Abstract
CtrA controls cell cycle programs of chromosome replication and genetic transcription. Phosphorylated CtrA∼P exhibits high affinity (dissociation constant [Kd], <10 nM) for consensus TTAA-N7-TTAA binding sites with “typical” (N = 7) spacing. We show here that ctrA promoters P1 and P2 use low-affinity (Kd, >500 nM) CtrA binding sites with “atypical” (N ≠ 7) spacing. Footprints demonstrated that phosphorylated CtrA∼P does not exhibit increased affinity for “atypical” sites, as it does for sites in the replication origin. Instead, high levels of CtrA (>10 μM) accumulate, which can drive CtrA binding to “atypical” sites. In vivo cross-linking showed that when the stable CtrAΔ3 protein persists during the cell cycle, the “atypical” sites at ctrA and motB are persistently bound. Interestingly, the cell cycle timing of ctrA P1 and P2 transcription is not altered by persistent CtrAΔ3 binding. Therefore, operator DNA occupancy is not sufficient for regulation, and it is the cell cycle variation of CtrA∼P phosphorylation that provides the dominant “activation” signal. Protein dimerization is one potential means of “activation.” The glutathione S-transferase (GST) protein dimerizes, and fusion with CtrA (GST-CtrA) creates a stable dimer with enhanced affinity for TTAA motifs. Electrophoretic mobility shift assays with GST-CtrA revealed cooperative modes of binding that further distinguish the “atypical” sites. GST-CtrA also binds a single TTAA motif in ctrA P1 aided by DNA in the extended TTAACCAT motif. We discuss how “atypical” sites are a common yet distinct category of CtrA regulatory sites and new implications for the working and evolution of cell cycle control networks.
The Caulobacter crescentus transcription regulator CtrA provides a new paradigm of cell cycle control (4, 18, 24). CtrA belongs to the large OmpR/PhoB response regulator (RR) family. Bacterial stimulus-response mechanisms often use RR proteins, which switch between phosphorylated and unphosphorylated states by contact with the cognate histidine kinase and histidine phosphotransfer proteins (38). Despite the apparent simplicity of this paradigm, the regulatory tasks of CtrA are exceptionally varied and complex. C. crescentus uses CtrA to divide asymmetrically and to produce distinct swarmer and stalked cells (Fig. 1). Cell cycle control also involves differentiation of swarmer cells to stalked cells, asymmetric remodeling that creates a new flagellated swarmer cell pole, and the regulation of chromosome replication so that it occurs in the stalked cells but not in the swarmer cells. Accordingly, CtrA controls genetic transcription at key stages of the cell cycle. CtrA also controls chromosome replication by binding five sites inside the replication origin (25, 31).
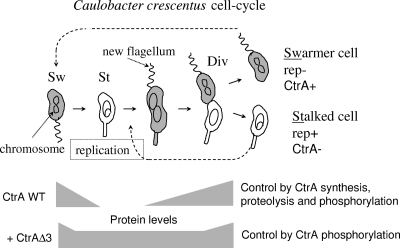
FIG. 1.
CtrA protein activity during the C. crescentus cell cycle. A nonreplicating but motile swarmer cell (Sw) differentiates into a replicating stalked cell (St). Growth of a predivisional cell (Div) produces a new flagellated swarmer pole. Segregating chromosomes are positioned in both the nonreplicating swarmer cell (rep−) and the replication-competent stalked cell (rep+). Shading indicates the temporal and spatial presence of CtrA. In wild-type (WT) cells, CtrA “activity” is controlled by cell cycle-programmed synthesis, proteolysis, and phosphorylation. In CtrAΔ3-containing cells, CtrA “activity” is controlled by cell cycle-programmed phosphorylation alone.
CtrA activity is controlled by cell cycle programs that adjust the CtrA concentration and phosphorylation state (5). In wild-type cells, cell cycle synthesis and proteolysis adjust the CtrA concentration (Fig. 1). CtrA is synthesized before cell division but after septum formation; the CtrA protein is degraded in a stalked cell while it is retained in a swarmer cell (Fig. 1). The CtrA protein is maintained in a swarmer cell until this cell differentiates into a stalked cell, where CtrA is rapidly degraded (5, 27). However, mutating the last three C-terminal amino acids blocks CtrA proteolysis. The resulting CtrAΔ3 protein is stable and maintained in all cell types. Surprisingly, the CtrAΔ3 protein fully complements a ctrA null allele (25), and CtrAΔ3-containing cells are indistinguishable from wild-type cells. Apparently, under laboratory conditions, CtrA synthesis and proteolysis are redundant controls for CtrA activity. In CtrAΔ3-containing cells (Fig. 1), the activity of CtrA is adjusted by cell cycle changes in its phosphorylation state alone (5). For example, when the CtrAΔ3 protein persists in stalked cells, it is poorly phosphorylated and presumably inactive. However, stable CtrAΔ3 becomes increasingly phosphorylated coincident with the requirement for it and coincident with the synthesis of wild-type CtrA protein in wild-type cells. An essential histidine kinase (10) and a phosphorelay system (4, 39) phosphorylate CtrA and thereby change its activity during the cell cycle.
Our studies address the specific basis of CtrA “activity” and the relative contributions of CtrA synthesis, proteolysis, and phosphorylation to cell cycle control (Fig. 1). How does phosphorylated CtrA∼P differ from unphosphorylated CtrA? One established biochemical “activity” of the phosphorylated CtrA∼P protein is increased affinity for the promoters of the C. crescentus fliQ, ccrM, and pilA genes (26, 34) and for the C. crescentus replication origin (31). The CtrA binding sites have the consensus sequence TTAA-N7-TTAA, whose N = 7 spacing is required for maximum CtrA-directed transcription (22). Over 50 transcription promoters are bound and regulated by CtrA (13), and it is not certain that all of their binding sites conform to the consensus described above. Below, we describe a distinct second category of weak CtrA binding sites that change previously untested assumptions about cell cycle transcription control.
MATERIALS AND METHODS
Strains and plasmids.
In all experiments requiring C. crescentus cultures, wild-type strain NA1000 (formerly CB15N), a synchronizable C. crescentus strain (8), was used. Synchronized swarmer cells were obtained by the density centrifugation method (8). All cultures were grown exponentially from an initial optical density at 600 nm (OD660) of 0.1 and were maintained at 30°C in liquid M2 medium supplemented with 0.2% glucose (M2G) (7). The lacZ transcription reporter plasmids pctrA-P1 and pctrA-P2 (6) and the ctrAΔ3-expressing plasmid pLS2747 (5) were mobilized into C. crescentus by conjugation with Escherichia coli strain S17-1 (33). The ctrAΔ3 allele which was used in these studies and which fully complements the ctrA null allele (25) was originally constructed by inserting the Omega antibiotic resistance cassette at the unique HpaI site of ctrA (5, 24). This manipulation replaced the last three natural amino acids (NAA) of CtrA with six new amino acids (GDPEID) from the Omega cassette (5). The ctrAΔ3 allele on plasmid pLS2747 was transcribed from the PxylX promoter, and in M2G it maintained the CtrAΔ3 protein throughout the cell cycle at levels approximately equal to the peak levels of wild-type CtrA protein.
In vitro CtrA binding to DNA.
DNase I protection footprint assays were done as previously described (31). A DNA fragment labeled with 32P at the 5′ end containing the P1 and P2 promoters of the ctrA gene (6) and a DNA fragment labeled with 32P end at the 5′ end containing the CtrA replication origin binding sites (binding sites a, b, c, d, and e) (31) were prepared as previously described. In our previous footprint studies (31), we used an N-terminal six-histidine-tagged protein (His-CtrA from plasmid pTRC7.4) and the same protein cleaved by enterokinase (EK-CtrA) to remove the six-histidine tag (Invitrogen). We also cloned the ctrA-containing DNA, the BamHI-EcoRI fragment of pTRC7.4, into plasmid pGEX-2T (GE Healthcare Biosciences), and we prepared a glutathione S-transferase (GST)-tagged protein (GST-CtrA from plasmid pGM2586) by affinity chromatography as recommended by the manufacturer. Purification of the CtrA protein and its conversion to the phosphorylated CtrA∼P form by E. coli EnvZ kinase have been described previously (26, 31). Previous experiments also demonstrated that more than 50% of the CtrA protein is phosphorylated by EnvZ (31). The phosphorylated CtrA∼P form has a half-life of approximately 1 h under the assay conditions used (data not shown). The binding assays in the experiments whose results are shown in Fig. 2B were completed within 10 min, and therefore there was not significant depletion of the phosphorylated CtrA∼P form.
