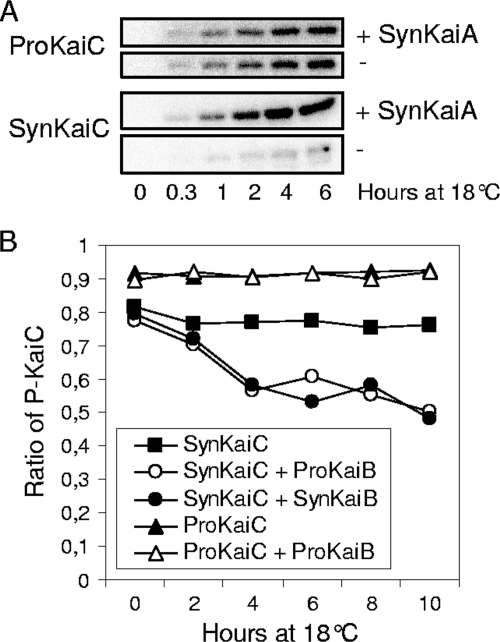
To determine whether ProKaiC possesses properties similar to those of the well-studied SynKaiC, we used the recombinant proteins for in vitro phosphorylation assays with [γ-32P]ATP. Interestingly, ProKaiC alone showed a high level of autophosphorylation activity (Fig. 2A). Moreover, the addition of SynKaiA had no influence on ProKaiC autophosphorylation, which is in strong contrast to SynKaiC exhibiting significant autophosphorylation only in the presence of SynKaiA (Fig. 2A). All experiments were performed at 18°C, a temperature also encountered by Prochlorococcus species in nature. Higher (30°C) and lower (13°C) temperatures gave similar results (data not shown).
FIG. 2.
(A) Pronounced autokinase activity of ProKaiC in comparison to SynKaiC. ProKaiC and SynKaiC were incubated with [γ-32]ATP either with (+) or without (−) SynKaiA. (B) No effect of KaiB on the dephosphorylation of ProKaiC was detected, but an effect on SynKaiC dephosphorylation was detected.
ProKaiB promotes dephosphorylation of SynKaiC.
KaiB sequences from Prochlorococcus sp. strain MED4 and S. elongatus are highly conserved (see Fig. S2 in the supplemental material), indicating that this protein may have maintained its circadian clock function. In order to obtain information regarding whether ProKaiB is able to promote the dephosphorylation of KaiC, we performed dephosphorylation assays with ProKaiB and SynKaiB as a control (Fig. 2B). Neither ProKaiB nor SynKaiB can enhance the dephosphorylation of ProKaiC. Thus, in our in vitro system, KaiC and KaiB from Prochlorococcus sp. strain MED4 are not sufficient to drive an autonomous phosphorylation-dephosphorylation cycle.
However, ProKaiB significantly enhances SynKaiC dephosphorylation activity in a manner similar to that of SynKaiB (Fig. 2B). Here, SynKaiC was preincubated with SynKaiA and [γ-32P]ATP and then mixed with recombinant SynKaiB or ProKaiB, respectively.
ATPase activity of ProKaiC is reduced by ProKaiB.
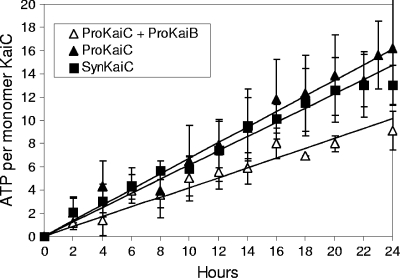
In the well-studied protein clock of S. elongatus, KaiC possesses another intrinsic enzymatic activity besides autokinase and autophosphatase: it also has a very weak ATPase activity. Interestingly, it was previously found that the circadian period correlates with ATPase activity in S. elongatus (26). We performed ATPase assays using a malachite green colorimetric method, which revealed a weak activity (16 molecules of ATP per day) (Fig. 3) of ProKaiC, which is comparable to that observed previously for S. elongatus (26). Moreover, the addition of ProKaiB to the assay mixture reduced the ATPase activity by about one-half (Fig. 3).
