Abstract
Proteins whose synthesis is enhanced by polyamines at the level of translation were identified with a polyamine-requiring mutant cultured in the presence of 0.1% glucose and 0.02% glutamate at 42°C. Polyamines had a greater effect on cell growth at 42°C than at 37°C. At 42°C, the synthesis of RpoE (σ24) and StpA, which are involved in the transcription of a number of heat shock response genes, was stimulated by polyamines at the level of translation. In the rpoE and stpA mRNAs, a Shine-Dalgarno (SD) sequence is located at 13 and 12 nucleotides, respectively, upstream of the initiation codon AUG. When the SD sequences were moved to the more common position 7 nucleotides upstream of the initiation codon AUG, the degree of polyamine stimulation was reduced, although the level of RpoE and StpA synthesis was markedly increased. The mechanism underlying polyamine stimulation of RpoE synthesis was then studied. Polyamine stimulation of RpoE synthesis was reduced by changing the bulged-out structure in the initiation site of rpoE mRNA, although the level of RpoE synthesis increased. A selective structural change of this bulged-out region induced by spermidine at 42°C was observed by circular dichroism. Polyamine stimulation of fMet-tRNA binding to ribosomes at 42°C also disappeared by changing the bulged-out structure in the initiation site of rpoE mRNA. The results suggest that polyamines enhance the synthesis of RpoE by changing the bulged-out structure in the initiation site of rpoE mRNA.
Polyamines (putrescine, spermidine, and spermine), aliphatic cations present in almost all living organisms, are necessary for normal cell growth (6, 16, 18). Because polyamines interact with nucleic acids and exist mostly as polyamine-RNA complexes in cells (29, 41), their proliferative effects are presumed to be caused by changes in RNA function. In this context, it has been reported that polyamines stimulate the synthesis of some proteins in vitro (3, 22), increase the fidelity of protein synthesis (19, 24), and induce in vivo assembly of 30S ribosomal subunits (10, 21), suggesting that polyamines regulate protein synthesis at several different steps.
Previously we found that translation of a defined set of proteins in polyamine-requiring mutant Escherichia coli MA261 is enhanced by polyamines (16). We proposed that a set of genes whose expression is enhanced by polyamines at the level of translation can be classified as a “polyamine modulon” (16). There are three different mechanisms underlying polyamine stimulation of the translation of various members of the polyamine modulon. First, polyamine stimulation of protein synthesis takes place when a Shine-Dalgarno (SD) sequence in the mRNA is obscure or is distant from the initiation codon AUG. Polyamines cause structural changes in a region of the SD sequence and the initiation codon AUG of the mRNA, facilitating formation of the initiation complex. This is the case for OppA, a periplasmic substrate binding protein of the oligopeptide uptake system (46); FecI σ factor (σ18), a σ factor for the iron transport operon (45); Fis, a global regulator of transcription of some growth-related genes, including genes for rRNA and some tRNAs (45); RpoN (σ54), a σ factor for the nitrogen metabolism genes (38); and H-NS, a positive regulator of expression of genes involved in flagellin synthesis and ribosomal protein synthesis (38). Second, polyamines enhance translation initiation from the inefficient initiation codon UUG or GUG, such as in cya mRNA encoding adenylate cyclase (43) or cra mRNA encoding a global transcription factor for a large number of genes involved in glycolysis and glyconeogenesis (38). Third, polyamines stimulate read-through of amber codon UAG-dependent Gln-tRNASupE on ribosome-associated rpoS mRNA encoding σ38, a σ factor for genes expressed at the stationary phase (44), or stimulate a +1 frameshift at the 26th UGA codon of prfB mRNA encoding a polypeptide release factor 2 (12).
In this study, we looked for additional genes that belong to the polyamine modulon under heat shock stress conditions by culturing E. coli MA261 cells at 42°C. We found that the rpoE and stpA genes are new members of the polyamine modulon and studied the mechanism of polyamine stimulation of RpoE and StpA synthesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A polyamine-requiring mutant of Escherichia coli, MA261 (speB speC gly leu thr thi) (7), and E. coli MA261 lacZ::Em (27) were cultured in medium A [0.1% glucose (5.6 mM) and 0.02% glutamic acid (1.4 mM), 40.2 mM K2HPO4, 22.1 mM KH2PO4, 1.7 mM sodium citrate, 7.6 mM (NH4)2SO4, 6 μM thiamine, 40 μM biotin, 0.8 mM leucine, 0.8 mM threonine, 0.7 mM methionine, 1 mM serine, 1 mM glycine, 0.6 mM ornithine (pH 6.8)] in the presence and absence of 100 μg/ml (0.6 mM) putrescine dihydrochloride. Where indicated, 0.4% (22.2 mM) glucose was added instead of 0.1% glucose and 0.02% glutamic acid as an energy source. Cell growth was monitored by measuring the absorbance at 540 nm.
Plasmids.
Total chromosomal DNA from E. coli W3110 was prepared according to the method of Wilson et al. (42). To make the rpoE-lacZ fusion gene, PCR was performed using total chromosomal DNA as template and 5′-TCTACCC-GGGTAATGCGATGTTCAGATTCT-3′ (P1) and 5′-CCAGCCCGGGATAGGCTTTAATAAAAGCTT-3′ (P2) as primers. The amplified rpoE gene (a 203-nucleotide 5′-upstream region and a 184-nucleotide open reading frame) was digested with XmaI and inserted into the same restriction site of the medium-copy-number vector pMC1871 (36) to make the pMCrpoE-lacZ fusion plasmid. For construction of pMWrpoE-lacZ, the SalI fragment containing the rpoE-lacZ gene of pMCrpoE-lacZ was inserted into the same restriction site of the low-copy-number vector pMW119 (Nippon Gene). Site-directed mutagenesis for the construction of a mutated fusion gene with modified SD sequences was performed by overlap extension using PCR (15). To make the rpoE(SD)-lacZ fusion gene, the initial PCR was performed using total chromosomal DNA from E. coli W3110 as template and P1, 5′-CCGAGGCTCCATCTAACCAAACCAAATT-TC-3′ (P3), 5′-GTTTGGTTAGATGGAGCCTCGGATGAGCGA-3′ (P4), and P2 as primers. Then the second PCR was performed using the initial PCR products as template and P1 and P2 as primers. Plasmid pMWrpoE(SD)-lacZ was prepared using the same method used for the construction of pMWrpoE-lacZ.