FIG. 2.
DNase I footprint analysis of CtrA and CtrA∼P binding to the ctrA P1 and P2 promoters and to the replication origin (Cori). (A) DNA sequence landmarks of the P1 and P2 promoters (6) and Cori (31). Bent arrows indicate RNA start sites. Cross-hatched and filled bars indicate the sequences protected in vitro from DNase I digestion by CtrA. P1 transcription is repressed in vivo, while P2 transcription is activated in vivo, and the minus and plus signs indicate that CtrA binding is repressive at P1 and activating at P2. (B) Simultaneous DNase I footprint assays for ctrA P1 and P2 and for CtrA Cori binding sites a and b. The diagram shows that the same protein mixture (His-CtrA and EnvZ kinase) was used (with or without 0.5 mM ATP) for all reactions. The binding reaction monomer concentrations of unphosphorylated (CtrA) and phosphorylated (CtrA∼P) proteins are indicated above the lanes. The DNA substrates (20,000 cpm per lane) were labeled with 32P at the 5′ end at unique XmnI (ctrA P1 and P2) and HindIII (Cori binding sites a and b) endonuclease sites.
Molecular weights of proteins and protein-DNA complexes.
We used the electrophoretic mobility shift assay (EMSA) methods of Orchard and May (21) to determine the molecular weights of native proteins (Table 1) and of protein-DNA complexes (Table 2). The GST-CtrA protein was bound to double-stranded oligonucleotides spanning the specified target sites (see Fig. 8). Examples of the results of EMSA experiments for the P1 and P2 promoters are shown in Fig. 6 and 7. GST-CtrA (0.3 nM) and the double-stranded oligonucleotide indicated (0.1 nM) were combined in 50 μl of binding buffer (10 mM Tris [pH 7.6], 10 mM MgCl2, 50 mM NaCl, 5% glycerol) and incubated on ice for 10 min. Portions (10 μl) were removed, quickly mixed with 1 μl of dye (0.2% bromophenol blue in binding buffer), and applied to a prerun polyacrylamide gel containing 0.5× Tris-borate-EDTA prepared with either 5.0, 5.5, 6.0, 6.5, or 7.0% polyacrylamide (ratio of acrylamide to bisacrylamide, 29:1); 7.0% polyacrylamide gels are shown in Fig. 6, 7, and 10. Following electrophoresis (100 V, 8 mA, 60 min), the gels were stained first with ethidium bromide and then with Coomassie blue (PageBlue; Fermentas); side-by-side photographs of gels are shown in Fig. 6 and 7.
TABLE 1.
Molecular weights of CtrA protein preparations
| Protein band | Mol wt (103) | Multiple (protein monomers) |
|---|---|---|
| Denaturing SDS-PAGE | ||
| His-CtrA | 27 | |
| GST-CtrA | 52 | |
| Native PAGE | ||
| His-CtrA | 29 ± 3 | 1.1 ± 0.1 |
| GST-CtrA | 100 ± 10 | 1.9 ± 0.2 |
TABLE 2.
Molecular weights of GST-CtrA protein-DNA complexes, as determined by EMSA
| Bound DNA | Mol wt of protein plus mol wt of DNA (103) | Multiple (protein monomers)a |
|---|---|---|
| Cori binding sites a and b, lower band | 132 ± 12 | 2.0 ± 0.3 |
| Cori binding sites a and b, upper band | 234 ± 8 | 3.9 ± 0.2 |
| Cori binding site c | 135 ± 14 | 2.1 ± 0.3 |
| Cori binding site e | 153 ± 17 | 2.3 ± 0.4 |
| Wild-type P1 | 140 ± 14 | 2.2 ± 0.2 |
| P1 m3 | 138 ± 15 | 2.2 ± 0.2 |
| Wild-type P2, lower band | 144 ± 15 | 2.3 ± 0.3 |
| Wild-type P2, upper band | 227 ± 20 | 3.9 ± 0.4 |
| P2 m3 | 135 ± 15 | 2.1 ± 0.2 |
| P2 N7 | 147 ± 15 | 2.3 ± 0.3 |
Multiples were determined as follows: [(molecular weight of protein + molecular weight of DNA) − molecular weight of DNA]/(52 × 103).
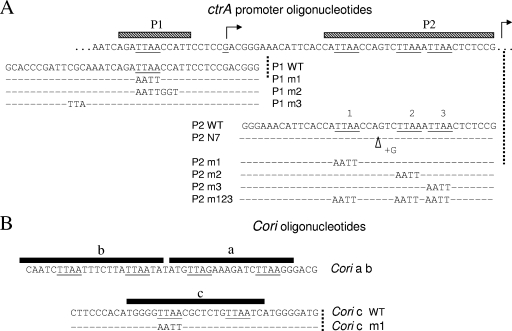
FIG. 8.
Double-stranded oligonucleotides (A) based on the ctrA P1 and P2 promoters and (B) based on the Cori replication origin. The bars indicate the established CtrA footprints. The full sequence of the wild type (unaltered DNA) is shown, and only the changes are shown below for the corresponding DNA molecules, which were used for the binding experiments whose results are shown in Fig. 6 and 7. WT, wild type.
FIG. 6.
Sample EMSA experiments illustrating GST-CtrA protein binding to P1 ctrA (promoter) and to Cori (replication origin) oligonucleotides. As described in Materials and Methods, following electrophoresis, the gel was first stained with ethidium bromide and then with Coomassie blue, and side-by-side photographs are shown. The bars above the ethidium bromide-stained lanes indicate the different oligonucleotides used (the black sections indicate the TTAA motifs). The following double-stranded oligonucleotides were used (their sequences are shown in Fig. 8): lane 1, P1 m1; lane 2, P1 m2; lane 4, wild-type P1; lane 5, P1 m3; lane 6, wild-type Cori binding site c; lane 7, Cori binding site c m1. Bovine serum albumin (2.0 μg) was also loaded in lane 3, and other protein molecular weight standards were loaded in flanking lanes (not shown). “Bound DNA” indicates bands in lanes 4 to 6 that were stained with both ethidium bromide and Coomassie blue. “Free DNA” indicates the oligonucleotides whose mobility was not altered by the protein, and “released” indicates the smear of DNA that presumably bound but disassociated during electrophoresis. “Free protein” indicates the positions of the GST-CtrA whose mobility was not altered by the DNA. WT, wild type; BSA, bovine serum albumin.
FIG. 7.
Sample EMSA experiments illustrating GST-CtrA protein binding to the ctrA P2 promoter and Cori (replication origin) oligonucleotides. Except for the different oligonucleotides, the experiments were identical to those whose results are shown in Fig. 6. The following oligonucleotides were used (their sequences are shown in Fig. 8): lane 1, wild-type Cori binding sites a and b; lane 2, P2 N7; lane 3, wild-type P2; lane 4, wild-type Cori binding site c; lane 5, P2 m1; lane 6, P2 m2; lane 7, P2 m3; lane 8, P2 m123. WT, wild type.
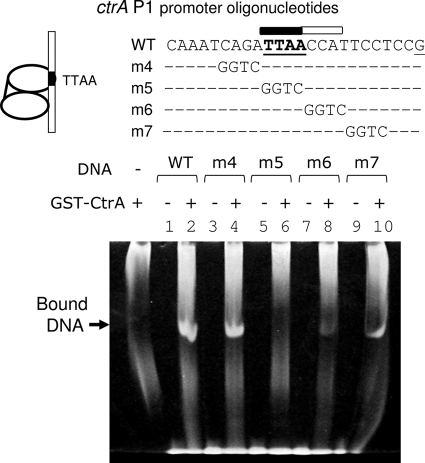
FIG. 10.
EMSA experiments using GST-CtrA protein and ctrA P1 promoter oligonucleotides with scanning mutations across the single TTAA motif. The experiments confirmed the primary importance of TTAA (m5), and they revealed the secondary importance of its flanking CCAT motif (m6) for affinity to GST-CtrA. The full-length wild-type P1 double-stranded oligonucleotide is shown in Fig. 8, and variations of this oligonucleotide (m4, m5, m6, and m7) are identical to wild-type P1 except at the positions of the GGTC blocks, as shown. Otherwise, the experimental conditions were the same as those described in the legends to Fig. 6 and 7. The GST-CtrA protein was added to the reaction mixtures in the alternate lanes. The GST-CtrA preparation also contained a significant amount of contaminating DNA from the E. coli chromosome (shown in the control lane on the left). Only the ethidium-bromide stained gel is shown. WT, wild type.
