Abstract
The respiratory chain of Escherichia coli is usually considered a device to conserve energy via the generation of a proton motive force, which subsequently may drive ATP synthesis by the ATP synthetase. It is known that in this system a fixed amount of ATP per oxygen molecule reduced (P/O ratio) is not synthesized due to alternative NADH dehydrogenases and terminal oxidases with different proton pumping stoichiometries. Here we show that P/O ratios can vary much more than previously thought. First, we show that in wild-type E. coli cytochrome bo, cytochrome bd-I, and cytochrome bd-II are the major terminal oxidases; deletion of all of the genes encoding these enzymes results in a fermentative phenotype in the presence of oxygen. Second, we provide evidence that the electron flux through cytochrome bd-II oxidase is significant but does not contribute to the generation of a proton motive force. The kinetics support the view that this system is as an energy-independent system gives the cell metabolic flexibility by uncoupling catabolism from ATP synthesis under non-steady-state conditions. The nonelectrogenic nature of cytochrome bd-II oxidase implies that the respiratory chain can function in a fully uncoupled mode such that ATP synthesis occurs solely by substrate level phosphorylation. As a consequence, the yield with a carbon and energy source can vary five- to sevenfold depending on the electron flux distribution in the respiratory chain. A full understanding and control of this distribution open new avenues for optimization of biotechnological processes.
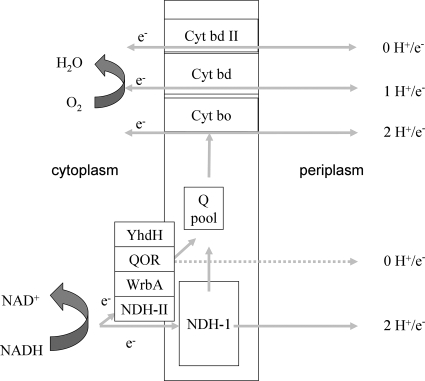
The aerobic respiratory chain of Escherichia coli can function with a variety of different membrane-bound NADH dehydrogenases, including NDH-I, NDH-II, and WrbA (8, 26-28), as well as YhdH and QOR (15, 38, 39), on the electron input side and three ubiquinol oxidases (cytochromes bd-I, bd-II, and bo) (12, 14, 19, 22, 29) on the output side (Fig. 1). The stoichiometry for the number of protons pumped for each two electrons transferred (H+/2e− ratio) has unequivocally been determined for NDH-I (H+/2e−, 4) and NDH-II (H+/2e−, 0) (10, 23, 41). Although no specific data are available for WrbA, YhdH, and QOR, it is generally assumed that these NADH:quinone oxidoreductases are not electrogenic because of the absence of (predicted) transmembrane alpha-helices (15, 38, 39). Similarly, the energy-conserving efficiencies of the cytochrome bd-I oxidase and the cytochrome bo oxidase are different; the cytochrome bd-I complex does not actively pump protons, but due to the oxidation of the quinol on the periplasmic side of the membrane and subsequent uptake of protons from the cytoplasmic side of the membrane, which are used in the formation of water, net electron transfer results in proton translocation with an H+/2e− stoichiometry of 2 (32). In contrast, the cytochrome bo complex actively pumps protons over the membrane, resulting in an H+/2e− stoichiometry of 4 (33, 42). The stoichiometry of proton translocation of the cytochrome bd-II complex is unknown.
FIG. 1.
Diagram of all NADH:quinone oxidoreductases and quinol:oxygen oxidoreductases in E. coli and their proton translocation properties. Cyt, cytochrome; Q, quinone.
Due to the differences in the H+/e− ratios of the dehydrogenases involved, two-electron transfer from NADH to the quinone pool may be accompanied by the translocation of any number of protons between 0 and 4, and subsequent reoxidation of the quinol pool may contribute to proton translocation again with a stoichiometry that depends on the relative activities of the terminal oxidases. The loose coupling between energy conservation and electron flow in respiration has been interpreted as a physiological means for the cell to cope with sudden changes in the rate of electron influx into the respiratory chain and/or in the availability of terminal electron acceptors on its terminal side (10). The fact that this energetic efficiency can vary is of great interest, both for understanding the physiological adaptive responses of the microbial cell and for biotechnological applications (e.g., synthesis of any oxidized compound with minimal biomass production). For this, it is important to quantify the flux distribution over and the efficiencies of the components of the respiratory machinery in relation to environmental conditions.
Previous studies (10) have shown that NDH-I, NDH-II, and the two well-characterized cytochrome oxidases contribute significantly to the overall electron flux and furthermore that the distribution of fluxes over these components depends on environmental conditions, such as the growth rate in glucose-limited chemostats (10). In addition, it has been suggested that the flux distribution over the terminal oxidases of E. coli is dependent on the culture pH (40). However, the cytochrome bd-II oxidase was not taken into account in these previous studies.
Here we present data that show that cytochrome bd-II oxidase participates significantly in oxygen reduction both during nonlimited growth in batch cultures and in glucose-limited chemostat cultures. For further quantification of the contribution of the respiratory chain to oxidative phosphorylation, it is essential to assess the in vivo H+/2e− stoichiometry of the cytochrome bd-II oxidase (4, 37). Essentially, the approach used in previous studies by Calhoun et al. (10) was followed: strains with respiratory chains that were modified such that their H+/2e− stoichiometry was fixed and known were grown under identical, glucose-limited conditions in chemostat culture. A flux analysis with respect to glucose catabolism and respiration allowed calculation of the rate of ATP synthesis for these strains. The data were then used as reference flux data for a strain that contained the cytochrome bd-II oxidase as the sole terminal oxidase. This strain showed a decreased yield with respect to oxygen and glucose. In this way we demonstrated that electron flow through the cytochrome bd-II oxidase does not contribute to the generation of a proton motive force. The results are discussed in view of the biochemical characterization of the enzyme and its physiological importance to adaptive responses by E. coli to an ever-changing environment.