The plasmids pMWrpoE[-8(U→A)]-lacZ, pMWrpoE[-20(ΔU)]-lacZ, pMWrpoE[-20(U→A)]-lacZ, pMWrpoE[-42(+U)]-lacZ, and pMWrpoE[-46(ΔG)]-lacZ were prepared, as described above, with site-directed mutagenesis by overlap extension using PCR (15). A list of oligonucleotide primers used for mutagenesis is available from the authors upon request.
To make the stpA-lacZ fusion gene, PCR was performed using total chromosomal DNA as template and 5′-GAGCCCCGGGCTGTTCACCAATAATCT-AAA-3′ (P5) and 5′-CAACCCCGGGTTTTTCGAGCATTTCTTCAA-3′ (P6) as primers. The amplified stpA gene (a 220-nucleotide 5′-upstream region and a 106-nucleotide open reading frame) was digested with XmaI and inserted into the same restriction site of pMC1871 to make the pMCstpA-lacZ fusion plasmid. For construction of pMWstpA-lacZ, the SalI fragment containing the stpA-lacZ gene of the pMCstpA-lacZ fusion plasmid was inserted into the same restriction site of pMW119. Plasmid pMWstpA(SD)-lacZ was prepared as described above. The initial PCR was performed using P5, P5(SD) (5′-CATAATAAACTCCAGGTTTTAACGCCAAAA-3′), P6(SD) (5′-CGTTAAAACCTGGAGTTTATTATGTCCGTA-3′), and P6 as primers. The nucleotide sequence of the plasmids was confirmed by the 3130 genetic analyzer (Applied Biosystems).
Dot blot and Northern blot analyses.
E. coli MA261 cells were cultured at an A540 of 0.05 in the presence and absence of putrescine and harvested at an A540 of 0.15. E. coli rpoE-deficient mutant CAG22216 (34), which was kindly supplied by C. A. Gross, University of California-San Francisco, was grown in modified Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, and 5 g NaCl per liter) and harvested at an A540 of 0.15. Total RNA was prepared from these cells by the method of Emory and Belasco (11). Dot blot and Northern blot analyses were performed according to the method of Sambrook et al. (35) using the ECL direct nucleic acid labeling and detection systems (GE Healthcare Bio-Sciences). Total RNA used for dot blotting and Northern blotting was 0.2 to 3 μg and 7.5 μg, respectively. Probes were made by PCR, and the sizes of the probes used for rpoE, rpoH, and stpA mRNAs were 598, 870, and 380 nucleotides, respectively. Chemical luminescence was detected by a LAS-3000 luminescent image analyzer (Fuji Film).
Western blot analysis.
Western blot analysis was performed using ECL Western blotting reagents (GE Healthcare Bio-Sciences). Antibodies to RpoD (σ70), RpoN (σ54), RpoS (σ38), RpoH (σ32), RpoF (σ28), RpoE (σ24), FecI (σ18), OppA, Cra, Fis, Cya, and StpA were prepared as described previously (2, 25, 26, 38, 46). Antibodies to RF2 (23) and H-NS (40) were kindly supplied by Y. Nakamura, University of Tokyo, and T. Mizuno, Nagoya University, respectively. Antibody to β-galactosidase (β-Gal) was obtained from Sigma-Aldrich. The level of protein was quantified with a LAS-3000 luminescent image analyzer (Fuji Film).
Measurement of RpoE-β-Gal and StpA-β-Gal synthesis in E. coli MA261 by immunoprecipitation of proteins labeled with [35S]methionine.
E. coli MA261 lacZ::Em cells containing pMWrpoE-lacZ or pMWstpA-lacZ were cultured in medium A containing 0.03 mM methionine instead of 0.7 mM, in the absence of putrescine at 42°C. At an A540 of 0.15, the culture was divided into 5-ml aliquots and continued to grow in the presence (100 μg/ml) and absence of putrescine for 10 min. Then, [35S]methionine (1 MBq) was added to each 5-ml aliquot, and the cells were allowed to grow for an additional 20 min. After the addition of unlabeled methionine at a final concentration of 20 mM, the cells were harvested, resuspended in 1 ml of buffer A (10 mM sodium phosphate, at pH 7.4; 100 mM NaCl; 1% Triton X-100; and 0.1% sodium dodecyl sulfate), and disrupted by grinding for 30 s at 2,500 rpm six times with a Multi-Beads Shocker (Yasui Kikai). Immunoprecipitation of [35S]methionine-labeled RpoE-β-Gal and StpA-β-Gal fusion proteins was carried out using whole-cell lysate containing 1,000,000 cpm of [35S]methionine-labeled proteins and antiserum against β-Gal, as described previously (43). After SDS-polyacrylamide gel electrophoresis, the radioactivity associated with fusion proteins was quantified using a BAS-1800II imaging analyzer (Fuji Film).
Measurement of polyamines in whole cells.
Polyamines in whole cells were extracted by treatment of the cells with 10% trichloroacetic acid at 70°C for 15 min, with occasional shaking. Polyamine content was determined by high-pressure liquid chromatography, as described previously (20). Protein content was determined by the method of Bradford (4).
Prediction of the secondary structure of RNA.
Optimal computer folding of mRNAs was performed by the method of Zuker (49). Free energy (ΔG) for the formation of the secondary structure was calculated on the basis of the data of Turner et al. (39).
Circular dichroism (CD) measurement of RNA.
Purified RpoE wild-type (WT) RNA (5′-UUUGGUUUGGGGAGACUUUACCUCG-3′), corresponding to the −26th to −2nd nucleotides upstream from the initiation codon AUG of rpoE mRNA, and RpoE −20(ΔU) RNA (5′-UUUGGUΔUGGGGAGACUUUACCUCG-3′), which lacks one uridine nucleotide at the −20th position of RpoE WT RNA, were obtained from Hokkaido System Science. CD spectra were recorded over 200 to 320 nm on a Jasco J-820 spectropolarimeter (Jasco International Co.) using a 0.1-cm-path-length cuvette at 37°C and 42°C (31). Scan speed was 100 nm/min, and CD samples contained 10 mM Tris-HCl (pH 7.5), 50 mM KCl, and 50 μM RNA. Where indicated, magnesium acetate and/or spermidine were added to the CD samples. Typical spectra at 42°C corresponded to the average of three scans.