FIG. 3.
ProKaiB reduces the ATPase activity of ProKaiC. ATPase assays were performed using the malachite green colorimetric method. Aliquots were sampled every 2 h. Assay buffer served as a negative control.
DISCUSSION
For the model organism S. elongatus, all three kai genes, kaiA, kaiB, and kaiC, are essential for circadian timing, and an inactivation of any of them abolishes it (7). The three Kai proteins have been demonstrated to constitute the S. elongatus core oscillator (17). KaiA stimulates the autophosphorylation activity of KaiC, whereas KaiB enhances its intrinsic dephosphorylation activity. In contrast, cyanobacteria belonging to the Prochlorococcus group do not harbor the complete set of kai genes. Here, kaiA became deleted during an evolutionary reduction process streamlining the Prochlorococcus genome. However, genes encoding KaiB and KaiC remained at the genome locus and exhibit a high level of sequence similarity to the homologous sequences of S. elongatus. Thus, we analyzed biochemically if KaiC as well as KaiB from kaiA-lacking Prochlorococcus sp. strain MED4 are functional proteins. Indeed, we found residues S427 and T428, homologous to S431 and T432, the sites of KaiC phosphorylation in the S. elongatus oscillator, to also be phosphorylated in KaiC of Prochlorococcus sp. strain MED4 (Fig. 1A). Thus, sequential phosphorylation reactions at S427 and T428 might also underlie the daily timing mechanism of Prochlorococcus sp. strain MED4 (21). However, KaiC from Prochlorococcus sp. strain MED4 showed pronounced autokinase activity without KaiA (Fig. 2A) and remained hyperphosphorylated even in the presence of KaiB (Fig. 2B); this finding is in contrast to S. elongatus KaiC, which is an autophosphatase in the absence of KaiA.
Recently, it was demonstrated for S. elongatus that KaiA is critical for the shift between the autophosphatase and autokinase activities of KaiC resulting in the phosphorylation-dephosphorylation rhythms of the 24-h period (12). KaiA stabilizes the C-terminal residues of KaiC, termed the A-loops. For Prochlorococcus species, which lack kaiA homologs, a different mechanism has been suggested. Here, ProKaiC has been predicted to possess a high steady-state level of phosphorylation caused by a dynamic equilibrium that favors the exposed state of the A-loop (12). Thus, KaiC of Prochlorococcus seems to be a natural variant that favors autokinase activity and, thereby, compensates for the genome deletion of kaiA. In S. elongatus, constitutive KaiC hyperphosphorylation (13) does not abolish in vivo circadian gene expression, although the KaiC phosphorylation rhythm is disrupted. This finding suggests that an alternative pacemaker exists in living cells. A transcriptional/translational feedback loop has been proposed to lead to independent oscillations in the cell (13), a situation that might be relevant for Prochlorococcus species as well.
In our experiments, ProKaiB was able to stimulate the dephosphorylation of the SynKaiC protein, as does KaiB from S. elongatus (Fig. 2B); however, it had no effect on ProKaiC autokinase or autophosphatase activities, raising the question of the likely function of KaiB in Prochlorococcus. We reasoned that KaiB may modulate the ATPase, rather than autokinase or autophosphatase, activities of KaiC in Prochlorococcus. It was previously proposed that ATP hydrolysis might be the most fundamental reaction that defines the 24-h period of the clock (26). We measured a weak ATPase activity of ProKaiC (16 molecules of ATP per day) (Fig. 3), comparable to that of S. elongatus (26), which was reduced after the addition of ProKaiB (Fig. 3). Thus, in the Prochlorococcus system, the main mechanism of KaiBC function might be related to the regulation of ATPase activity.