MATERIALS AND METHODS
Continuous cultures.
E. coli K-12 strain MG1655 and various deletion strains were grown in an Applikon 2-liter fermentor at a constant dilution rate of 0.15 ± 0.01 h−1. A defined simple salts medium described by Evans et al. (16) with nitriloacetic acid (2 mM) rather than citrate as a chelator was used. Selenite (30 μg/liter) and thiamine (15 mg/liter) were added to the medium. Glucose was used as the sole carbon and energy source at a final concentration of 50 mM. The dilution rate was set by adjusting the medium supply rate. The pH was maintained at 7.0 ± 0.1 by titration with sterile 4 M NaOH, and the temperature was controlled at 37°C with a stirring rate of 800 rpm. The air supply rate was set at 1 liter/min.
In all cultures the steady-state specific rates of fermentation product formation and glucose and O2 consumption were measured as described by Alexeeva et al. (3).
Batch cultures.
In batch cultures, the composition of the medium was similar to the composition described above, except that sodium phosphate (pH 7) was used at a concentration of 100 mM instead of 10 mM to increase its buffer capacity. In selected experiments, glycerol or succinate was added as a carbon and energy source at a final concentration of 20 mM instead of glucose. Aeration of cultures during aerobic growth was accomplished by shaking 10-ml cultures in 100-ml flasks designed for aerobic cultivation. For anaerobic growth 30-ml cultures in sealed bottles (30 ml) were used. Cultures were inoculated from LB plates. The strains were maintained in vials in LB medium with 30% (wt/vol) glycerol at −70°C.
Construction of deletion mutants.
The single-deletion strains were ordered from the KEIO collection (5, 13). In order to construct strains with multiple deletions, the kanamycin marker was first removed as described by Datsenko and Wanner (13). Mutants with double and triple deletions were constructed by P1 phage transduction of the desired mutations (Table 1). Mutants were checked by PCR.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Cytochrome(s) present | Reference(s) |
|---|---|---|---|
| BW25113 | K-12 wild type | All cytochromes | 5 |
| MB20 | JW0421 ΔcyoB, kanamycin marker removed | Cytochromes bd-I and bd-II | 5; this study |
| MB21 | JW0961 ΔappB, kanamycin marker removed | Cytochromes bd-I and bo | 5; this study |
| MB28 | JW0723 ΔcydB, kanamycin marker removed | Cytochromes bo and bd-II | 5; this study |
| MB30 | BW25113 ΔcyoB ΔappB ΔnuoB, kanamycin marker removed | Cytochrome bd-I | This study |
| MB34 | BW25113 ΔcydB ΔappB ΔnuoB, kanamycin marker removed | Cytochrome bo | This study |
| MB37 | BW25113 ΔcyoB ΔcydB ΔnuoB, kanamycin marker removed | Cytochrome bd-II | This study |
| MB44 | BW25113 ΔcydB::kan ΔcyoB ΔappB ΔnuoB | None | This study |
| MG1655 | K-12 wild type | All cytochromes | |
| TBE016 | K-12 wild type, ΔappB | Cytochromes bd-I and bo | This study |
Quinone analysis.
Culture samples (2 ml) were quenched with 6 ml of ice-cold methanol. Then 6 ml of petroleum ether (boiling point, 40 to 60°C) was added rapidly to each mixture, and the mixture was vortexed for 1 min. After centrifugation of the mixture (900 × g, 2 min), the upper petroleum ether phase was removed and transferred to a test tube under a flow of nitrogen. Then 3 ml of petroleum ether was added to the lower phase, and the vortexing and centrifugation steps were repeated. The upper phases were combined. After evaporation under a flow of nitrogen to dryness, the extracts were stored. That extracts could be kept for at least 7 days under nitrogen at −20°C without changes in the quinone-quinol content was checked. Immediately before use, the extracted quinone-quinol mixture was resuspended using a glass rod in 80 μl ethanol and fractionated by high-performance liquid chromatography (HPLC) (Pharmacia LKB 2249 gradient pump system with an LKB 2151 variable-wavelength monitor) using a reversed-phase Lichrosorb (Chrompack, Bergen op Zoom, The Netherlands) RP10 C18 column (size, 4.6 mm; interna1 diameter, 250 mm). The column was equilibrated with ethanol-methanol (1:1, vol/vol) as the mobile phase. The flow rate was set at 1 ml/min at 20°C. Detection of the eluate was performed at 290 nm for ubiquinones and at 248 nm for menaquinones. The amount of each quinone species was calculated from the relevant peak area using ubiquinone-10 and menaquinone-4 as standards and the method described by Shestopolov et al. (35). Methanol, ethanol, and petroleum ether were analytical grade.
Peaks were identified by UV/visible and tandem mass spectrometry mass spectral analysis. A UV/visible spectrum of demethylmenaquinone (DMK) was kindly provided by A. V. Bogachev (Moscow University, Moscow, Russia). For mass spectral analysis, fractions collected from the HPLC were evaporated under nitrogen and redissolved in 89% acetonitrile, 10% water, 1% formic acid (LC grade; Merck, Frankfurt, Germany). The fractions were analyzed by off-line electrospray mass spectrometry using coated Picotips (Econo12; New Objective, Woburn, MA) and an electrospray ionization quantitative time of flight mass spectrometer (Micromass, Waters, Manchester, United Kingdom). Ions selected for tandem mass spectrometry collided with argon in a hexapole collision cell.
Analysis of carbon fluxes.