Preparation of ribosomes, [3H]fMet-tRNA, and mRNAs.
E. coli Q13 (rna pnp) ribosomes and [3H]fMet-tRNA were prepared as described previously (46). For preparation of RpoE WT (166-mer) RNA and RpoE −20(ΔU) (165-mer) RNA, including the initiation region of 130- and 129-mer of RpoE mRNA and RpoE −20(ΔU) mRNA, respectively, PCR was performed using pMWrpoE-lacZ and pMWrpoE[-20(ΔU)]-lacZ as templates and 5′-GCTAAAAGCTTGGCGTTTC-GATAGCGCGTG-3′ and 5′-CCAGTAGGATCCAGGCTTTCTGATCTCCC-T-3′ as primers. The PCR products were digested with HindIII and BamHI and inserted into the same restriction site of the pBluescript II SK+ (Toyobo, Japan). The plasmid DNA was linearized by BamHI, and RNA was synthesized by a ScriptMAX Thermo T7 transcription kit (TOYOBO, Japan), according to the manufacturer's protocol. RpoE mRNAs were purified by MicroSpin G-25 columns (GE Healthcare).
Assay for fMet-tRNA binding to ribosomes.
The reaction mixture (0.1 ml) containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM dithiothreitol, 1 mM GTP, 200 μg (74 pmol) of ribosomes, 75 pmol of [3H]fMet-tRNA, 90 pmol of RpoE WT (166-mer), or RpoE −20(ΔU) (165-mer) RNA, and either various concentrations of Mg2+ in the presence and absence of 1 mM spermidine or various concentrations of spermidine in the presence of 3 mM Mg2+ was incubated at 37°C or 42°C for 2 min. The reaction mixture was passed through a cellulose nitrate filter (pore size, 0.45 μm) (Advantec), and radioactivity on the filter was measured as described previously (46).
RESULTS
Stimulation of cell growth by polyamines under heat shock conditions.
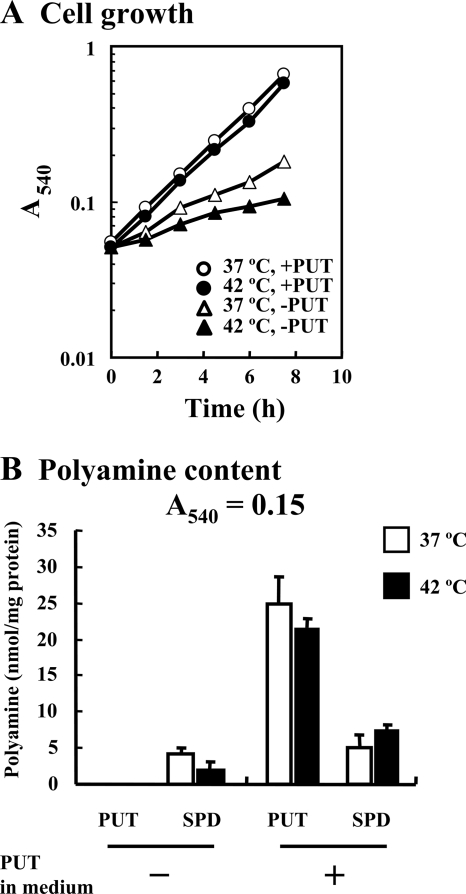
The MA261 strain of E. coli is unable to synthesize putrescine, and its growth slows down in the absence of exogenous putrescine (7). When putrescine is added to the culture medium, it is taken up into cells (17) and spermidine is synthesized from putrescine, leading to recovery of cell growth. To study the effects of polyamines under heat shock conditions, growth of E. coli MA261 was compared at 37°C and 42°C in the presence of 0.1% glucose and 0.02% glutamate. As shown in Fig. 1A, the rate of cell growth of E. coli MA261 in the presence of putrescine was much higher than that in its absence at both 37°C and 42°C. However, growth of E. coli MA261 in the absence of putrescine at 42°C was slower than that at 37°C. Putrescine was efficiently transported into cells at both 42°C and 37°C (Fig. 1B). Similar results were obtained when cells were cultured in the presence of 0.4% glucose instead of 0.1% glucose and 0.02% glutamate (data not shown). These results indicate that the polyamine requirement for cell growth is more evident at elevated temperatures.
FIG. 1.
Growth and polyamine content of E. coli MA261 at 37°C and 42°C. (A) Culture of E. coli MA261 cells with or without 100 μg/ml putrescine. (B) Polyamine content in cells harvested at an A540 of 0.15 was measured as described in Materials and Methods. Data are shown as means ± standard errors. PUT, putrescine; SPD, spermidine.
Stimulation of synthesis of RpoE by polyamines at the level of translation at 42°C.