In experiments with synchronized Prochlorococcus cultures, it was previously shown that robust 24-h rhythms of DNA replication and gene expression found under alternating 12-h-light and 12-h-dark periods rapidly damp under continuous light (6). Thus, it was previously proposed that a core timing mechanism and output apparatus are functional in Prochlorococcus but that they work in an hourglass-like fashion, requiring a daily resetting, rather than as a self-sustained oscillator (6). The biochemical properties that we have observed are consistent with such a daily timing mechanism for Prochlorococcus and might be important for other prokaryotes, e.g., species of proteobacteria and archaebacteria, possessing kaiB and kaiC genes but also lacking the KaiA component (4).
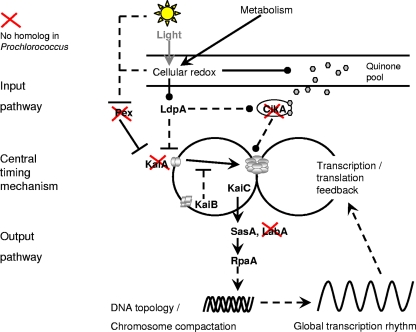
The reduced timing mechanism of Prochlorococcus might be part of a similarly reduced network combining input and output pathways. Compared to the molecular mechanism suggested previously for S. elongatus (2), only one input pathway and one output pathway would remain, taking into account the homologous components present in Prochlorococcus sp. strain MED4 only (Fig. 4). All other genes have been deleted during evolution. In the reduced network of Prochlorococcus, an input signal might be transmitted via the LdpA homolog, PMM1560, which is likely sensitive to the redox state of the cell (8), into the central timer consisting of KaiC and KaiB. The central timing mechanism might be stabilized by an outer transcription/translation feedback loop, where the amount of phosphorylated KaiC regulates its own transcription (16). The output pathway via homologs to SasA, PMM1077, and RpaA, PMM0128, could forward the timing signal to the DNA and/or influence global gene expression.
FIG. 4.
Scheme illustrating a putative molecular mechanism for the circadian clock in Prochlorococcus sp. strain MED4 in comparison to S. elongatus. The scheme is adapted from data described previously by Dong and Golden (2), with permission of the publisher. The homologous components that are present or absent (red crosses) in Prochlorococcus sp. strain MED4 are indicated. In the input pathway, CikA and Pex are missing in Prochlorococcus sp. strain MED4, whereas a gene, PMM1560, homologous to LdpA was found by a BLASTp search against the genome. The central timing mechanism of Prochlorococcus sp. strain MED4 lacks the KaiA component. In the output pathway, SasA (PMM1077) and RpaA (PMM0128) might influence DNA topology by underlying global gene expression rhythms. An alternative pathway via LabA was recently suggested for S. elongatus (25) but is missing in Prochlorococcus sp. strain MED4. Solid lines indicate direct effects, and dotted lines indicate indirect or unknown mechanisms. Arrows indicate the direction of signaling. Blunt ends represent inhibition, and circle ends indicate an unspecific direction.
The temperature-compensated posttranslational oscillator made of KaiA, KaiB, and KaiC might provide additional robustness when conditions are changing. Natural Prochlorococcus populations are highly synchronized to a stable environment in specific niches of the Earth's oceans; they divide exactly once a day and at the same time (6). Thus, an extremely robust oscillator like that in S. elongatus might not be required for the growth of marine cyanobacteria and, thus, was reduced during evolution.
Supplementary Material
Acknowledgments
We thank Laurence Garczarek for providing genomic DNA of Prochlorococcus sp. strain MED4 and Takao Kondo for the E. coli BL21 strains and Kai antisera. We thank Wolfgang R. Hess for critical reading of the manuscript and Susan Golden for stimulating discussions.
This work was supported by the European Commission (BioSim Network grant SHB-CT-2004.005137) and by the BMBF (FORSYS-Partner Project, grant 0315294).