Steady-state bacterial dry weights were determined as described previously (1). Glucose, pyruvate, lactate, formate, acetate, succinate, and ethanol contents were determined by HPLC (LKB) with a REZEX organic acid analysis column (Phenomenex) at 45°C with 7.2 mM H2SO4 as the eluent, using an RI 1530 refractive index detector (Jasco) and AZUR chromatography software for data integration. All data have a carbon balance of 96% ± 4% as calculated from the glucose consumption and product formation rates.
Determination of cytochrome bd contents.
The cytochrome bd contents were determined by UV/visible spectroscopy using whole cells or purified membranes (20 mM potassium phosphate, pH 7) with an Olis upgraded DW2000 spectrophotometer and 1-mm cuvettes to reduce scatter. Reduced-versus-oxidized difference spectra were used for quantification of the cytochrome bd contents with wavelengths of 629 to 652 nm. For membranes, an extinction coefficient of 24 mM−1 cm−1 (629 to 652 nm) was used for the reduced-minus-oxidized spectra for cytochrome bd-I and bd-II oxidases (20). For whole cells, the reduced spectrum was used to calculate the cytochrome bd content after correction for scatter. The extinction coefficient for reduced cytochrome bd is 17.2 mM−1 cm−1 for 631 nm minus 657 nm.
Determination of enzymological parameters for cytochromes bd-I and bd-II.
Ubiquinol oxidase activities (37°C) were determined amperometrically using a Clark electrode and decyl-ubihydroquinone as the substrate with membranes purified from strains MB30 (containing only cytochrome bd-I) and MB37 (containing only cytochrome bd-II). The buffer was 20 mM potassium phosphate (pH 7.0). The Km and Vmax values were determined by the integrated rate method using initial substrate concentrations between 100 and 250 μM; the traces showed no indication of product inhibition by decyl-ubiquinone. Values for turnover or Vmax (s−1) were calculated from the oxidase activities and cytochrome bd concentrations determined optically. The maximal solubility of decyl-ubihydroquinone is approximately 250 μM. Therefore, the Km value determined and indicated below for cytochrome bd-II (250 ± 45 μM) in conjunction with a Vmax of 1,637 ± 150 s−1 represents the Km-Vmax pair with the lowest Km value compatible with the fit of the experimental traces.
Calculation of specific ATP synthesis rates and H+/e− ratios.
The rate of substrate level ATP synthesis [qATP (SLP aerobic)] is stoichiometrically coupled to the rate of CO2 production for complete oxidation of glucose as follows (see Fig. S1 in the supplemental material): 1Glc + 6O2 + 4ADP + 4Pi → 6CO2 + 4ATP + 6H2O. Hence, qATP (SLP aerobic) = 2/3·qCO2, where qCO2 is the rate of CO2 synthesis.
For strains or conditions that involve acetate and/or ethanol production, the additional amount of CO2 produced stoichiometrically per acetate and/or ethanol was taken into account as it is assumed that under the conditions used in this study pyruvate formate lyase is not active (21). In addition, lactate production is accompanied by ATP synthesis with a stoichiometry of 1:
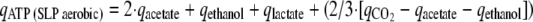 |
or
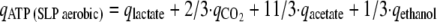 |
(1) |
where qacetate, qethanol, qlactate, and 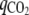 are the rates of synthesis of acetate, ethanol, lactate, and CO2, respectively.
are the rates of synthesis of acetate, ethanol, lactate, and CO2, respectively.
The rate of ATP synthesis with oxidative phosphorylation [qATP (ETP)] is related to the rate of oxygen reduction, to the number of protons translocated per electron transferred to oxygen, and to the stoichiometry of the number of ATP molecules synthesized per proton flowing back through the ATPase. Assuming that this stoichiometry is 4 in E. coli (36) and taking into account the fact that four electrons are needed to reduce molecular oxygen to water, it follows that
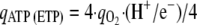 |
(2) |
where qO2 is rate of synthesis of O2. Combining equations 1 and 2 and reducing the yields results in:
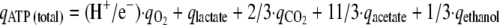 |
where qATP (total) is the total rate of ATP synthesis. This formula was used for calculating the qATP (total) of strains MB30 and MB34. To determine the H+/e− of MB37 it was rewritten as follows:
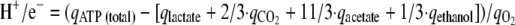 |
For qATP (total) the mean value for qATP (total) calculated for strains MB30 and MB34 was used.
RESULTS
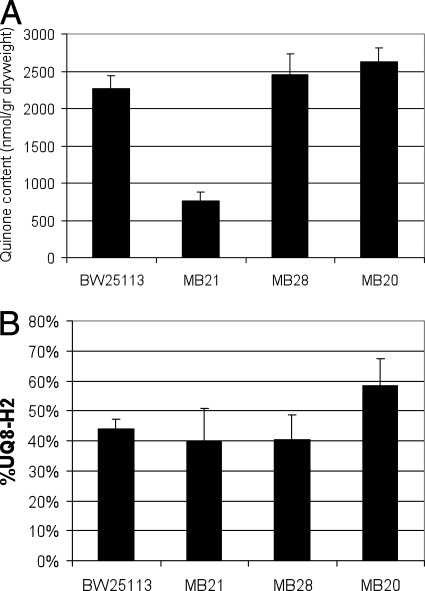
A comparative study was carried out with a number of isogenic E. coli strains that were modified in their respiratory chains. Strain MB20 lacks cytochrome bo oxidase, strain MB21 lacks cytochrome bd-II oxidase, and strain MB28 lacks cytochrome bd-I oxidase. These strains were grown aerobically in batch cultures on a minimal medium with glucose as the sole energy and carbon source. Samples were taken at mid-log phase and assayed to determine their redox state and the contents of their ubiquinone pool (Fig. 2A and 2B) (see Materials and Methods). Figure 2 shows that deletion of cyoB (strain MB20) resulted in a significant increase in the reduction level of the ubiquinone pool, whereas the total ubiquinone pool content remained unchanged. In contrast, the cydB deletion (strain MB28) did not result in a phenotype either with respect to the redox state or with respect to the cellular amount of total ubiquinone. This absence of a phenotype suggested that under these conditions cytochrome bd-I oxidase does not make a major contribution. Finally, the absence of the cytochrome bd-II complex (strain MB21) had a completely different effect on the ubiquinone pool; in strain MB21, the ubiquinone content was decreased almost threefold to 750 nmol/g (dry weight) cells, but no change in the redox state of the ubiquinone pool compared to the wild type was observed.