To measure molecular weights, the same binding reaction mixtures were run on a series of 5.0 to 7.0% polyacrylamide gels. As described by Orchard and May, protein molecular weight standards (MWND500-1KT; Sigma) were run in adjacent lanes. A gel retardation coefficient (Kr) was obtained for each band. Kr is the linear slope for the plotted relative mobility versus the polyacrylamide concentration. A first-order approximation is that the Kr is a linear function of molecular weight. The molecular weight of a DNA-protein complex is obtained by comparing its Kr to the Kr values for the molecular weight standards. Table 2 summarizes the molecular weights and the calculated multiples of bound protein (after subtraction of the oligonucleotide molecular weight) obtained from an extended series of similar EMSA experiments.
Transcription reporter assays.
The synchronized cultures with the lacZ reporter plasmids (see Fig. 4) were periodically sampled, and 1.0-ml portions were pulse-labeled with 15 μCi of [35S]methionine for 10 min at room temperature. Cell protein lysates were prepared by treatment with 10 mg/ml lysozyme in 50 mM Tris (pH 8), 450 mM NaCl, 0.5% Triton X-100 (TNT buffer), followed by preclearing with protein A-coated agarose beads (Sigma-Aldrich). Approximately 2 × 106 cpm of 35S-labeled protein lysate and 0.5 μl of polyclonal rabbit anti-β-galactosidase antibody (Sigma-Aldrich) were used for each immunoprecipitation (IP) and pull-down reaction with protein A-coated agarose beads (Sigma-Aldrich). The beads were washed three times with TNT buffer and finally suspended in 4 packed bead volumes of sodium dodecyl sulfate (SDS) protein loading buffer (2% SDS, 2% β-mercaptoethanol). The immunoprecipitated 35S-labeled LacZ protein samples were resolved on 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels, and LacZ band radioactivity was measured by phosphorimaging (Molecular Dynamics).
FIG. 4.
(A) Cell cycle-regulated transcription monitored independently for the ctrA P1 promoter and for the ctrA P2 promoter in wild-type cells (filled squares) and in CtrAΔ3-containing cells (open diamonds). The lacZ transcription reporter plasmids pctrA-P1 and pctrA-P2 were individually introduced into wild-type strain (ctrA WT) cells and into CtrAΔ3-containing strain (+ctrAΔ3) cells, which were also used in the experiments whose results are shown in Fig. 3. These cells were synchronized similarly and sampled at the indicated times in the swarmer (Sw), stalked (St), and asymmetric division phases of the cell cycle. To measure transcription from the lacZ transcription reporters, the sampled cells were pulse-labeled for 10 min with [35S]methionine, the LacZ protein was immunoprecipitated from equal amounts of cell extracts, the radiolabeled LacZ protein was resolved by SDS-PAGE, and the radioactivity was measured by phosphorimaging. The results are expressed as percentages of the peak LacZ signal in wild-type cells. (B) Immunoblot analysis of CtrA protein levels in wild-type cells (CtrA WT) and in CtrAΔ3-containing cells (+ CtrAΔ3) from the cultures used for two experiments described above (left side in panel A). Equal amounts (optical density, 0.1) of cells were assayed at the indicated times during the cell cycles.
CtrA immunoblotting.
CtrA protein levels were measured using standard SDS-PAGE resolving conditions, followed by transfer to polyvinylidene difluoride membranes (HyBond; Amersham Biosciences). These membranes were blocked in 5% nonfat skim milk in Tris-buffered saline. The CtrA protein was detected using a 1:5,000 dilution of the CtrA rabbit polyclonal antibody serum, which was provided by Lucy Shapiro, Stanford, CA (24). For signal detection and measurement, a West Pico chemiluminescent substrate kit (Pierce) was used as specified by the manufacturer.
ChIP assays.
Synchronized C. crescentus cells were released into fresh M2G at an OD660 of 0.2 and allowed to progress through the cell cycle. Our chromatin IP (ChIP) protocol was adapted from a previously described protocol (13). For each time point, approximately 2 OD660 units of cells was incubated with 1% formaldehyde buffered in 100 mM sodium phosphate (pH 7.6) at room temperature for 10 min. Next, the cells were placed on ice for 30 min, collected by centrifugation, and washed twice with sodium phosphate buffer (0.1 M, pH 7.6) to remove excess formaldehyde. Cells were also washed with 2× IP wash buffer (100 mM Tris [pH 7], 300 mM NaCl, 2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) and incubated for an additional 10 min at 37°C. The BugBuster cell lysis reagent (Novagen) was then applied as specified by the manufacturer. Complete cell disruption was confirmed by microscopy, and the preparations were subsequently sonicated on ice to break the genomic DNA into 500- to 1,000-bp fragments. Samples were centrifuged and split into equal IP and mock IP portions. Approximately 1 μl of CtrA antiserum (24) was added to each IP portion and incubated overnight at 4°C. Next, 25 μl of protein A Sepharose beads (Sigma-Aldrich) was added to both the IP and mock IP portions, which were then incubated for 1 h at room temperature with gentle rocking. The Sepharose beads were washed five times with 1× IP wash buffer and twice with Tris-EDTA buffer (10 mM Tris, 1 mM EDTA; pH 8.0) and finally suspended in 50 μl Tris-EDTA buffer. The formaldehyde cross-linkage was removed by heating preparations for 6 h at 65°C. Following centrifugation, a PCR analysis was performed using the DNA in the supernatant without further processing. The FailSafe PCR premix system (Epicentre) was used as specified by the manufacturer. A 157-bp fragment overlapping the P1 and P2 promoters of the ctrA gene was PCR amplified from serial (1:2) dilutions of immunoprecipitated and mock-immunoprecipitated DNA using the CTRA-Forward (5′-CGC TGT CAT CCT CGA TCA AC-3′) and CTRA-Reverse (5′-CTC CGA CGG GAA ACA TTC AC-3′) oligonucleotide primers. Similarly, a 308-bp fragment overlapping the motB promoter was PCR amplified using the MOTB-Forward (5′-ATC ATC GCC GTC ACG AAG T-3′) and MOTB-Reverse (5′-GGC CTG CGG CCT TCA GGT-3′) oligonucleotide primers. The PCR DNA bands were resolved on agarose gels, stained with ethidium bromide, and measured using the Kodak GelDoc system.
RESULTS
Phosphorylation of the CtrA protein does not increase its low affinity for two promoters.
A previous report demonstrated that CtrA bound and regulated two distinct promoters (P1 and P2) directly upstream of the ctrA gene (6). However, the experiments described in that report did not address how CtrA phosphorylation affects CtrA binding to P1 and P2. The previous results are summarized in Fig. 2A, which shows the zones of DNase I protection caused by pure CtrA protein binding in an in vitro “footprint” assay. The TTAA motifs indicated in Fig. 2A are presumably the contact sites of CtrA, but these motifs lack the consensus TTAA-N7-TTAA 7-bp spacing seen in the chromosome replication origin, Cori (Fig. 2A). Instead, P2 has N6 and N1 spacing, and P1 contains only one TTAA motif. Our previous studies of mutated CtrA binding sites demonstrated that CtrA could bind to an isolated TTAA motif. However, the binding was weak (with dissociation constant [Kd] values between 300 nM and 700 nM CtrA), and phosphorylation of the CtrA protein did not stimulate its binding to an isolated TTAA motif (31).
We demonstrated that phosphorylation of the CtrA protein does not stimulate CtrA binding to the P1 and P2 promoters, while the identical biochemical conditions do increase CtrA binding to the C. crescentus replication origin at least 50-fold (Fig. 2B). Concurrent DNase I protection in vitro “footprint” assays were performed using 32P-end-labeled DNA containing the ctrA P1 and P2 promoters and also using 32P-end-labeled DNA containing the C. crescentus replication origin (Cori binding sites a and b). To facilitate comparison, the unphosphorylated CtrA preparation was “mock phosphorylated” since it differed from the phosphorylated CtrA∼P preparation only by omission of 0.5 mM ATP from the CtrA (and kinase EnvZ) phosphorylation reaction (31). The phosphorylation of the CtrA protein was very efficient, as judged by increased affinity to Cori CtrA binding sites (Fig. 2B). For example, 5 nM phosphorylated CtrA∼P fully occupied Cori binding site b, while 200 nM unphosphorylated CtrA only partially occupied this site. Similarly, 10 nM CtrA∼P fully occupied Cori binding site a, while 200 nM unphosphorylated CtrA did not even partially occupy this site (Fig. 2B). In contrast to Cori DNA, the ctrA P1 and P2 DNA binding sites were not discriminated by identical phosphorylated CtrA∼P and unphosphorylated CtrA protein preparations (Fig. 2B). The ctrA P1 and P2 promoters were occupied equally by CtrA and CtrA∼P at CtrA protein concentrations of 200 nM, 500 nM, and 1,000 nM. Also, unlike the findings for the Cori binding sites, even these relatively high CtrA protein concentrations failed to fully occupy the ctrA P1 and P2 promoters. Therefore, the CtrA binding sites in the ctrA P1 and P2 promoters are both qualitatively and quantitatively different from those in Cori (31) and from the previously studied CtrA binding sites (which also gain affinity upon CtrA∼P phosphorylation) in the fliQ, ccrM, and pilA cell cycle-regulated promoters (26, 34).