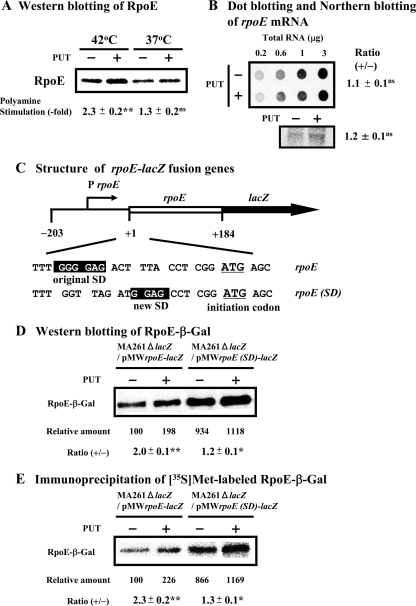
To clarify the mechanisms underlying the more pronounced effects of polyamines at 42°C than at 37°C, the effects of polyamines on synthesis of specific kinds of proteins were examined. RpoE is one of the sigma factors for transcription of a set of heat shock response genes (14). Since the predicted SD sequence of rpoE mRNA is distant from the initiation codon AUG (33), we determined whether synthesis of RpoE is influenced by polyamines at 42°C. When E. coli MA261 cells were cultured at 37°C, the addition of putrescine slightly stimulated the synthesis of RpoE (Fig. 2A). However, synthesis of the RpoE protein was enhanced 2.3-fold by polyamines (i.e., by addition of putrescine) at 42°C (Fig. 2A), whereas the level of rpoE mRNA was slightly enhanced (1.1- to 1.2-fold) by polyamines (Fig. 2B) probably because of the enhancement of transcription of its own gene by RpoE (32). No distinct band for rpoE mRNA was observed in rpoE-deficient mutant CAG22216 by Northern blotting (data not shown). The results indicate that RpoE is one of the target proteins that are stimulated by polyamines at the level of translation at 42°C. To determine whether the distance of the SD sequence from the initiation codon AUG of rpoE mRNA is important for polyamine stimulation of RpoE synthesis, the original SD sequence was replaced by a typical SD sequence seven nucleotides upstream of the initiation codon AUG in an rpoE-lacZ fusion mRNA (Fig. 2C). The synthesis of the RpoE-β-Gal fusion protein produced from this construct was measured by Western blotting and measurement of the [35S]methionine-labeled RpoE-β-Gal fusion protein. Essentially the same results were obtained by measuring the protein level and synthetic activity of the RpoE-β-Gal fusion protein. Synthesis of an RpoE-β-Gal fusion protein from mRNA containing the original SD sequence was stimulated by polyamines by 2.0- to 2.3-fold, whereas the polyamine stimulation was reduced 1.2- to 1.3-fold after replacement with the typical SD sequence, even though the basal level of protein synthesis in the absence of polyamines was enhanced 8.7- to 9.3-fold (Fig. 2D and E). The results indicate that the synthesis of RpoE was enhanced by polyamines at the level of translation due to the existence of a unique SD sequence in the rpoE mRNA.
FIG. 2.
Effect of polyamines on synthesis of RpoE in E. coli MA261 cells at 42°C. (A) Cells were harvested at an A540 of 0.15. Western blotting of RpoE was performed using 20 μg of protein of cell lysate. (B) Dot blot and Northern blot analyses of rpoE mRNA were performed as described in Materials and Methods. (C) Schematic of the rpoE-lacZ fusion genes. The rpoE gene containing a 203-nucleotide 5′-upstream region with an unmodified or modified SD sequence and a 184-nucleotide open reading frame was fused to the lacZ gene. (D) Western blotting of RpoE-β-Gal fusion protein was performed using 10 μg of protein of cell lysate. (E) Immunoprecipitation of [35S]methionine-labeled protein by antibody against β-Gal was performed as described in Materials and Methods. Values are means ± standard errors of triplicate determinations. Student's t test was performed for the value obtained in the presence of putrescine versus that in the absence of putrescine. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01. PUT, putrescine.
Effect of polyamines on the synthesis of other sigma factors and proteins encoded by polyamine modulon genes at 42°C.
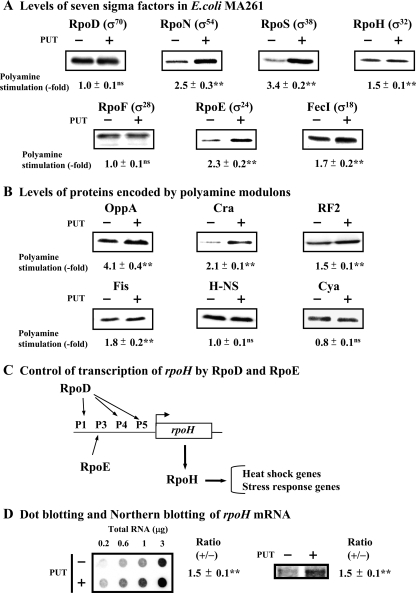
We next determined whether the synthesis of other sigma factors is stimulated by polyamines at 42°C. As shown in Fig. 3A, synthesis of RpoN, RpoS, RpoH, and FecI was enhanced by polyamines together with RpoE. However, synthesis of RpoD and RpoF was not stimulated by polyamines. Synthesis of RpoF was enhanced at 37°C at the level of transcription due to an increase in the level of cAMP caused by the translational stimulation of Cya synthesis (43) or stimulation of H-NS synthesis (38). Thus, we tested the effects of polyamines at 42°C on the synthesis of proteins encoded by previously identified members of the polyamine modulon. As shown in Fig. 3B, the synthesis of OppA, Cra, RF2, and Fis was enhanced by polyamines, but the synthesis of Cya and H-NS was not enhanced by polyamines. Polyamine stimulation of Cya synthesis was not seen when cells were cultured in the presence of 0.1% glucose and 0.02% glutamate instead of 0.4% glucose as an energy source (38), but the polyamine effect of H-NS synthesis disappeared when cells were cultured at 42°C, regardless of the energy source. As RpoF is involved in the transcription of genes for flagellin synthesis, the results are in accordance with the finding that flagellin is not synthesized effectively at 42°C (1). Stimulation of RpoH synthesis by polyamines was observed at the level of transcription (Fig. 3C and D), because transcription of the rpoH gene is controlled by RpoE (8).
FIG. 3.
Levels of seven sigma factors and proteins encoded by polyamine modulons in E. coli MA261 cells at 42°C. Cells were harvested at an A540 of 0.15. (A) Western blotting was performed using 10 μg protein of cell lysate for RpoD; 20 μg protein of cell lysate for RpoN, RpoS, RpoH, RpoF, and RpoE; and 50 μg protein of cell lysate for FecI. (B) Western blotting was performed using 1 μg of protein of cell lysate for OppA and 20 μg of protein of cell lysate for Cra, RF2, Fis, H-NS, and Cya. (C) Schematic of control of transcription of rpoH by RpoD and RpoE. (D) Dot blot and Northern blot analyses of rpoH mRNA were performed as described in Materials and Methods. Values are means ± standard errors of triplicate determinations. ns, P > 0.05; **, P ≤ 0.01. PUT, putrescine.
Stimulation of synthesis of StpA by polyamines at 42°C.