FIG. 2.
Biomass-specific ubiquinone-8 content (A) and redox state (B) during exponential growth in minimal medium on glucose in a shake flask culture for different respiratory deletion mutants: ubiquinone pool during aerobic batch growth in minimal media containing 50 mM glucose for strains BW25113 (wild type), MB21 (ΔappB), MB28 (ΔcydB), and MB20 (ΔcyoB). The error bars indicate standard deviations (n = 3). UQ8, ubiquinone-8.
Growth in steady-state chemostat cultures allows quantitative analysis of specific oxygen consumption rates and NADH/NAD cycling (2, 3). To assess qualitatively the contribution of cytochrome bd-II to respiration in fully aerobic conditions, glucose-limited chemostats set at a dilution rate of 0.2 h−1 (corresponding to a doubling time of approximately 3.6 h) were analyzed to determine specific catabolic rates. Wild-type MG1655 cells had a specific respiration rate that was approximately 47% higher than that of mutant strain TBE016 (similar to MB21 in an MG1655 background instead of BW25113) lacking cytochrome bd-II oxidase (8.7 ± 0.7 mmol g [dry weight]−1 h−1 and 5.9 ± 0.4 mmol g [dry weight]−1 h−1, respectively). Also (Fig. 2A), the total ubiquinone content was decreased threefold in the absence of cytochrome bd-II (TBE016) compared to the wild-type strain. These changes in the phenotype indicate that under glucose-limited aerobic conditions cytochrome bd-II oxidase is both present and active in the wild-type strain.
To further assess the functionality of the cytochrome bd-II complex as a quinol:oxygen oxidoreductase, three deletion mutants that contained only a single terminal oxidase and also lacked the coupled NADH dehydrogenase were constructed (Table 1). These three mutants, designated MB34 (containing cytochrome bo as the sole terminal oxidase), MB30 (containing cytochrome bd-I as the sole terminal oxidase), and MB37 (containing cytochrome bd-II as the sole terminal oxidase), were tested to determine their growth on the nonfermentable carbon sources succinate and glycerol. All three strains were able to grow well on both glucose (with acetate as the sole overflow product) and the nonfermentable carbon source glycerol as a carbon source. The same was true for MB34 and MB30 when succinate was supplied as the sole carbon source. However, MB37 could not be cultured on succinate. Another control strain (MB44), which lacked all three above-mentioned quinol:oxygen oxidoreductases (Table 1), could grow only on glucose and catabolized this energy source by full homolactic fermentation; i.e., all of the glucose consumed could be accounted for by lactate and biomass production. In accordance with the observed homolactic behavior, this strain expressed no significant respiratory activity. When this control strain was grown under anaerobic conditions, homolactic fermentation was not observed; instead, the cells produced amounts of acetate and ethanol usually observed for wild-type cells carrying out mixed fermentations (data not shown). In the absence of oxygen, pyruvate formate lyase was apparently functional. The inactivity of pyruvate formate lyase under aerobic conditions (9) prevents mixed acid fermentation, and therefore strain MB44 exhibits homolactic fermentation in aerobic conditions. It should be mentioned that the absence of respiration in a strain lacking the three terminal oxidases contrasts with the findings reported by Portnoy et al. (30), who found that oxygen consumption occurred after deletion of all three quinol oxidases.
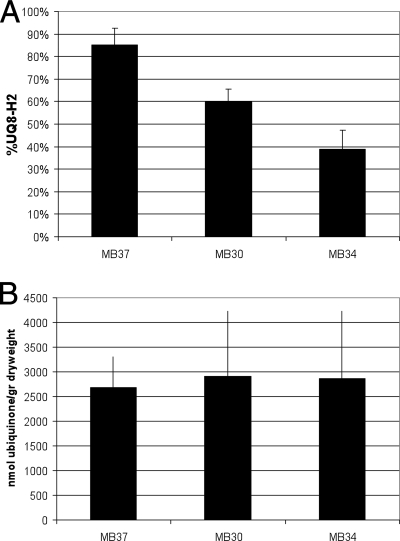
Growth in steady-state chemostat cultures allows quantitative analysis of specific oxygen consumption rates and NADH/NAD cycling (2, 3). Glucose-limited chemostats set at a dilution rate of 0.15 h−1 (corresponding to a doubling time of approximately 4.6 h) were analyzed to determine specific catabolic rates. In contrast to the wild-type strain, which oxidized glucose completely to carbon dioxide (2), the mutant strains that contain a respiratory chain with a fixed H+/e− stoichiometry produced various amounts of acetate as an overflow metabolite (Table 2). This suggests that the overall capacity for NADH oxidation is not sufficient to cope with the specific NADH production rate due to glycolytic and tricarboxylic acid activity. The highest level of acetate production, which was accompanied by minor but significant lactate production, was observed for strain MB37 (Table 2). All strains with a linear respiratory chain exhibited high oxygen consumption rates (Table 2) and, accordingly, increased glycolytic fluxes compared to the wild type. It should be emphasized that these strains, which lack the coupled NADH dehydrogenase (ΔnuoB), have a lower efficiency of energy conservation (i.e., a smaller amount of ATP synthesized per oxygen molecule reduced [P/O ratio]) and therefore have to increase their catabolic rates to fulfill the energetic demands for growth and maintenance under the prevailing conditions. Apparently this is due in part to increased activity of the respiratory chain and in part to increased substrate level phosphorylation. Fig. 3A shows that the ubiquinone pool of strain MB37 was much more reduced than those of strains MB34 and MB30. No significant differences in the ubiquinone content were observed (Fig. 3B), although it should be mentioned that very high standard deviations were obtained for unknown reasons. Throughout these experiments, the oxygen tension in the chemostats varied between 80 and 90% air saturation, and therefore the observed differences in the redox state of the ubiquinone pool cannot be explained by changes in oxygen availability or ubiquinone content (6).