Abundant cellular CtrA protein is theoretically sufficient to fill low-affinity sites.
The high Kd values (300 nM to 600 nM) that we observed for CtrA binding to the ctrA P1 and P2 promoters (Fig. 2B and data not shown) suggest that the cellular concentrations of CtrA should be greater than 600 nM to allow binding in swarmer and predivisional cells. We therefore measured the number of CtrA molecules per cell in synchronous cultures of wild-type strain NA1000 by comparing immunoblots of C. crescentus whole-cell lysates with immunoblots of pure CtrA protein. We found 9,000 ± 1,000 molecules of CtrA/cell in swarmer cells and a peak level of 18,000 ± 2,000 molecules of CtrA/cell in predivisional cells (data not shown). These values agree very well with values in a previous report (28). If we assume that the C. crescentus cell volume is 1 × 10−15 to 2 × 10−15 liter (12), then a CtrA level of 1 × 104 to 2 × 104 protein molecules per cell corresponds to a theoretical CtrA protein concentration of 10 to 30 μM. Since this concentration range is 30- to 100-fold greater than the Kd values (300 nM to 600 nM) for ctrA P1 and P2, these promoters should be occupied by CtrA in vivo. Unexpectedly, such high concentrations of the CtrA protein also indicate that CtrA phosphorylation is not needed to drive promoter occupancy during the cell cycle.
CtrA binding to the P1 and P2 promoters follows the cell cycle flux of CtrA protein.
We performed a ChIP assay that used formaldehyde to cross-link CtrA protein to DNA in live cells. As described in Materials and Methods, during the ChIP protocol, the chromosome DNA is fragmented, and the specific DNA-protein cross-linked material is recovered by IP with anti-CtrA protein serum. In a previous study, a similar ChIP protocol and the same anti-CtrA protein serum identified 55 C. crescentus operons that are regulated by the CtrA protein (13). In our experiments, the ctrA P1 and P2 DNA was measured using quantitative PCR. The CtrA ChIP signal shown in Fig. 3 accurately measured ctrA P1 and P2 occupancy by CtrA, because it was a comparative measure. The PCR band intensities obtained for CtrA IP were always compared to an otherwise identical “mock” precipitation in which only the anti-CtrA serum was omitted. Figures 3 and 5 show the ratios of the ChIP and “mock” precipitation signals.
FIG. 3.
ChIP analysis of CtrA protein binding to the ctrA P1 and P2 promoters of synchronized C. crescentus cells. Wild-type (WT) C. crescentus strain NA1000 cells or CtrAΔ3-containing (Δ3) C. crescentus strain NA1000 cells with plasmid pLS2747 were grown in M2G. Synchronized swarmer cells (Sw) were isolated (zero time) and allowed to proceed through the stalked (St) (45 min) and asymmetric division (Div) (100 min) stages, as indicated. The ChIP protocol was used for each of the cell samples, as described in Materials and Methods. The bars indicate the CtrA ChIP signals which were derived from PCR analysis as described in Materials and Methods.
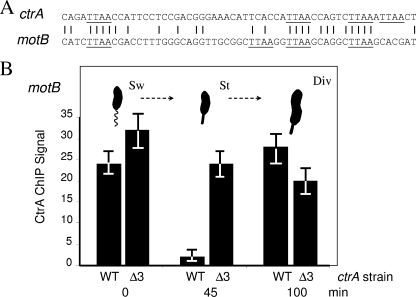
FIG. 5.
(A) The promoter TTAA motifs of motB match those of ctrA P1 and P2. Both promoters also lack the typical N = 7 spacing. (B) ChIP analysis of CtrA protein binding to the motB promoter of synchronized C. crescentus cells. Wild-type (WT) C. crescentus strain NA1000 cells or CtrAΔ3-containing (Δ3) C. crescentus cells (strain NA1000 with plasmid pLS2747) were grown, sampled, and analyzed as described in the legend to Fig. 3, except that the motB-specific primers were used in the PCR analysis, as described in Materials and Methods. Sw, swarmer cells; St, stalked cells; Div, cells in the asymmetric division stage.
To track CtrA binding, synchronized swarmer cells were isolated and allowed to proceed through the developmental cell cycle. Samples of these cells were then used for ChIP analysis at the swarmer cell (zero time), stalked cell (45 min), and asymmetrically dividing cell (100 min) stages (compare Fig. 1 with Fig. 3). In the wild-type strain culture, swarmer cells produced a significant CtrA ChIP signal, but when they differentiated into stalked cells, the ChIP signal decreased to the background (mock IP) levels. Still later, when the stalked cells started asymmetric division (100 min), a significant ChIP signal was observed (Fig. 3). These ChIP signals coincide with the presence of CtrA protein, as previously reported (5) and as shown in our immunoblot experiments (Fig. 4B).
Persistent CtrA binding to the P1 and P2 promoters throughout the cell cycle.
Since CtrA protein phosphorylation does not increase CtrA affinity for the ctrA P1 and P2 promoters (Fig. 2B), the results shown in Fig. 3 imply that the cell cycle variations in CtrA occupancy at these promoters are not driven by variations in CtrA protein phosphorylation. Instead, the variations in CtrA protein concentration alone appear to determine CtrA occupancy of the ctrA P1 and P2 promoters in wild-type cells. This hypothesis predicts that constant levels of CtrA protein force persistent occupancy throughout the cell cycle. In particular, maintaining CtrA in stalked cells, where CtrA becomes unphosphorylated (5), should maintain CtrA binding to the ctrA promoters.
To test this prediction, we introduced a plasmid expressing a stable form of the CtrA protein (CtrAΔ3) into wild-type cells. The ctrAΔ3 allele changes the three C-terminal amino acids that tag the CtrA protein for proteolysis (5, 28). Under our growth conditions in M2G, approximately equal amounts of the wild-type CtrA and CtrAΔ3 proteins were present in swarmer cells, and while the wild-type CtrA protein was degraded in stalked cells, most of the CtrAΔ3 protein remained. (For example, the presence of stable CtrAΔ3 in stalked cells was demonstrated by the immunoblot experiment whose results are shown in Fig. 4B.) The ChIP experiment whose results are shown in Fig. 3 also demonstrated that CtrAΔ3 cross-links to the ctrA P1 and P2 promoters in stalked cells (at 45 min). Consistent with the comparable level of the CtrA protein, the strength of the CtrAΔ3 ChIP signal is comparable to that of the wild-type CtrA protein ChIP signal in the swarmer cells and predivisional cells (Fig. 3). Therefore, a continuous supply of ∼10,000 molecules of the CtrAΔ3 protein per cell is sufficient to continuously occupy the ctrA P1 and P2 promoters.
Persistent CtrA binding to the ctrA promoters does not alter their timing.
Wild-type C. crescentus can alter promoter DNA occupancy by altering CtrA protein affinity (through phosphorylation) and by altering the CtrA protein concentration (through synthesis and proteolysis [Fig. 1]). However, the first means of regulating CtrA binding does not apply to the ctrA P1 and P2 promoters (Fig. 2B), and the second means is blocked when CtrAΔ3 is present and persistently bound (Fig. 3). If CtrA binding to DNA alone were sufficient to regulate transcription, then transcription from the P1 and P2 promoters would not be properly regulated when CtrAΔ3 is present.