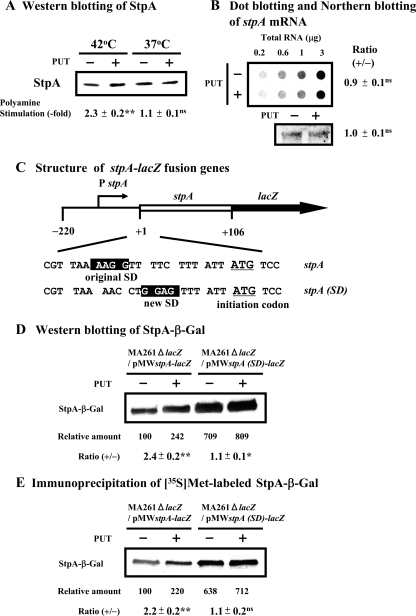
Two nucleoid-associated DNA binding proteins, H-NS and StpA, function similarly (37, 48), but the levels of these two proteins change differentially, depending on the culture temperature. The level of H-NS was reported to be decreased but that of StpA to be increased at 42°C compared to levels at 37°C (30, 37). Since the predicted SD sequence of stpA mRNA is located distantly from the initiation codon AUG (47), we determined whether the synthesis of StpA is altered by polyamines. The level of StpA was significantly enhanced by polyamines at 42°C but not at 37°C (Fig. 4A). However, polyamines did not affect the stpA mRNA level (Fig. 4B), suggesting that, at 42°C, polyamines stimulate the synthesis of StpA at the level of translation. The effects of polyamines on the synthesis of an StpA-β-Gal fusion protein were compared with those of stpA-lacZ fusion mRNAs having the original SD sequence and the typical SD sequence seven nucleotides upstream from the initiation codon AUG (Fig. 4C). Essentially the same results were obtained by Western blotting and measurement of the [35S]methionine-labeled StpA-β-Gal protein (Fig. 4D and E). The synthesis of the StpA-β-Gal fusion protein from mRNA containing the original SD sequence was stimulated 2.2- to 2.4-fold by polyamines, whereas the polyamine stimulation was reduced to 1.1-fold after replacement with the typical SD sequence, even though the basal level of protein synthesis was enhanced 6.4- to 7.1-fold (Fig. 4D and E). The results indicate that the synthesis of StpA was enhanced by polyamines at the level of translation due to the existence of a unique SD sequence in the stpA mRNA.
FIG. 4.
Effect of polyamines on synthesis of StpA in E. coli MA261 cells at 42°C. (A) Cells were harvested at an A540 of 0.15. Western blotting of StpA was performed using 20 μg of protein of cell lysate. (B) Dot blot and Northern blot analyses of stpA mRNA were performed as described in Materials and Methods. (C) Schematic of the stpA-lacZ fusion genes. The stpA gene containing a 220-nucleotide 5′-upstream region with an unmodified or modified SD sequence and a 106-nucleotide open reading frame was fused to the lacZ gene. (D) Western blotting of StpA-β-Gal fusion protein was performed using 10 μg of protein of cell lysate. (E) Immunoprecipitation of [35S]methionine-labeled protein by antibody against β-Gal was performed as described in Materials and Methods. Values are means ± standard errors of triplicate determinations. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01. PUT, putrescine.
Mechanism of polyamine stimulation of the synthesis of RpoE at 42°C.
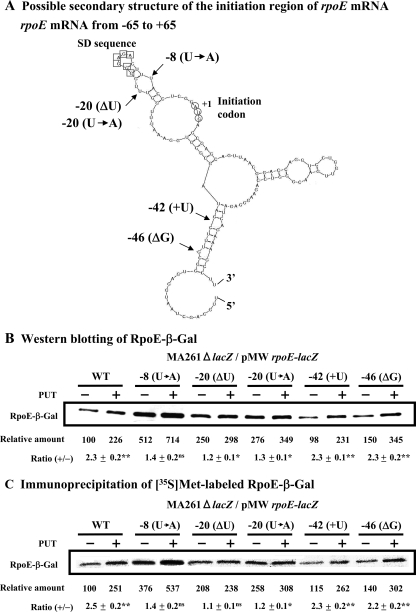
We have previously reported that polyamine stimulation of OppA synthesis and rat liver Ile-tRNA formation likely involves a structural change of the bulged-out region of double-stranded RNA (28, 46) and have recently shown that a selective structural change of the bulged-out region of oppA mRNA and rat liver tRNA is important for polyamine stimulation (13). Thus, to study the mechanism of polyamine stimulation of the synthesis of RpoE at 42°C, a possible secondary structure of the initiation region of rpoE mRNA (−65 to +65) was constructed by the method of Zuker (49), and it was found that the bulged-out region existed (Fig. 5A). The nucleotide sequence of the bulged-out region of double-stranded RNA, near the initiation codon AUG and the SD sequence of rpoE mRNA, was modified to make three different forms of the double-stranded RNA. Those mRNAs are rpoE[-8(U→A)] mRNA, rpoE[-20(ΔU)] mRNA, and rpoE[-20(U→A)] mRNA. As controls, two other alternative forms, rpoE[-42(+U)] mRNA and rpoE[-46(ΔG)] mRNA, were constructed to remove the bulged-out region of another double-stranded region that is located distantly from the initiation site of rpoE mRNA. The predicted secondary structure of the initiation region (positions −65 to +65) of five mutants of rpoE mRNA was essentially the same as that of wild-type rpoE mRNA (data not shown).
FIG. 5.
Effect of polyamines on the synthesis of RpoE-β-Gal fusion protein derived from mutated rpoE-lacZ mRNA in the 5′ untranslated region. (A) Possible secondary structure of the initiation region of rpoE mRNA was constructed by the method of Zuker (49). The positions in which the sequence was mutated are shown. (B) Effect of polyamines on RpoE-β-Gal fusion protein synthesis was tested by Western blot analysis using 10 μg of protein of cell lysate. The levels of rpoE-lacZ mRNA in cells cultured with or without 100 μg/ml putrescine were nearly equal, judging from dot blot analysis. (C) Immunoprecipitation of [35S]methionine-labeled protein by antibody against β-Gal was performed as described in Materials and Methods. Values are means ± standard errors of triplicate determinations. Δ, removal of the nucleotide; +, addition of the nucleotide; →, the replacement of the nucleotide; ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; PUT, putrescine.