TABLE 2.
Properties of various E. coli mutants that have a linear respiratory chain grown aerobically in glucose-limited chemostat conditions at a dilution rate of 0.15 h−1a
| Mutant | qO2 {mmol (g [dry wt])−1 h−1} | qglucose {mmol (g [dry wt])−1 h−1} | Yglucose (g [dry wt]) (g Glc)−1 | qacetate {mmol (g [dry wt])−1 h−1} | qATP {mmol (g [dry wt])−1 h−1} | H+/2e− |
|---|---|---|---|---|---|---|
| MB30 (cytochrome bd-I) | 8.8 ± 1.3 | 3.2 | 0.26 | 1.5 ± 0.5 | 16.3 ± 2.2 | 2 |
| MB37 (cytochrome bd-II) | 11.1 ± 0.6 | 6.0 | 0.14 | 5.7 ± 0.7 | 16.6 | 0.2 ± 0.1 |
| MB34 (cytochrome bo) | 6.4 ± 0.4 | 2.3 | 0.36 | 0.4 ± 0.4 | 17.0 ± 0.4 | 4 |
qATP values were calculated as described in Materials and Methods with an H+/2e− stoichiometry of 2 for strain MB30 and of 4 for strain MB34, respectively. The average qATP value was then used to calculate the H+/2e− stoichiometries for strain MB37. Other fluxes were determined as described in Materials and Methods. Strain MB37 produced lactate with a qlactate of 0.8 mmol g (dry weight)−1 h−1. The carbon balances were 96% ± 4% (n = 3). qglucose, rate of synthesis of glucose; Yglucose, yield on glucose.
FIG. 3.
Analysis of the ubiquinone pool during continuous growth in glucose-limited chemostat conditions for different respiratory deletion mutants: redox state (A) and content (B) of the ubiquinone pool of MB30, MB37, and MB34 during aerobic glucose-limited chemostat growth in minimal media containing 50 mM glucose. The error bars indicate standard deviations (n = 3).
The results clearly show that the lack of two of the three terminal oxidases affected the balance between glycolytic and tricarboxylic acid fluxes on the one hand and the respiratory flux on the other. In order to determine whether this was due to the capacity of the oxidases or to differences in the affinity for ubiquinol, the oxidase contents of glucose-limited MB30 and MB37 cells were determined, along with their Vmax, Km (QH2), and Km (O2) values (see Fig. S2 in the supplemental material). The data for the kinetics of cytochrome bd-II oxidase and all of the kinetic parameters calculated are summarized in Table 3. Whereas the cellular contents of the mutants are similar, the Vmax, Km, O2 and Km, QH2 values of the two cytochrome bd oxidases differ. The cytochrome bd-I oxidase has a fourfold-lower maximal turnover rate but a threefold-higher affinity for ubiquinol and a 10- to 20-fold-higher affinity for oxygen. From the specific respiration rates and the cellular contents, it was calculated that the specific in vivo activities of the oxidases are not significantly different under the conditions tested (Table 3) and that the rates for these enzymes are approximately 27% (cytochrome bd-I oxidase) and 9% (cytochrome bd-II oxidase) of the maximal rates attainable.
TABLE 3.
Enzymatic characteristics of the two cytochromes bd from E. colia
| Cytochrome | Vmax (mol O/mol cytochrome bd/s) | Km (O2) at pH 7 (μM) | Km (UQ-H2) at pH 7 (μM) | Cellular content (nmol/g protein) | In vivo sp act (mol O2/mol cytochrome bd/s) |
|---|---|---|---|---|---|
| Cytochrome bd-I | 218 ± 20 | 0.3b | 85 ± 5 | 90 ± 29 | 58 ± 11 |
| Cytochrome bd-II | 818 ± 75 | 2.0 ± 0.3 | 250 ± 45 | 91 ± 32 | 70 ± 12 |
| Cytochrome bo | 225c | 6.0d | 47e | ND | ND |
In vitro Vmax and Km were determined as described in Materials and Methods. For the in vivo specific activity of the two quinol oxidases, the oxygen fluxes measured in continuous cultures (see Table 2) were divided by the measured cytochrome bd content per gram of cells (n = 3). ND, not determined.
Data from reference 34.
Data from reference 25.
Data from reference 34.