To test this hypothesis, we assayed P1 and P2 transcription timing under the persistent binding conditions (Fig. 3) with the following cell cycle timing experiments. A P1 transcription-reporter plasmid (pctrA-P1) and a separate P2 transcription-reporter plasmid (pctrA-P2) were introduced into wild-type C. crescentus and into CtrAΔ3-containing cells. In a previous report (6), an analysis with the same transcription-reporter plasmids demonstrated that transcription from P1 is repressed by CtrA, while transcription from P2 is activated by CtrA. Our results with wild-type cells confirmed these results (Fig. 4). For example, when wild-type swarmer cells proceeded synchronously through the cell cycle, transcription from P1 peaked only in stalked cells (Fig. 4A) when CtrA protein was absent (Fig. 4B). Under the same conditions, transcription from P2 peaked later in flagellated predivisional cells (Fig. 4A) when new CtrA protein returned (Fig. 4B).
These distinct cell cycle patterns of transcription were maintained in the CtrAΔ3 cells (Fig. 4A) when the CtrAΔ3 protein was continuously present at all stages of the cell cycle (Fig. 4B). The steady level of CtrAΔ3 is comparable to the maximum level of wild-type CtrA, and this was also demonstrated by the uniformly high levels of CtrA binding to the P1 and P2 promoters (Fig. 3). Therefore, CtrA binding to DNA is not sufficient to repress transcription from P1 or to activate transcription from P2. CtrA must acquire additional properties, presumably by phosphorylation, that account for cell cycle variation in transcription activity.
CtrA binds the motB promoter, whose DNA resembles that of the ctrA promoters.
CC1573 encodes the C. crescentus homologue of the E. coli chemotaxis protein MotB (20). The MotB/CC1573 and CC1574 genes are positioned divergently, implying that the intervening DNA (114 bp) contains two promoters. Accordingly, while CC1574 transcription remains constant, MotB/CC1573 transcription varies during the cell cycle (13, 14). The transcription of MotB/CC1573 peaks in dividing stalked cells coincident with the transcription of genes encoding other flagellar and chemotaxis proteins that are regulated by CtrA (11). The DNA immediately preceding MotB/CC1573 resembles the DNA containing the ctrA P1 and P2 promoters (Fig. 5A). Both sequences contain four TTAA CtrA binding motifs, and both have N6 spacing instead of the N7 consensus spacing.
To confirm that the CtrA protein binds to the proposed motB promoter in vivo, we reexamined the CtrA formaldehyde cross-linking (ChIP) experiments whose results are shown in Fig. 3 using PCR primers that amplify the motB promoter DNA (Fig. 5B). The pattern of cell cycle binding of CtrA to motB DNA was essentially the same as the pattern for CtrA binding to ctrA P1 and P2. For example, motB showed a strong CtrA ChIP signal (∼20-fold greater than the mock control) only when CtrA was present in the wild-type swarmer (zero time) and late predivisional (100 min) cells. Similarly, continuous presence of CtrAΔ3 resulted in a strong motB ChIP signal during all three stages of the cell cycle. The comparable ChIP signals (compare Fig. 3 with Fig. 5B) suggest that ctrA P1 and P2 and motB DNA (Fig. 5A) have comparable affinities for the CtrA protein. Since CtrAΔ3 is not phosphorylated in stalked cells, this result also implies that in vivo CtrA concentrations are sufficient to bind motB DNA without the aid of CtrA phosphorylation.
The artificial GST-CtrA fusion protein has higher affinity for target DNA.
In our previous footprint studies (31), we used an N-terminally six-histidine-tagged protein (His-CtrA) and the same protein cleaved by enterokinase (EK-CtrA) to remove the six-histidine tag. In the present studies, we also prepared an N-terminal GST protein fused to the full-length CtrA protein (GST-CtrA from plasmid pGM2586, as described in Materials and Methods). When used in DNase I footprint experiments, all three proteins, His-CtrA, EK-CtrA, and GST-CtrA, protected the same base pairs of all five Cori binding sites (binding sites a, b, c, d, and e), indicating that all three proteins have the same DNA recognition abilities (data not shown). However, the GST-CtrA protein has a significantly higher affinity for the binding sites. Compared to the unphosphorylated His-CtrA protein (or to the EK-CtrA protein), approximately 50-fold less GST-CtrA protein is required to produce comparable DNase I protection at Cori binding sites a b, c, d, and e (data not shown). This higher DNA affinity suggests that the GST-CtrA protein is in an “active state” compared with both the His-CtrA and EK-CtrA proteins, because both of the latter proteins require phosphorylation to acquire comparably high affinity for Cori binding sites a b, c, d, and e (31). Interestingly, the high-affinity GST-CtrA protein cannot be phosphorylated under the same EnvZ kinase conditions (data not shown).
The GST-CtrA fusion protein is a stable dimer in solution.
We used PAGE techniques, as described in Materials and Methods, to measure and compare the molecular weights of the His-CtrA and GST-CtrA proteins under conditions that denature them and conditions that preserve their native states (Table 1). Denaturing SDS-PAGE analysis showed the expected polypeptide molecular weights for both proteins (Table 1). Native PAGE analysis showed approximate multiples of one and two for His-CtrA and for GST-CtrA (Table 1). Therefore, while His-CtrA is a monomer, GST-CtrA is a dimer, and its dimerization may account for its higher affinity for DNA.
Molecular weights of stable GST-CtrA protein-DNA complexes.
The high stability of GST-CtrA protein dimers is supported by the results of native PAGE experiments with Coomassie blue-stained gels (Fig. 6 and 7). Such gels show that “free” GST-CtrA protein migrates as a single band without significant smearing and therefore without dissociation during a ∼1-h gel run. The high-affinity and very stable GST-CtrA dimers provided a significant technical advantage in EMSA. As described in Materials and Methods, we used the EMSA methods of Orchard and May (21) to determine how many GST-CtrA molecules bind to double-stranded oligonucleotides (Tables 2). The sequences of the DNA molecules are shown in Fig. 8. Either ∼2 or ∼4 GST-CtrA proteins bound per DNA molecule for all oligonucleotides tested (Table 2).
How do the atypical DNA sequences of ctrA promoters P1 and P2 alter CtrA binding?
Figure 2 shows that the atypically spaced TTAA motifs have poor affinity for CtrA, yet Fig. 3 to 5 show that the poor in vitro affinity allows in vivo CtrA binding and cell cycle regulation. Figure 2 also shows that unlike Cori DNA, ctrA P1 and P2 DNA bound CtrA and CtrA∼P with equal affinity. To account for these differences, CtrA might bind and arrange itself differently on ctrA P1 and P2 than it does on Cori DNA. EMSA can distinguish different molecular arrangements that otherwise have the same molecular weights. In the following comparative EMSA experiments the stable and high-affinity GST-CtrA protein was used, since six-histidine-tagged CtrA failed to produce discretely shifted ctrA P1 or P2 DNA bands (data not shown).
GST-CtrA binding requires one TTAA motif in ctrA promoter P1 but two TTAA motifs in Cori binding site c.
The EMSA experiments (Fig. 6) demonstrated that the single TTAA motif in P1 binds two molecules of GST-CtrA (or one GST-CtrA dimer). The wild-type P1 and wild-type Cori binding site c oligonucleotides each produced only one GST-CtrA-bound band (Fig. 6, lanes 4 and 6). The two bands comigrated in 5% to 7% polyacrylamide gels (Fig. 6 and data not shown), indicating that they have the same molecular weight. This comigration also suggests that P1 binds two GST-CtrA molecules, because the two DNAs have the same molecular weight and we predicted that wild-type Cori binding site c DNA binds two GST-CtrA molecules (since Cori binding site c contains two N7-spaced TTAA motifs). This inference was confirmed by Orchard-May EMSA molecular weight measurements (determined using multiple gel runs) shown in Table 2. GST-CtrA dimer binding requires the single TTAA motif in P1, because changing this motif to AATT (P1 m1) eliminated binding (Fig. 6, lane 1). One TTAA motif is sufficient, because a second TTAA motif (with N7 spacing) did not improve binding (Fig. 6, lane 5; Table 2). In contrast, GST-CtrA dimer binding requires both TTAA motifs in Cori binding site c, because changing one motif to AATT (Cori binding site c m1) completely eliminated binding (Fig. 6, lane 7).
GST-CtrA protein binds the three TTAA motifs in ctrA promoter P2 as a tetramer.