The effects of polyamines on the synthesis of an RpoE-β-Gal fusion protein were studied using these various rpoE-lacZ fusion mRNAs. As shown in Fig. 5B and C, polyamine stimulation of the synthesis of RpoE-β-Gal fusion protein from rpoE[-8(U→A)]-lacZ, rpoE[-20(ΔU)]-lacZ, and rpoE[-20(U→A)]-lacZ mRNAs was reduced to 1.1- to 1.4-fold from 2.3- to 2.5-fold seen with wild-type rpoE-lacZ mRNA, even though the basal level of protein synthesis in the absence of polyamines was enhanced. In the case of rpoE[-8(U→A)]-lacZ mRNA, the basal level of protein synthesis in the absence of polyamines was enhanced 3.8- to 5.1-fold, suggesting that an SD-like sequence may be created since two adenine residues are located at the position of a typical SD sequence. When polyamine stimulation of the synthesis of the RpoE-β-Gal fusion protein from rpoE[-42(+U)]-lacZ mRNA and rpoE[-46(ΔG)]-lacZ mRNA was examined as controls, the degree of polyamine stimulation was nearly equal to that of synthesis from wild-type rpoE-lacZ mRNA. The results support the idea that a structural change of the bulged-out region of double-stranded RNA close to the initiation codon AUG and the SD sequence of rpoE mRNA is involved in polyamine stimulation of RpoE synthesis.
Selective structural change of the bulged-out region of rpoE RNA by spermidine at 42°C.
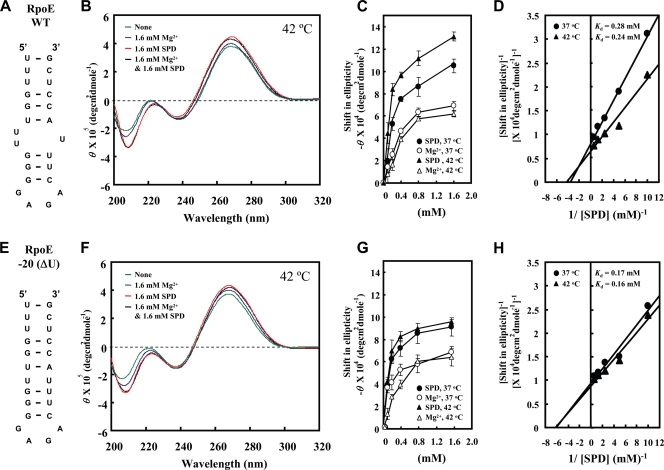
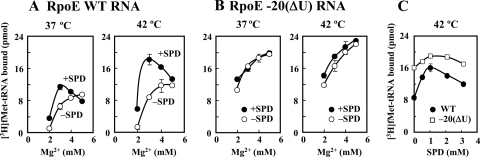
Structural changes of the bulged-out region induced by spermidine were then studied using synthetic RNAs containing the wild type or a mutated initiation region of the rpoE mRNA. Structural changes induced by spermidine of RpoE WT RNA (corresponding to the −26th to −2nd nucleotides upstream from the initiation codon AUG) (Fig. 6A) and of RpoE −20(ΔU) RNA, which lacks one uridine nucleotide at the −20 position (Fig. 6E), were analyzed in the presence of 10 mM Tris-HCl (pH 7.5) and 50 mM KCl at 42°C. A substantial increase in the relative intensity of the negative band at 208 nm in CD reflects stabilization (or an increase) of A-form double-stranded RNA (31). There was a marked increase in the relative intensity of the negative band at 208 nm induced by 1.6 mM spermidine in RpoE WT RNA, including the bulged-out region in double-stranded RNA, and this was greater than the increase in RpoE −20(ΔU) RNA with or without Mg2+ (Fig. 6B and F). In contrast, 1.6 mM Mg2+ had a much smaller effect than spermidine on RpoE WT RNA and RpoE −20(ΔU) RNA. Concentration-dependent shifts of RpoE WT RNA and RpoE −20(ΔU) RNA induced by spermidine or Mg2+ at 208 nm were then measured at 37°C and 42°C (Fig. 6C and G). With RpoE WT RNA, the increase induced by spermidine in the shift at 208 nm was greater at 42°C than at 37°C, but in RpoE −20(ΔU) RNA the increases were nearly equal at 37°C and 42°C. The apparent dissociation constant (Kdapp) values for spermidine of RpoE WT RNA were 0.28 mM and 0.24 mM at 37°C and 42°C, respectively, and those for spermidine of RpoE −20(ΔU) RNA were 0.17 mM and 0.16 mM at 37°C and 42°C, respectively (Fig. 6D and H). These values indicate that spermidine binds to double-stranded RNA with greater affinity than double-stranded RNA containing bulged-out nucleotides. We next studied the effect of spermidine on RpoE WT (166-mer) RNA- and RpoE −20(ΔU) (165-mer) RNA-dependent [3H]fMet-tRNA binding to ribosomes. As shown in Fig. 7, RpoE WT RNA-dependent [3H]fMet-tRNA binding to ribosomes at 42°C was significantly stimulated by 1 mM spermidine, but RpoE −20(ΔU) RNA-dependent [3H]fMet-tRNA binding was not. These results suggest that the bulged-out region located at the initiation site of rpoE mRNA is stabilized and that initiation complex formation is then enhanced if a suitable amount of spermidine is present.
FIG. 6.
CD spectra of RpoE WT RNA and RpoE −20(ΔU) RNA. (A and E) Structure of RpoE WT RNA and RpoE −20(ΔU) RNA. (B and F) CD spectra were recorded as described in Materials and Methods. Green line, no addition; blue line, 1.6 mM Mg2+; red line, 1.6 mM spermidine; black line, 1.6 mM Mg2+ and 1.6 mM spermidine. (C and G) Concentration-dependent shifts induced by Mg2+ at 37°C, Mg2+ at 42°C, spermidine at 37°C, or spermidine at 42°C in magnitude at 208 nm are shown. Values are means ± standard deviations for three determinations. (D and H) The Kdapp values of spermidine for RpoE WT RNA and RpoE −20(ΔU) RNA at 37°C and 42°C were determined according to the double-reciprocal equation plot. SPD, spermidine.
FIG. 7.
Effect of spermidine on RpoE WT (166-mer) and RpoE −20(ΔU) (165-mer) RNA-dependent [3H]fMet-tRNA binding to ribosomes. The binding of [3H]fMet-tRNA to ribosomes in the presence of RpoE WT RNA (A) or RpoE −20(ΔU) RNA (B) was measured in the presence of various concentrations of Mg2+ with (•) or without (○) 1 mM spermidine at 37°C or 42°C. (C) The binding of [3H]fMet-tRNA to ribosomes with RpoE WT RNA (•) or RpoE −20(ΔU) RNA (□) was measured in the presence of various concentrations of spermidine and 3 mM Mg2+ at 42°C. Values are means ± standard errors of triplicate determinations. SPD, spermidine.