The quantitative results for the catabolic fluxes can be used to assess the in vivo contribution of the cytochrome bd-II oxidase to the buildup of a proton motive force. Previously, we used a similar method to determine H+/2e− stoichiometries in vivo by growing E. coli cultures in glucose-limited chemostats (10); however, the alternative cytochrome bd-II oxidase and the alternative NAD(P)H:quinone oxidoreductases WrbA, QOR, and YhdH were not taken into account. Here we did take the presence of these enzymes into account. Essentially, this method uses the rate of ATP synthesis calculated for strains with linear respiratory chains composed of mechanistically well-characterized components as a reference value to calculate the rate in a mutant with the uncharacterized cytochrome bd-II oxidase. It was assumed that the ATP demands for growth and maintenance are identical in the three strains since the growth conditions (and hence the growth rates) were identical. The equations used for the calculations are shown in Materials and Methods. The validity of the method is shown by the fact that for strains MB30 and MB34 (containing oxidases well characterized with respect to H+/2e− stoichiometries, which are 0 for NDH-II, 2 for the cytochrome bd-I oxidase, and 4 for the cytochrome bo oxidase [2, 10, 32, 33]) similar values for the specific ATP synthesis rates were obtained (16.3 ± 2.4 mmol g [dry weight]−1 h−1 and 17.0 ± 0.4 mmol g [dry weight]−1 h−1, respectively). With an average value of 16.6 mmol g (dry weight)−1 h−1 as a reference, it was then calculated that in MB37 all ATP is synthesized at the level of substrate phosphorylation and that consequently the cytochrome bd-II oxidase does not translocate protons electrogenically; i.e., an H+/2e− ratio as low as 0.2 ± 0.1 was calculated. These results are summarized in Table 2.
DISCUSSION
Two of the three quinone:oxygen oxidoreductases present in E. coli have previously been shown to be active in batch cultures (11, 17), as well as during continuous glucose-limited growth (10). So far, the role of the third quinone:oxygen oxidoreductase, cytochrome bd-II oxidase, in an actively growing and respiring cell has not been clarified in vivo.
We elucidated this role with respect to both the relative contribution to respiration and energy conservation under glucose-excess and glucose-limited conditions. For these conditions, we first concluded that the cytochrome bd-II oxidase is the third and final significant catabolic oxygen-reducing system since strain MB44, which lacks all three quinol:oxygen oxidoreductases (Table 1), is not able to reduce oxygen and behaves like a homolactic fermentative organism. The reason that this mutant strain does not carry out mixed fermentation must be determined in the absence of functional pyruvate formate lyase, as this enzyme is reversibly inactivated by oxygen (21, 43). This is consistent with the observation that under anaerobic conditions strain MB44 carries out a normal mixed-acid fermentation. Interestingly, it has recently been reported (30) that a similar mutant is able to respire. Whether this is due to differences in growth conditions or to differences in strains awaits further study.
Second, our data justify the conclusion that, in accordance with previous suggestions (4, 12, 37), cytochrome bd-II oxidase functions as a proper quinol:oxygen oxidoreductase. This conclusion is further supported by the observation that the expression of cytochrome bd-II oxidase as the sole terminal oxidase allows growth on a highly reduced substrate like glycerol. The phenotype of a strain that lacks this oxidase (MB21) includes a smaller ubiquinone pool when the strain is grown under glucose-excess conditions. Furthermore, such a strain has a much decreased oxygen flux in glucose-limited chemostat conditions. These results strongly suggest that cytochrome bd-II oxidase contributes significantly to the overall respiratory electron flux under standard growth conditions.
Under the conditions tested, all strains that contain only a single terminal oxidase were not able to oxidize glucose to completion. The shift to overflow and fermentative catabolism was most clearly observed for strain MB37, in which only cytochrome bd-II oxidase is functional. This finding seems to be compatible with the fact that this oxidase has the lowest affinity for ubiquinol (Table 2), but its high Vmax value counteracts this characteristic. However, one key to a proper kinetic analysis is reliable estimates of apparent substrate concentrations, and since such estimates cannot be made for processes taking place in the membrane, a straightforward kinetic analysis cannot be carried out. Therefore, we can present only a qualitative interpretation, namely, that the cytochrome bd-II oxidase is a low-affinity system that, under the conditions tested, has activity that is approximately 10% of the activity measured in vitro. As we have shown that the cytochrome bd-II oxidase is nonelectrogenic, strain MB37 is energetically compromised and therefore must have a high catabolic flux (i.e., a high rate of NADH synthesis) in order to maintain a sufficient ATP synthesis rate. Together with the impeded electron flux to oxygen, this hampers complete oxidation of glucose and may give rise to an increased NADH pool; the resulting lower tricarboxylic acid cycle and pyruvate dehydrogenase complex activity then leads to increased intracellular pyruvate levels, allowing lactate dehydrogenase, despite its relative low affinity for pyruvate, to compete successfully with the pyruvate dehydrogenase complex.
The conclusion that cytochrome bd-II oxidase is nonelectrogenic is fully compatible with the observation that strain MB37 was unable to grow on succinate. It has been reported (37) that a strain with cytochrome bd-II oxidase as the sole terminal oxidase can grow on succinate. However, in the study of Sturr et al. (37) the coupled NADH dehydrogenase (NDH-I) was present, and apparently sufficient energy conservation could proceed via this complex. In MB37 NDH-I is not active, and therefore no proton motive force can be formed by respiration. Hence, ATP synthesis completely depends on substrate level phosphorylation; it should be noted that succinate catabolism yields only one ATP molecule per succinate molecule and, in addition, that succinate import requires two protons per molecule (18, 24). Without oxidative phosphorylation, the net ATP gain is clearly too small to support growth on this substrate.
An attractive, but very speculative, hypothesis to explain why cytochrome bd-II does not form a proton motive force results from the fact that the protons and electrons used for generation of water from oxygen are taken up from the periplasmic space, whereas in cytochrome bd-I the protons are taken up from the cytoplasm (32). The di-heme center of cytochrome bd-I is thought to be located near the periplasm (7), and sequence comparison has suggested that this is the case for E. coli cytochrome bd-I, as well as cytochrome bd-II. One could assume that the overall transmembrane and heme topologies of all cytochrome bd oxidases are essentially the same. However, changes in only a few amino acid residues in the di-heme region of the protein may lead to changes in the vectorial process.