EMSA experiments (Fig. 7 and Table 2) demonstrated that GST-CtrA binds the three TTAA motifs of P2 as a tetramer and that the mode of binding is also distinct from that of the Cori binding sites. The wild-type P2 oligonucleotide (Fig. 8) produced two GST-CtrA bands (Fig. 7, lane 3), as did Cori binding sites a and b (Fig. 7, lane 1). We predicted that GST-CtrA binds Cori binding sites a and b as a tetramer (31) and that GST-CtrA would bind to the stronger site (binding site b) as a dimer before binding to both binding sites a and b as a tetramer (as also implied by footprints shown in Fig. 2B). This binding pattern easily explains the Cori binding site a and b lower and upper bands (Fig. 7, lane 1), and dimer and tetramer binding was confirmed by the Orchard-May EMSA molecular weights shown in Table 2. Similar measurements demonstrated that the wild-type P2 DNA is bound by a GST-CtrA dimer (lower band) and by a GST-CtrA tetramer (Fig. 7, lane 3, upper band; Table 2). (Also note that the lower bands comigrated with the Cori binding site c GST-CtrA dimer bands in Fig. 7, lane 4.) Interestingly, while the wild-type P2 and Cori binding site a and b lower bands comigrated, the wild-type P2 upper band was significantly retarded compared to the Cori binding site a and b upper band (Fig. 7, lanes 1 and 3). The molecular weight of the DNA cannot account for this difference (the P2 DNA was in fact 5 kDa smaller, and this did not visibly alter the migration of the lower band). This observation suggests that the (wild-type P2)-(tetramer GST-CtrA) complex acquires a distinct shape that also retards its migration in the gel matrix.
Hierarchical and cooperative binding to TTAA motifs “1,” “2,” and “3” in ctrA promoter P2.
EMSA experiments with mutant P2 DNA also indicated that GST-CtrA binds P2 in a distinct way and that cooperative protein-protein contacts are required for tetramer GST-CtrA binding. We designated the P2 TTAA motifs “1,“ “2,” and “3” (Fig. 8) and changed them to AATT individually (motifs m1, m2, and m3) and together (motif m123). Compared to wild-type P2, the P2 m3 mutation eliminated the upper band but strengthened the lower band (Fig. 7, compare lanes 3 and 7). Comigration with established dimer bands (for example, comigration with Cori binding site c in lane 4) indicates that GST-CtrA also binds P2 m3 DNA as a dimer, and Orchard-May EMSA molecular weight measurements confirmed dimer binding (Table 2). In contrast, replacing wild-type P2 with either P2 m1 or P2 m2 eliminated both the upper and lower bands, but weak bands with altered mobility remained (Fig. 7, lanes 5 and 6). (Molecular weight measurements for these weak bands were not reliable but suggested that two or three GST-CtrA molecules were bound, consistent with their retarded mobilities.) Therefore, all three TTAA motifs are required for GST-CtrA tetramer binding. Also, there is a distinct binding hierarchy, since P2 m3 is the least severe mutation (it retains strong dimer binding), followed by P2 m2 and then P2 m1 (which is as severe as the triple m123 mutation [Fig. 7, lane 8]).
Our mutation analysis also indicated that there are cooperative protein-protein contacts. For example, the P2 m3 mutation (Fig. 7, lane 7) also shows that P2 TTAA motifs 1 and 2 (with N6 spacing) support dimer binding (Table 2). This finding implies that the second dimer must bind P2 TTAA motif 3. Apparently, GST-CtrA recognizes the single P2 motif 3, as it does the single P1 TTAA motif (Fig. 6, lanes 1 and 4). But if P2 m1 completely prevents binding (Fig. 7, lane 5), then GST-CtrA at P2 TTAA motif 1 must communicate with GST-CtrA at P2 TTAA motif 3 (11 bp away). Protein-to-protein contacts through GST-CtrA bound at P2 TTAA motif 2 seem to be required for this communication.
The atypical N = 6 spacing is required for tetramer GST-CtrA binding in ctrA promoter P2.
Further evidence for cooperative protein-protein contacts between two GST-CtrA dimers comes from the P2 N7 mutation (Fig. 8). When the N6 spacing between P2 TTAA motifs 1 and 2 is changed to N7 spacing, only dimer GST-CtrA binding is observed (Fig. 7, lane 2; Table 2). Apparently, the N6-to-N7 change “stretches” the binding (between motifs 1 and 2) and “breaks” a line of communication (between motifs 2 and 3). This interpretation readily explains the apparently identical EMSA results for the P2 N7 mutation (Fig. 7, lane 2) and P2 m3 (Fig. 7, lane 7), and this is discussed below in relation to binding site organization (Fig. 9).
FIG. 9.
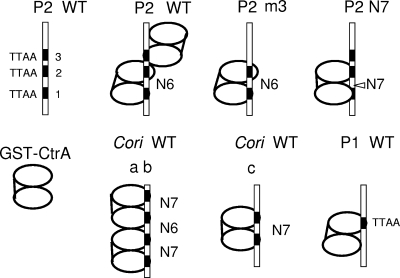
Cartoons comparing and contrasting GST-CtrA binding to ctrA P1 and P2 and GST-CtrA binding to Cori. The bars indicate the different double-stranded DNA oligonucleotides described in Fig. 6 to 8. The pairs of ovals represent GST-CtrA dimer molecules apparently held together by the N-terminal GST domain. The different binding configurations are discussed in the text. WT, wild type.
DISCUSSION
Unexpected significance of weak CtrA binding sites.
Our experiments unexpectedly revealed the physiological importance of weak CtrA binding sites. Our previous studies demonstrated that CtrA can bind to an isolated TTAA motif (when its partner motif is mutated) and that phosphorylation of the CtrA protein does not increase its affinity for isolated TTAA motifs (31). The in vitro binding is weak (the Kd values are between 300 nM and 700 nM CtrA), and in this artificial context, it was judged to be apparently not significant. Although the CtrA and CtrA∼P affinities for ctrA P1 and P2 are comparably weak (Fig. 2), they are nonetheless clearly physiologically important. The ctrA gene is essential (24), and its transcription is autoregulated by CtrA binding at P1 and P2 (6). These transcripts are precisely timed during every cell cycle (Fig. 4), and they become sufficiently abundant to replenish very high CtrA protein concentrations (10 to 30 μM). Therefore, weak CtrA binding is clearly sufficient to cause a significant regulatory response.
Previous studies demonstrated that weak binding sites significantly influence regulatory protein binding to strong target sites (37). Weak nonspecific DNA binding can constrain random diffusion and thereby rapidly guide the regulatory protein to its destination (improve kinetic parameters). In contrast, weak but specific DNA binding sites compete with and reduce protein binding to the strong target sites (decrease steady-state parameters). However, our results demonstrate another, simpler principle. The weak binding sites in ctrA P1 and P2 are the target sites through which CtrA effects transcription.
Implied structural distinctions between weak and strong CtrA binding sites.
Our EMSA experiments suggest additional structural distinctions between the “typical” binding sites in Cori and the “atypical” weak binding sites in ctrA P1 and P2. These structural distinctions are summarized in Fig. 9. Note that either the N6 spacing or the N7 spacing would position the centers of the TTAA motifs (N10 or N11) on the same side of a B-form DNA helix. Accordingly, the two GST-CtrA molecules at Cori binding site c and the four GST-CtrA molecules at Cori binding sites a and b were positioned on the same side of the DNA helix (Fig. 9). In contrast, the N1 spacing between P2 TTAA motifs 2 and 3 positions their centers (N5) on opposite sides of the DNA helix. Accordingly, the first GST-CtrA dimer that binds P2 TTAA motifs 1 and 2 was located on the side of the DNA helix opposite the second CtrA dimer that binds motif 3. A comparison of the GST-CtrA tetramer bound to Cori binding sites a and b with the tetramer bound to wild-type P2 (Fig. 9) indicates that, despite having practically identical molecular weights (Table 2), these tetramers have significantly different shapes. This hypothesis readily explains their different upper-band mobilities (Fig. 7, lanes 1 and 3) and their identical lower-band mobilities.
These different GST-CtrA tetramer arrangements also agree with the ascribed physiological functions of these binding sites. CtrA Cori binding sites a and b repress a strong promoter in the swarmer cells, and the second Cori binding site, binding site a, should position the CtrA dimer directly over the −10 promoter element to block RNA polymerase contact (17). In contrast, P2 promoter transcription is activated by CtrA (6). The mechanism for P2 activation is not known, but it is interesting that the second CtrA dimer would be positioned on the opposite face of the DNA helix, thereby leaving the −10 promoter element accessible to RNA polymerase.