DISCUSSION
To study the role of polyamines on cell growth at 42°C, a polyamine-requiring mutant, MA261, was used (7). The doubling time of the mutant cultured in the synthetic medium A in the presence of 0.6 mM putrescine was 120 min, while that of the wild-type strain W3110 cultured in the absence of putrescine was 90 min. Polyamine content in E. coli W3110 (33.5 and 12.4 nmol/mg protein of putrescine and spermidine, respectively) was higher than that in E. coli MA261 cultured in the presence of 0.6 mM putrescine (Fig. 1). Thus, it is thought that polyamine content in cells is correlated with the rate of cell growth. In the present study, changes in polyamine levels in polyamine-requiring mutant MA261 at least partially mimic the polyamine effects in normal cells. Actually, the level of RpoE protein per rpoE mRNA in E. coli W3110, i.e., an indicator of stimulation of RpoE synthesis at the level of translation, was nearly equal to that in E. coli MA261 cultured in the presence of putrescine. Furthermore, the rate of cell growth of E. coli W3110 cultured in the synthetic medium A did not change significantly at 37°C and 42°C, which was similar to that of E. coli MA261 cultured in the presence of putrescine. However, cell growth of E. coli MA261 in the absence of putrescine was slower at 42°C than at 37°C. The results suggest that polyamines function more effectively at high temperature.
It is known that polyamines exist mostly as polyamine-RNA complexes in E. coli (29). Thus, we looked for proteins whose synthesis is enhanced by polyamines at the level of translation at 42°C, as we have previously seen with cells grown at 37°C (16, 45), in order to clarify the role of polyamines under heat shock conditions. We found that synthesis of RpoE, one of the sigma factors involved in the heat shock response (14), and of StpA, a nucleoid-associated DNA binding protein (37, 48), is enhanced by polyamines at the level of translation at 42°C. These two proteins are important proteins for E. coli to grow at 42°C, so a greater effect of polyamines on cell growth at 42°C than at 37°C is at least partially explained by polyamine stimulation of the synthesis of RpoE and StpA. It has been reported that polyamines decrease E. coli outer membrane permeability (9). However, this change in membrane permeability occurs at very high concentrations of polyamines (9), so it is unlikely that changes in membrane permeability are involved in the enhanced effects of polyamines on cell growth at 42°C. It has also been reported that growth of mammalian cells is inhibited by an increase in decarboxylated S-adenosylmethionine, the aminopropyl donor for spermidine and spermine synthesis produced by S-adenosylmethionine decarboxylase (5). The activity of S-adenosylmethionine decarboxylase in E. coli MA261 cultured in the absence of putrescine was lower than that cultured in the presence of putrescine, suggesting that toxicity caused by decarboxylated S-adenosylmethionine is negligible in E. coli MA261 cultured in the absence of putrescine.
Data on polyamine stimulation of the synthesis of RpoE and StpA were obtained with cells harvested at an A540 of 0.15. In this case, cells cultured in the presence of putrescine were harvested at an earlier time than cells cultured in the absence of putrescine. Polyamine stimulation of the synthesis of these proteins was also observed when cells were cultured in the presence and absence of putrescine at 8 h (data not shown). The results indicate that polyamines stimulate the synthesis of RpoE and StpA regardless of growth phase and incubation time.
It is shown that a selective structural change by polyamines of the bulged-out region in the initiation site of rpoE mRNA is involved in polyamine stimulation of RpoE synthesis. A predicted secondary structure of the initiation region of rpoE mRNA was constructed using the initiation region of rpoE mRNA by the method of Zuker (49). The predicted secondary structure of the initiation region of intact rpoE mRNA was exactly the same as that of rpoE mRNA consisting of 130 nucleotides (data not shown). The results support the idea that the bulged-out structure actually exists in the initiation site of rpoE mRNA.
The results, taken together, support our previous hypothesis concerning polyamine stimulation of cell growth (16). That is, polyamines enhance the synthesis of some key regulators of gene expression at the level of translation through interactions with specific regions of their cognate mRNAs. These regulatory proteins in turn enhance the expression of approximately 300 kinds of mRNAs, rRNA, and some kinds of tRNAs which are important for cell growth.
Acknowledgments
We thank K. Williams and A. J. Michael for their help in preparing the manuscript. Thanks are also due to Y. Nakamura, T. Mizuno, W. K. Maas, and C. A. Gross for the kind supply of antibodies to RF2 and H-NS, E. coli MA261, and E. coli CAG22216.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture, and Technology, Japan, and by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Adler, J., and B. Templeton. 1967. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46175-184. [DOI] [PubMed] [Google Scholar]
- 2.Ali Azam, T., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1816361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkins, J. F., J. B. Lewis, C. W. Anderson, and R. F. Gesteland. 1975. Enhanced differential synthesis of proteins in a mammalian cell-free system by addition of polyamines. J. Biol. Chem. 2505688-5695. [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Claverie, N., and P. S. Mamont. 1989. Comparative antitumor properties in rodents of irreversible inhibitors of l-ornithine decarboxylase, used as such or as prodrugs. Cancer Res. 494466-4471. [PubMed] [Google Scholar]
- 6.Cohen, S. S. 1998. A guide to polyamines, p. 1-543. Oxford University Press, Oxford, United Kingdom.