Our observation of a noncoupled terminal oxidase in E. coli is in agreement with previous suggestions concerning the importance of an uncoupled respiratory chain component as a tool for the cell to respond appropriately to environmental changes, such as a sudden influx of electrons at the starting point of catabolism (e.g., relief of energy source limitation). We now may view the respiratory chain of E. coli as a device with several input and output channels for electrons (NADH dehydrogenases and terminal oxidases) that are or are not sensitive to back pressure from the proton motive force. Such a device leads to greater metabolic (and regulatory) flexibility than highly “rigid” but more efficient systems.
Glucose-excess conditions presumably are not energy-limited conditions, and microbial species like E. coli are usually loosely coupled and do not tend to maximize energetic efficiency under these conditions. This is exemplified by the overflow metabolism phenomenon. Thus, cytochrome bd-II oxidase may have a role in oxidation of ubiquinol at a high oxidative flux, which is not hampered by back pressure from the proton motive force (6). On the other hand, under steady-state glucose-limited conditions (i.e., presumably energy-restricted conditions), the contribution of the cytochrome bd-II oxidase is likely to be relatively low due to the more oxidized ubiquinone pool (6% [6]), coupled with the poor affinity of the oxidase for ubiquinol. However, upon relief of the limitation, increased NADH synthesis and hence increased ubiquinol pools can be compensated for by the oxidase, thus preventing the cell from becoming metabolically compromised. The low affinity of cytochrome bd-II oxidase is compatible with its role as an uncoupling device.
Our results expand the variability and flexibility of the respiratory chain of E. coli further than has been assumed so far (10). Indeed, it is feasible that there is an electron flux distribution that results in the respiratory chain being virtually nonelectrogenic (i.e., NDH-II → cytochrome bd-II) at the one extreme and being fully coupled (i.e., NHD-1 → cytochrome bo) at the other extreme. Taking into account substrate level phosphorylation and redistribution of glucose over anabolism and catabolism, a fivefold difference in the yield on glucose between these extremes can be obtained. For apparent H+/ADP → ATP stoichiometries lower than the stoichiometry of 4 that was assumed here (36), a sevenfold change could be obtained. The implications and benefits for applications are obvious. Further insight into the regulation of the flux distribution should provide the tools to control the efficiency of carbon source conversion over a broad range. Thus, induction of a highly efficient respiratory activity during the growth phase, followed by a switch to a low-efficiency activity during the phase in which the carbon source is to be converted to an end product, should contribute to optimization of a production process by minimization of ATP (and therefore biomass) synthesis in the production phase.
Supplementary Material
Acknowledgments
This work was supported by NWO-ALW project 812.05.004. A. Ter Beek contributed to this study as a postdoctoral fellow for the SysMo-SUMO project supported by NWO-SysMo 826.06.002.
We thank Jorine Zandhuis and Marc J. F. Strampraad (TUD) for their contributions to this study.
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 1824934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2002. Quantitative assessment of oxygen availability: perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J. Bacteriol. 1841402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlung, T., and L. Brondsted. 1994. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J. Bacteriol. 1765414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bekker, M., G. Kramer, A. F. Hartog, M. J. Wagner, C. G. de Koster, K. J. Hellingwerf, and M. J. de Mattos. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 1531974-1980. [DOI] [PubMed] [Google Scholar]
- 7.Belevich, I., V. B. Borisov, J. Zhang, K. Yang, A. A. Konstantinov, R. B. Gennis, and M. I. Verkhovsky. 2005. Time-resolved electrometric and optical studies on cytochrome bd suggest a mechanism of electron-proton coupling in the di-heme active site. Proc. Natl. Acad. Sci. USA 1023657-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorklof, K., V. Zickermann, and M. Finel. 2000. Purification of the 45 kDa, membrane bound NADH dehydrogenase of Escherichia coli (NDH-2) and analysis of its interaction with ubiquinone analogues. FEBS Lett. 467105-110. [DOI] [PubMed] [Google Scholar]
- 9.Broderick, J. B., T. F. Henshaw, J. Cheek, K. Wojtuszewski, S. R. Smith, M. R. Trojan, R. M. McGhan, A. Kopf, M. Kibbey, and W. E. Broderick. 2000. Pyruvate formate-lyase-activating enzyme: strictly anaerobic isolation yields active enzyme containing a [3Fe-4S]+ cluster. Biochem. Biophys. Res. Commun. 269451-456. [DOI] [PubMed] [Google Scholar]
- 10.Calhoun, M. W., K. L. Oden, R. B. Gennis, M. J. de Mattos, and O. M. Neijssel. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 1753020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 1726333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dassa, J., H. Fsihi, C. Marck, M. Dion, M. Kieffer-Bontemps, and P. L. Boquet. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol. Gen. Genet. 229341-352. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Mello, R., S. Hill, and R. K. Poole. 1996. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 142755-763. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, K. J., J. M. Thorn, J. A. Daniher, N. E. Dixon, and D. L. Ollis. 1994. Crystallization and preliminary X-ray diffraction studies on a soluble Escherichia coli quinone oxidoreductase. J. Mol. Biol. 240501-503. [DOI] [PubMed] [Google Scholar]
- 16.Evans, C. G. T., D. Herbert, and D. W. Tempest. 1970. The continuous culture of microorganisms. Construction of a chemostat, vol. 2. Academic Press, London, United Kingdom.