Our EMSA results also suggest that N6 spacing and N7 spacing differentially position GST-CtrA molecules on the DNA. It is certainly remarkable that a 1-bp insertion (P2 N7) can block tetramer binding while enhancing dimer binding (Fig. 7, lanes 2 and 3). Figure 9 shows our interpretation, that wild-type P2 allows CtrA bound at TTAA motif 2 to contact and aid GST-CtrA binding to TTAA motif 3. We also speculate that P2 N7 causes CtrA bound at TTAA motif 1 to reposition CtrA bound at TTAA motif 2, thereby drawing it away from TTAA motif 3, apparently “stretching” and “breaking” a line of protein-protein communication (Fig. 9). An interesting alternative interpretation is that CtrA recognizes wild-type P2 not as TTAA-N6-TTAA-N1-TTAA but as TTAA-N7-TAAATTAA. This alternative pattern is therefore a hybrid between typically N7-spaced motifs and atypically (adjacently) spaced TTAA motifs. In either case, our data indicate that GST-CtrA binding to motif 3 is aided by GST-CtrA binding to motifs 1 and 2. This idea is also reinforced by a hierarchy for GST-CtrA dimer binding (P2 m1 < P2 m2 < P2 m3) (Fig. 7, lanes 6 to 8).
CtrA affinity for a single TTAA motif is substantially aided by adjacent sequences.
In Fig. 9, we interpret GST-CtrA binding to P1 as a dimer anchored by a single TTAA motif, while, for example, GST-CtrA binding to Cori binding site c is anchored by two TTAA motifs. This interpretation is supported by the relatively short (12 bp) footprint at P1 (Fig. 2A) compared with the longer footprints (>20 bp) at typical sites (for example, Cori binding site c). Changing the single TTAA in P1 to AATT (P1 m1) eliminated binding (Fig. 6, lane 1), and changing one of two TTAA motifs in Cori binding site c also eliminated binding (Fig. 6, lane 7). Therefore, one TTAA motif in P1 is sufficient to bind two GST-CtrA molecules, while two TTAA motifs are required in Cori binding site c.
These results also indicate that the CtrA binding modes at ctrA P1 and Cori binding site c are distinct and that additional DNA sequences are needed to enhance CtrA binding to a single TTAA motif. The TTAAC-N6-TTAAC sequence has also been proposed as a consensus CtrA binding site (39). The TTAACCAT sequence present in P1 and in C. crescentus flagellar promoters was also proposed as an alternative CtrA binding site (13). Our previous results also indicate that the TTAA-N7-TTAA sequence is important but not sufficient for maximal CtrA binding, but the same studies did not support the significance of the modified TTAAC-N6-TTAAC and TTAACCAT sequences (22).
The EMSA experiments (Fig. 10) demonstrated that 3′ flanking DNA sequences (CCAT) substantially aid GST-CtrA binding to a single TTAA motif. From the wild-type P1 oligonucleotide (Fig. 6) we derived P1 m4 to m7 by scanning a 4-bp (GGTC) block of mutations from 5′ to 3′ across the TTAA motif. The wild-type P1 and 5′ P1 m4 DNAs showed comparable affinities for GST-CtrA (Fig. 10, lanes 1 to 4). As expected (Fig. 6, lanes 1 and 2), the TTAA motif is essential, since P1 m5 completely eliminated binding (Fig. 10, lanes 5 and 6). Interestingly, the 3′ P1 m6 and m7 mutations also substantially reduced binding (Fig. 10, lanes 7 to 10). In comparable EMSA experiments in which protein concentrations were varied, we observed that ∼8-fold less GST-CtrA is required with wild-type P1 to produce a band intensity comparable to that produced with P1 m6 (data not shown). Therefore, the 3′ flanking CCAT motif increases the Kd of wild-type P1 by a factor of almost 10. Presumably, 3′ DNA aids cell cycle regulation when only one TTAA motif is present.
Mechanism of CtrA “activation.”
How CtrA gains “activity” and how it communicates with other proteins are not known. Our experiments also address the mechanism of CtrA “activation,” an important but vaguely defined concept that previously emphasized enhanced DNA binding upon CtrA∼P phosphorylation. CtrA belongs to the OmpR/PhoB RR family, and crystal structures for the representative members PhoB (1) and ArcA (36) suggest a common mechanism of active-state dimer formation across the highly conserved alpha4-beta5-alpha5 face. Based on the salt bridge and hydrophobic dimer-forming amino acids observed in ArcA, an alignment with the CtrA protein shows that CtrA has 10 of 11 potential dimer-forming contacts (data not shown). Our experiments also suggest that the active form of the CtrA protein is a dimer. GST-CtrA dimers have substantially higher affinity for DNA than His-CtrA monomers without phosphorylation. Since the GST protein of pGEX-2T forms a GST dimer in solution (35), it is tempting to propose that tethering CtrA by the GST domains is sufficient for CtrA activation. Such a “recruitment” model can often explain biological control (23). Recruitment models emphasize molecular adhesion and the synergy of multiple weak binding contacts that bring molecules together without changing their individual structures or their individual biochemical activities.
Before our experiments, one could argue that CtrA∼P phosphorylation need only stimulate DNA binding, which might then simply “recruit” RNA polymerase to the promoter. However, to account for transcription by the ctrA P1 and P2 promoters, this simple model must be modified. Persistent binding of CtrA to these promoters (Fig. 3) does not significantly alter their regulation during the cell cycle (Fig. 4). Therefore, the DNA binding (operator site occupancy) is not sufficient, and transcription regulation requires additional cell cycle signals. When phosphorylated, the receiver/regulatory domains of RR proteins have an altered surface that changes protein-protein contacts. However, it is difficult to extend established examples to CtrA, because different RR proteins use different parts of their receiver/regulatory surfaces for diverse phosphorylation-dependent regulatory interactions (38).
We propose that phosphorylated CtrA∼P also acquires altered or “activated” protein surfaces and that CtrA thereby signals through new protein-protein contacts. For example, the RR protein OmpR contacts the alpha subunit (30) and RR PhoB contacts the sigma subunit of RNA polymerase (16). If unphosphorylated CtrA is bound to DNA, it would still lack signaling “activity” because its surface would lack productive protein-protein contacts. Both DNA binding and CtrA phosphorylation signals are required for transcription regulation, but phosphorylation must be the dominant signal when high in vivo CtrA concentrations alone are sufficient to drive DNA binding. This view also explains how the CtrAD51E mutant protein, which cannot be phosphorylated, can still have partial “phospho-mimic” activity without having affinity for TTAA-N7-TTAA binding sites (32). Other RR proteins provide an interesting contrast to CtrA. For example, some RR proteins do not require a phosphorylation signal, and their in vivo activities may be regulated simply by protein abundance (15, 29). In contrast, C. crescentus uses both CtrA phosphorylation and protein abundance to communicate with the P1 and P2 ctrA and perhaps other cell cycle-controlled promoters.
Potential promoters resembling ctrA P1 and P2.
The results of whole-genome CtrA-DNA cross-linking and top-to-bottom signal rank analysis (13) imply that one-quarter or more of the C. crescentus intergenic DNA is bound by the CtrA protein in vivo. This proportion greatly exceeds the estimated number of promoters regulated by CtrA (13), but we speculate that among the lower-affinity sites are true regulatory sites, such as the sites 5′ to the ctrA and motB gene (Fig. 5). The ctrA P1 promoter appears to use only one TTAA motif to bind CtrA and block transcription (Fig. 2A). Similarly, fliX gene transcription is regulated by CtrA in vivo, yet an in vitro CtrA footprint experiment has suggested that there is binding to only one TTAA motif (19). In both the ctrA P1 promoter and the fliX promoter, the length of the CtrA footprint over the one TTAA motif is about one-half the length of the CtrA footprint over a consensus TTAA-N7-TTAA site (24, 25, 31). Also, the major C. crescentus chemotaxis operons have two promoters that appear to use only single TTAA motifs (11). A genome-wide ChIP assay also suggested that some C. crescentus genes might use atypical or isolated TTAA motifs in their promoters (13). These results suggest that there is wide use of weak CtrA binding sites where phosphorylation does not signal DNA binding. Weak CtrA binding sites are certainly sufficient; indeed, strong CtrA binding sites seem to be superfluous, considering that the CtrA protein accumulates at sufficiently high concentrations (10 to 30 μM) in wild-type C. crescentus cells.
Weak CtrA binding sites may explain the evolution of global regulation.