- 7.Cunningham-Rundles, S., and W. K. Maas. 1975. Isolation, characterization, and mapping of Escherichia coli mutants blocked in the synthesis of ornithine decarboxylase. J. Bacteriol. 124791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 27620866-20875. [DOI] [PubMed] [Google Scholar]
- 9.Dela Vega, A. L., and A. H. Delcour. 1996. Polyamines decrease Escherichia coli outer membrane permeability. J. Bacteriol. 1783715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echandi, G., and I. D. Algranati. 1975. Defective 30S ribosomal particles in a polyamine auxotroph of Escherichia coli. Biochem. Biophys. Res. Commun. 671185-1191. [DOI] [PubMed] [Google Scholar]
- 11.Emory, S. A., and J. G. Belasco. 1990. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 1724472-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higashi, K., K. Kashiwagi, S. Taniguchi, Y. Terui, K. Yamamoto, A. Ishihama, and K. Igarashi. 2006. Enhancement of +1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli. J. Biol. Chem. 2819527-9537. [DOI] [PubMed] [Google Scholar]
- 13.Higashi, K., Y. Terui, A. Suganami, Y. Tamura, K. Nishimura, K. Kashiwagi, and K. Igarashi. 2008. Selective structural change by spermidine in the bulged-out region of double-stranded RNA and its effect on RNA function. J. Biol. Chem. 28332989-32994. [DOI] [PubMed] [Google Scholar]
- 14.Hiratsu, K., M. Amemura, H. Nashimoto, H. Shinagawa, and K. Makino. 1995. The rpoE gene of Escherichia coli, which encodes σE, is essential for bacterial growth at high temperature. J. Bacteriol. 1772918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi, K., and K. Kashiwagi. 2006. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. 13911-16. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344633-642. [PMC free article] [PubMed] [Google Scholar]
- 18.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271559-564. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi, K., K. Kashiwagi, R. Aoki, M. Kojima, and S. Hirose. 1979. Comparative studies on the increase by polyamines of fidelity of protein synthesis in Escherichia coli and wheat germ cell-free systems. Biochem. Biophys. Res. Commun. 91440-448. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi, K., K. Kashiwagi, H. Hamasaki, A. Miura, T. Kakegawa, S. Hirose, and S. Matsuzaki. 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J. Bacteriol. 166128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi, K., K. Kashiwagi, K. Kishida, Y. Watanabe, A. Kogo, and S. Hirose. 1979. Defect in the split proteins of 30-S ribosomal subunits and under-methylation of 16-S ribosomal RNA in a polyamine-requiring mutant of Escherichia coli grown in the absence of polyamines. Eur. J. Biochem. 93345-353. [DOI] [PubMed] [Google Scholar]
- 22.Igarashi, K., K. Sugawara, I. Izumi, C. Nagayama, and S. Hirose. 1974. Effect of polyamines of polyphenylalanine synthesis by Escherichia coli and rat-liver ribosomes. Eur. J. Biochem. 48495-502. [DOI] [PubMed] [Google Scholar]
- 23.Ito, K., M. Uno, and Y. Nakamura. 1998. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc. Natl. Acad. Sci. USA 958165-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelenc, P. C., and C. G. Kurland. 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. USA 763174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 954953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1785447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashiwagi, K., R. Watanabe, and K. Igarashi. 1994. Involvement of ribonuclease III in the enhancement of expression of the speF-potE operon encoding inducible ornithine decarboxylase and polyamine transport protein. Biochem. Biophys. Res. Commun. 200591-597. [DOI] [PubMed] [Google Scholar]
- 28.Kusama-Eguchi, K., S. Watanabe, M. Irisawa, K. Watanabe, and K. Igarashi. 1991. Correlation between spermine stimulation of rat liver Ile-tRNA formation and structural change of the acceptor stem by spermine. Biochem. Biophys. Res. Commun. 177745-750. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto, S., K. Kashiwagi, K. Ito, S. Watanabe, and K. Igarashi. 1993. Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch. Biochem. Biophys. 30063-68. [DOI] [PubMed] [Google Scholar]
- 30.Müller, C. M., U. Dobrindt, G. Nagy, L. Emödy, B. E. Uhlin, and J. Hacker. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 1885428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano, S., T. Kanzaki, and N. Sugimoto. 2004. Influences of ribonucleotide on a duplex conformation and its thermal stability: study with the chimeric RNA-DNA strands. J. Am. Chem. Soc. 1261088-1095. [DOI] [PubMed] [Google Scholar]
- 32.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO J. 141043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezuchova, B., and J. Kormanec. 2001. A two-plasmid system for identification of promoters recognized by RNA polymerase containing extracytoplasmic stress response σE in Escherichia coli. J. Microbiol. Methods 45103-111. [DOI] [PubMed] [Google Scholar]
- 34.Rouvière, P. E., A. De Las Peñas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J. 141032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Extraction, purification, and analysis of mRNA from eukaryotic cells. In J. Sambrook and D. W. Russell (ed.), Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Shapira, S. K., J. Chou, F. V. Richaud, and M. J. Casadaban. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of β-galactosidase. Gene 2571-82. [DOI] [PubMed] [Google Scholar]
- 37.Sonden, B., and B. E. Uhlin. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 154970-4980. [PMC free article] [PubMed] [Google Scholar]
- 38.Terui, Y., K. Higashi, S. Taniguchi, A. Shigemasa, K. Nishimura, K. Yamamoto, K. Kashiwagi, A. Ishihama, and K. Igarashi. 2007. Enhancement of the synthesis of RpoN, Cra, and H-NS by polyamines at the level of translation in Escherichia coli cultured with glucose and glutamate. J. Bacteriol. 1892359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, D. H., N. Sugimoto, and S. M. Freier. 1988. RNA structure prediction. Annu. Rev. Biophys. Chem. 17167-192. [DOI] [PubMed] [Google Scholar]
- 40.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263149-162. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, S., K. Kusama-Eguchi, H. Kobayashi, and K. Igarashi. 1991. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 26620803-20809. [PubMed] [Google Scholar]
- 42.Wilson, K., F. M. Ausubel, R. Brent, and R. E. Kingston. 1987. Miniprep of bacterial genomic DNA, p. 2.4.1-2.4.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. E. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 43.Yoshida, M., K. Kashiwagi, G. Kawai, A. Ishihama, and K. Igarashi. 2001. Polyamine enhancement of the synthesis of adenylate cyclase at the translational level and the consequential stimulation of the synthesis of the RNA polymerase σ28 subunit. J. Biol. Chem. 27616289-16295. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, M., K. Kashiwagi, G. Kawai, A. Ishihama, and K. Igarashi. 2002. Polyamines enhance synthesis of the RNA polymerase σ38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 27737139-37146. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, M., K. Kashiwagi, A. Shigemasa, S. Taniguchi, K. Yamamoto, H. Makinoshima, A. Ishihama, and K. Igarashi. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 27946008-46013. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, M., D. Meksuriyen, K. Kashiwagi, G. Kawai, and K. Igarashi. 1999. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon AUG in OppA mRNA. J. Biol. Chem. 27422723-22728. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, A., and M. Belfort. 1992. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 206735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 151340-1349. [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]