- 17.Georgiou, C. D., T. J. Dueweke, and R. B. Gennis. 1988. Regulation of expression of the cytochrome d terminal oxidase in Escherichia coli is transcriptional. J. Bacteriol. 170961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutowski, S. J., and H. Rosenberg. 1975. Succinate uptake and related proton movements in Escherichia coli K12. Biochem. J. 152647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingledew, W. J., and R. K. Poole. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48222-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junemann, S., and J. M. Wrigglesworth. 1995. Cytochrome bd oxidase from Azotobacter vinelandii. Purification and quantitation of ligand binding to the oxygen reduction site. J. Biol. Chem. 27016213-16220. [DOI] [PubMed] [Google Scholar]
- 21.Knappe, J., and G. Sawers. 1990. A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol. Rev. 6383-398. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, C., R. K. Poole, I. Salmon, and B. Chance. 1985. The oxygen reaction of the cytochrome d-terminated respiratory chain of Escherichia coli at sub-zero temperatures. Kinetic resolution by EPR spectroscopy of two high-spin cytochromes. FEBS Lett. 190227-231. [DOI] [PubMed] [Google Scholar]
- 23.Leif, H., V. D. Sled, T. Ohnishi, H. Weiss, and T. Friedrich. 1995. Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur. J. Biochem. 230538-548. [DOI] [PubMed] [Google Scholar]
- 24.Lo, T. C., M. K. Rayman, and B. D. Sanwal. 1972. Transport of succinate in Escherichia coli. I. Biochemical and genetic studies of transport in whole cells. J. Biol. Chem. 2476323-6331. [PubMed] [Google Scholar]
- 25.Mason, M. G., M. Shepherd, P. Nicholls, P. S. Dobbin, K. S. Dodsworth, R. K. Poole, and C. E. Cooper. 2009. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 594-96. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi, Y., Y. Nakai, N. Shimba, H. Toyosaki, Y. Kawahara, S. Sugimoto, and E. Suzuki. 2004. The energetic conversion competence of Escherichia coli during aerobic respiration studied by 31P NMR using a circulating fermentation system. J. Biochem. 136509-515. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi, T., V. D. Sled, N. I. Rudnitzky, B. W. Jacobson, Y. Fukumori, S. W. Meinhardt, M. W. Calhoun, R. B. Gennis, H. Leif, T. Friedrich, et al. 1994. Biophysical and biochemical studies of bacterial NADH:quinone oxidoreductase (NDH-1). Biochem. Soc Trans. 2270S. [DOI] [PubMed] [Google Scholar]
- 28.Patridge, E. V., and J. G. Ferry. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 1883498-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, R. K., and G. M. Cook. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43165-224. [DOI] [PubMed] [Google Scholar]
- 30.Portnoy, V. A., M. J. Herrgard, and B. O. Palsson. 2008. Aerobic fermentation of d-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain. Appl. Environ. Microbiol. 747561-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pouvreau, L. A., M. J. Strampraad, S. Van Berloo, J. H. Kattenberg, and S. de Vries. 2008. NO, N2O, and O2 reaction kinetics: scope and limitations of the Clark electrode. Methods Enzymol. 43697-112. [DOI] [PubMed] [Google Scholar]
- 32.Puustinen, A., M. Finel, T. Haltia, R. B. Gennis, and M. Wikstrom. 1991. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 303936-3942. [DOI] [PubMed] [Google Scholar]
- 33.Puustinen, A., M. Finel, M. Virkki, and M. Wikstrom. 1989. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 249163-167. [DOI] [PubMed] [Google Scholar]
- 34.Sato-Watanabe, M., T. Mogi, H. Miyoshi, and Y. Anraku. 1998. Characterization and functional role of the QH site of bo-type ubiquinol oxidase from Escherichia coli. Biochemistry 375356-5361. [DOI] [PubMed] [Google Scholar]
- 35.Shestopalov, A. I., A. V. Bogachev, R. A. Murtazina, M. B. Viryasov, and V. P. Skulachev. 1997. Aeration-dependent changes in composition of the quinone pool in Escherichia coli. Evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett. 404272-274. [DOI] [PubMed] [Google Scholar]
- 36.Steigmiller, S., P. Turina, and P. Graber. 2008. The thermodynamic H+/ATP ratios of the H+-ATPsynthases from chloroplasts and Escherichia coli. Proc. Natl. Acad. Sci. USA 1053745-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturr, M. G., T. A. Krulwich, and D. B. Hicks. 1996. Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a Δcyo Δcyd strain complemented by genes from Bacillus firmus OF4. J. Bacteriol. 1781742-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sulzenbacher, G., V. Roig-Zamboni, F. Pagot, S. Grisel, A. Salomoni, C. Valencia, V. Campanacci, R. Vincentelli, M. Tegoni, H. Eklund, and C. Cambillau. 2004. Structure of Escherichia coli YhdH, a putative quinone oxidoreductase. Acta Crystallogr. D Biol. Crystallogr. 601855-1862. [DOI] [PubMed] [Google Scholar]
- 39.Thorn, J. M., J. D. Barton, N. E. Dixon, D. L. Ollis, and K. J. Edwards. 1995. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 249785-799. [DOI] [PubMed] [Google Scholar]
- 40.Verkhovskaya, M., M. Verkhovsky, and M. Wikstrom. 1992. pH dependence of proton translocation by Escherichia coli. J. Biol. Chem. 26714559-14562. [PubMed] [Google Scholar]
- 41.Weidner, U., S. Geier, A. Ptock, T. Friedrich, H. Leif, and H. Weiss. 1993. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J. Mol. Biol. 233109-122. [DOI] [PubMed] [Google Scholar]
- 42.Wikstrom, M., A. Bogachev, M. Finel, J. E. Morgan, A. Puustinen, M. Raitio, M. Verkhovskaya, and M. I. Verkhovsky. 1994. Mechanism of proton translocation by the respiratory oxidases. The histidine cycle. Biochim. Biophys. Acta 1187106-111. [DOI] [PubMed] [Google Scholar]
- 43.Yamada, T., S. Takahashi-Abbe, and K. Abbe. 1985. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 47129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.