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong, G., and S. S. Golden. 2008. How a cyanobacterium tells time. Curr. Opin. Microbiol. 11541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 10010020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvornyk, V., O. Vinogradova, and E. Nevo. 2003. Origin and evolution of circadian clock genes in prokaryotes. Proc. Natl. Acad. Sci. USA 1002495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goericke, R., and N. A. Welschmeyer. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res. 402283-2294. [Google Scholar]
- 6.Holtzendorff, J., F. Partensky, D. Mella, J. F. Lennon, W. R. Hess, and L. Garczarek. 2008. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23187-199. [DOI] [PubMed] [Google Scholar]
- 7.Ishiura, M., S. Kutsuna, S. Aoki, H. Iwasaki, C. R. Andersson, A. Tanabe, S. S. Golden, C. H. Johnson, and T. Kondo. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 2811519-1523. [DOI] [PubMed] [Google Scholar]
- 8.Ivleva, N. B., M. R. Bramlett, P. A. Lindahl, and S. S. Golden. 2005. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 241202-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki, H., Y. Taniguchi, M. Ishiura, and T. Kondo. 1999. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 181137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquet, S., F. Partensky, D. Marie, R. Casotti, and D. Vaulot. 2001. Cell cycle regulation by light in Prochlorococcus strains. Appl. Environ. Microbiol. 67782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kettler, G. C., A. C. Martiny, K. Huang, J. Zucker, M. L. Coleman, S. Rodrigue, F. Chen, A. Lapidus, S. Ferriera, J. Johnson, C. Steglich, G. M. Church, P. Richardson, and S. W. Chisholm. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, Y. I., G. Dong, C. W. Carruthers, Jr., S. S. Golden, and A. LiWang. 2008. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA 10512825-12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitayama, Y., T. Nishiwaki, K. Terauchi, and T. Kondo. 2008. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 221513-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 10095-97. [DOI] [PubMed] [Google Scholar]
- 15.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 16.Murayama, Y., T. Oyama, and T. Kondo. 2008. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J. Bacteriol. 1901691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima, M., K. Imai, H. Ito, T. Nishiwaki, Y. Murayama, H. Iwasaki, T. Oyama, and T. Kondo. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308414-415. [DOI] [PubMed] [Google Scholar]
- 18.Nishiwaki, T., Y. Satomi, Y. Kitayama, K. Terauchi, R. Kiyohara, T. Takao, and T. Kondo. 2007. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 264029-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishiwaki, T., Y. Satomi, M. Nakajima, C. Lee, R. Kiyohara, H. Kageyama, Y. Kitayama, M. Temamoto, A. Yamaguchi, A. Hijikata, M. Go, H. Iwasaki, T. Takao, and T. Kondo. 2004. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. USA 10113927-13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 4241042-1047. [DOI] [PubMed] [Google Scholar]
- 21.Rust, M. J., J. S. Markson, W. S. Lane, D. S. Fisher, and E. K. O'Shea. 2007. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlosser, A., J. T. Vanselow, and A. Kramer. 2007. Comprehensive phosphorylation site analysis of individual phosphoproteins applying scoring schemes for MS/MS data. Anal. Chem. 797439-7449. [DOI] [PubMed] [Google Scholar]
- 23.Schlosser, A., J. T. Vanselow, and A. Kramer. 2005. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal. Chem. 775243-5250. [DOI] [PubMed] [Google Scholar]
- 24.Sweeney, B. M., and M. B. Borgese. 1989. A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH7803. J. Phycol. 25183-186. [Google Scholar]
- 25.Taniguchi, Y., M. Katayama, R. Ito, N. Takai, T. Kondo, and T. Oyama. 2007. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2160-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terauchi, K., Y. Kitayama, T. Nishiwaki, K. Miwa, Y. Murayama, T. Oyama, and T. Kondo. 2007. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA 10416377-16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turgay, K., L. W. Hamoen, G. Venema, and D. Dubnau. 1997. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11119-128. [DOI] [PubMed] [Google Scholar]
- 28.Vaulot, D., D. Marie, R. J. Olson, and S. W. Chisholm. 1995. Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial pacific ocean. Science 2681480-1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.