Regulation by CtrA obviously implies the acquisition (through mutation) of a selective CtrA binding site, yet a strong selective binding site requires multiple changes in base pairs that rarely occur simultaneously. Sequential changes in base pairs are more likely, but the first changes are not preserved unless they confer at least a partial function and thereby a selective advantage. It is comparatively easy to form first one TTAA motif, then two TTAA motifs with atypical spacing (N ≠ 7), and finally a precisely spaced consensus TTAA-N7-TTAA binding site. The ctrA P1 (one TTAA motif) and ctrA P2 (atypical N = 6, N = 1 spacing) promoters appear to illustrate these transition steps.
CtrA is a global regulator found in many alphaproteobacteria (9). The Sinorhizobium meliloti ctrA gene also has two transcription promoters that are bound by CtrA (2). These S. meliloti promoters show both typical (N = 7) and atypical spacing between their TTAA motifs. Likewise, the Brucella abortus ctrA gene has two transcription promoters that also bind CtrA (3). Interestingly, in these B. abortus promoters CtrA binds two typically spaced (N = 7) sites and one “orphan” TTAA motif. We therefore suggest that C. crescentus and its close relatives S. meliloti and B. abortus exhibit all three binding site transitions. They use CtrA binding sites with the simplest solo TTAA motif, with two TTAA motifs atypically spaced (N ≠ 7), and with precisely spaced consensus TTAA-N7-TTAA binding sites.
Weak CtrA binding sites may explain the need for CtrA proteolysis.
The significance of programmed (stalked cell-specific) CtrA proteolysis has always been obscure, because the constant presence of the CtrAΔ3 protein, even at levels significantly higher than the levels that we used in our studies, does not alter the C. crescentus cell cycle (5). It was therefore argued that programmed proteolysis is a redundant system working in parallel with programmed CtrA∼P phosphorylation. Weak CtrA binding sites, where phosphorylation does not signal DNA binding, require another mechanism to control DNA binding. The wild-type cell cycle flux of the CtrA protein can certainly drive the binding of CtrA protein at ctrA P1 and P2 (Fig. 3) and at motB (Fig. 5). Although the constant presence of the CtrAΔ3 protein does not significantly alter timed transcription from the ctrA P1 and P2 promoters (Fig. 4), other promoters may require CtrA removal. In such cases, programmed CtrA proteolysis would be the only means to regulate CtrA binding.
Significance of phosphorylation-dependent binding to typical TTAA-N7-TTAA sites.
The normally high level of CtrA protein drives wild-type CtrA binding to low-affinity TTAA sites at ctrA P1 and P2 (Fig. 3), but typical TTAA-N7-TTAA sites also have comparable or moderately higher affinities for unphosphorylated CtrA protein (31). Therefore, paradoxically, phosphorylation is also not needed for CtrA binding to typical TTAA-N7-TTAA sites during most of the cell cycle. It is possible that the CtrA phosphorylation signal is communicated to the genome mostly by protein conformation changes and not, as we previously thought, by DNA binding (shifting between bound and unbound states). Phosphorylation-dependent CtrA binding is superfluous except when CtrA protein concentrations become sufficiently low. In wild-type C. crescentus, these conditions occur at the key cell cycle transition stages, when the CtrA protein is removed or replenished (Fig. 1). These stages are also the times in the cell cycle when phosphorylation-dependent CtrA binding might be most critical for timing the start of chromosome replication and the start of asymmetric cell division. Perhaps this explains why the replication origin and the flagellar promoter CtrA binding sites conform to the phosphorylation-responsive binding consensus TTAA-N7-TTAA. Creating such critical and discriminatory concentrations of the CtrA protein provides yet another rationale for programmed CtrA synthesis and proteolysis that was not apparent from previous studies.
Acknowledgments
G.T.M. wrote the text, created the illustrations, and performed all experiments whose results are shown in Fig. 6 and 7 and Tables 1 and 2. M.C.O. performed the experiments whose results are shown in Fig. 2 and prepared the CtrA protein. R.S. performed preliminary footprint experiments that established the conclusions derived from the results shown Fig. 2. W.S. performed the experiments whose results are shown Fig. 3 to 5 and Western blotting (not shown). D.P.B. performed the experiments whose results are shown in Fig. 10. We thank James Taylor for critically reading the manuscript.
This work was funded by grant MT-13453 from the Canadian Institutes for Health Research.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Bachhawat, P., G. V. T. Swapna, G. T. Montelione, and A. M. Stock. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 131353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., D. Y. Hung, A. Reisenauer, L. Shapiro, and S. R. Long. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol. 1833204-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellefontaine, A.-F., C. E. Pierreux, P. Mertens, J. Vandenhaute, J.-J. Letesson, and X. De Bolle. 2002. Plasticity of a transcriptional regulatory network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43945-960. [DOI] [PubMed] [Google Scholar]
- 4.Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444899-904. [DOI] [PubMed] [Google Scholar]
- 5.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1 to S transition in a bacterial cell cycle. Cell 90415-424. [DOI] [PubMed] [Google Scholar]
- 6.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell cycle regulator. Proc. Natl. Acad. Sci. USA 966648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 8.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallez, R., A. F. Bellefontaine, J. J. Letesson, and X. De Bolle. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12361-365. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97111-120. [DOI] [PubMed] [Google Scholar]
- 11.Jones, S. E., N. L. Ferguson, and M. R. K. Alley. 2001. New member of the ctrA regulon: the major chemotaxis operon in Caulobacter is CtrA dependent. Microbiology 147949-958. [DOI] [PubMed] [Google Scholar]
- 12.Judd, E. M., K. R. Ryan, W. E. Moerner, L. Shapiro, and H. H. McAdams. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. USA 1008235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 994632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laub, M. T., H. H. McAdams, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 2902144-2148. [DOI] [PubMed] [Google Scholar]
- 15.Lejona, S., M. E. Castelli, M. L. Cabeza, L. J. Kenney, E. Garcia Vescovi, and F. C. Soncini. 2004. PhoP can activate its target genes in a PhoQ-independent manner. J. Bacteriol. 1862476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makino, K., M. Amemura, T. Kawamoto, S. Kimura, H. Shinagawa, A. Nakata, and M. Suzuki. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 25915-26. [DOI] [PubMed] [Google Scholar]
- 17.Marczynski, G. T., K. Lentine, and L. Shapiro. 1995. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 91543-1557. [DOI] [PubMed] [Google Scholar]
- 18.Marczynski, G. T., and L. Shapiro. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56625-656. [DOI] [PubMed] [Google Scholar]
- 19.Mohr, C., J. K. MacKichan, and L. Shapiro. 1998. A membrane-associated protein, FliX, is required for an early step in Caulobacter flagellar assembly. J. Bacteriol. 1802175-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 984136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orchard, K., and G. E. May. 1993. An EMSA-based method for determining the molecular weight of a protein-DNA complex. Nucleic Acids Res. 213335-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouimet, M.-C., and G. T. Marczynski. 2000. Analysis of a cell-cycle promoter bound by a response regulator. J. Mol. Biol. 302761-775. [DOI] [PubMed] [Google Scholar]
- 23.Ptashne, M., and A. Gann. 1997. Transcription activation by recruitment. Nature 386569-577. [DOI] [PubMed] [Google Scholar]
- 24.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 8483-93. [DOI] [PubMed] [Google Scholar]
- 25.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 1812430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan, K. R., S. Huntwork, and L. Shapiro. 2004. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. USA 1017415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan, K. R., E. M. Judd, and L. Shapiro. 2002. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J. Mol. Biol. 324443-455. [DOI] [PubMed] [Google Scholar]
- 29.Schar, J., A. Sickmann, and D. Beier. 2005. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 1873100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharif, T. R., and M. M. Igo. 1993. Mutations in the alpha subunit of RNA polymerase that affect the regulation of porin gene transcription in Escherichia coli K-12. J. Bacteriol. 1755460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 191138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siam, R., and G. T. Marczynski. 2003. Glutamate at the phosphorylation site of response regulator CtrA provides essential activities without increasing DNA binding. Nucleic Acids Res. 311775-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Puler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 34.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 193223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, D. B., and L. M. Corcoran. 2001. Expression and purification of glutathione-S-transferase fusion proteins. Curr. Protoc. Mol. Biol. 2001 May; Chapter 16:Unit16.7. [DOI] [PubMed]
- 36.Toro-Roman, A., T. R. Mack, and A. M. Stock. 2005. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J. Mol. Biol. 34911-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Hippel, P. H., and O. G. Berg. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264675-678. [PubMed] [Google Scholar]
- 38.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 39.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA 951443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]