Abstract
Most bacteriophages (phages) release their progeny through the action of holins that form lesions in the cytoplasmic membrane and lysins that degrade the bacterial peptidoglycan. Although the function of each protein is well established in phages infecting Streptococcus pneumoniae, the role—if any—of the powerful bacterial autolysin LytA in virion release is currently unknown. In this study, deletions of the bacterial and phage lysins were done in lysogenic S. pneumoniae strains, allowing the evaluation of the contribution of each lytic enzyme to phage release through the monitoring of bacterial-culture lysis and phage plaque assays. In addition, we assessed membrane integrity during phage-mediated lysis using flow cytometry to evaluate the regulatory role of holins over the lytic activities. Our data show that LytA is activated at the end of the lytic cycle and that its triggering results from holin-induced membrane permeabilization. In the absence of phage lysin, LytA is able to mediate bacterial lysis and phage release, although exclusive dependence on the autolysin results in reduced virion egress and altered kinetics that may impair phage fitness. Under normal conditions, activation of bacterial LytA, together with the phage lysin, leads to greater phage progeny release. Our findings demonstrate that S. pneumoniae phages use the ubiquitous host autolysin to accomplish an optimal phage exiting strategy.
Streptococcus pneumoniae (pneumococcus), a common and important human pathogen, is characterized by the high incidence of lysogeny in isolates associated with infection (34, 44). Pneumococcal bacteriophages (phages) share with the majority of bacteriophages infecting other bacterial species the “holin-lysin” system to lyse the host cell and release their progeny at the end of the lytic cycle. Genes encoding both holins and lysins (historically termed “endolysins”) are indeed found in the genomes of all known pneumococcal phages (8, 28, 31, 37). Supporting this mechanism, a lytic phenotype in the heterologous Escherichia coli system was achieved only by the simultaneous expression of the Ejh holin and the Ejl endolysin of pneumococcal phage EJ-1 (8). When these proteins were independently expressed, cellular lysis was not perceived. Similar results were shown for pneumococcal phage Cp-1, not only in E. coli, but also in the pneumococcus itself (28).
Phage lysins destroy the pneumococcal peptidoglycan network due to their muralytic activity, whereas holins have been shown in S. pneumoniae to form nonspecific lesions (8), most likely upon a process of oligomerization in the cytoplasmic membrane, as observed for the E. coli phage λ (13, 14, 43). It was generally proposed that holin lesions allow access of phage lysins to the cell wall (52, 54), as the majority of phage lysins, including the pneumococcal endolysins, lack a typical N-terminal secretory signal sequence and transmembrane domains (8). However, recent evidence also highlights the possibility for a holin-independent targeting of phage lysins to the cell wall, where holin lesions seem to be crucial for the activation of the already externalized phage lysins (42, 50, 51). Regardless of the mechanism operating in S. pneumoniae to activate phage lysins, holin activity compromises membrane integrity.
Pneumococcal cells present their own autolytic activity, mainly due to the presence of a powerful bacterial cell wall hydrolase, LytA (an N-acetylmuramoyl-l-alanine-amidase), responsible for bacterial lysis under certain physiological conditions (47). Although other bacterial species also encode peptidoglycan hydrolases, the extensive lysis shortly after entering stationary phase caused by LytA is a unique feature of S. pneumoniae. Interestingly, LytA is translocated across the cytoplasmic membrane to the cell wall—where it remains inactive—in spite of the absence of a canonical N-terminal sequence signal (7). In the cell wall, autolysin activities are tightly regulated by mechanisms that seem to be related to the energized state of the cell membrane. In fact, depolarizing agents are able to trigger autolysis in Bacillus subtilis (16, 17), and bacteriocin-induced depletion of membrane potential triggers autolysis of some species of the genera Lactococcus and Lactobacillus, closely related to streptococci (29). It is therefore possible that the holin-inflicted perturbations of the S. pneumoniae cytoplasmic membrane upon the induction of the lytic cycle may trigger not only the lytic activity of the phage lysin, but also that of inactive LytA located in the cell wall. Accordingly, LytA could also participate in the release of phage particles at the end of the infectious cycle, especially considering its powerful autolytic activity. Previous studies have suggested a role for the host autolytic enzyme in the release of phage progeny (11, 38), but in fact, the evidence is unclear and dubious, considering that the existence of phage-encoded lysins was unknown or very poorly understood and some of the experimental conditions used to show a role of LytA could have also affected the activity of the phage lysin (38).
To clarify the possible role of the bacterial autolysin in host lysis, we used the S. pneumoniae strain SVMC28, lysogenic for the SV1 prophage (34), which contains a typical “holin-lysin” cassette, and a different host strain lysogenized with the same SV1 phage. Our results show that LytA is activated by the holin-induced membrane disruption, just like the phage endolysin. In the absence of the endolysin, LytA is capable of mediating host lysis, releasing functional phage particles able to complete their life cycle. Still, sole dependence on LytA results in an altered pattern of phage release that may reduce phage fitness. Importantly, we also show that, together with the endolysin, the concurrent LytA activation is critical for optimal phage progeny release.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae strains SVMC28 and R36A were obtained from the Rockefeller University collection (A. Tomasz). R36A ΔlytA was kindly provided by S. Filipe. SVMC28 is a clinical isolate lysogenic for phage SV1. All S. pneumoniae strains were grown in a casein-based semisynthetic medium (C+Y) at 37°C without aeration (21) or in tryptic soy agar (Oxoid, Basingstoke, England) supplemented with 5% (vol/vol) sterile sheep blood and incubated at 37°C in 5% CO2. Pneumococcal mutant strains were grown in the presence of 2 μg/ml erythromycin or 4 μg/ml chloramphenicol (Sigma, Steinheim, Germany), or both, as appropriate. E. coli strains were usually grown in LB medium (Difco, MD). When required, the medium was supplemented with 100 μg/ml ampicillin (Sigma, Steinheim, Germany), 1 mg/ml erythromycin, or 20 μg/ml chloramphenicol for plasmid selection. M9 minimal medium agar containing thiamine (1 mM; Sigma, Steinheim, Germany) was used for JM109 growth prior to the preparation of competent cells.
TABLE 1.
Bacterial strains, plasmids, and DNA constructs used in this study
| Strain, plasmid, or DNA construct | Relevant characteristicsa | Use in this study | Sourceb or reference |
|---|---|---|---|
| S. pneumoniae | |||
| SVMC28 | Lysogenic for phage SV1; parental strain susceptible to Ery and Cm | Expression of LytA and Svl | 34 |
| SVMC28 ΔlytA | SVMC28 ΔlytA::erm(B); Eryr | Expression of Svl, absence of LytA expression | SVMC28 × aLTA |
| SVMC28 Δsvl | SVMC28 Δsvl::cat; Cmr | Expression of LytA, absence of Svl expression | SVMC28 × aLS1 |
| SVMC28 Δsvl ΔlytA | SVMC28 Δsvl::cat ΔlytA::erm(B); Cmr Eryr | Absence of LytA and Svl expression | SVMC28ΔSV1 × aLTA |
| CP1500 | Novr | Donor of point markers, control in transformation assays | 30 |
| R36A | Laboratory strain; nonlysogenic; susceptible to Ery and Cm | Recipient for phage infection | Rockefeller University Collection |
| R36A ΔlytA | R36A lytA::(pJDC9)::lytA; Eryr | Recipient for phage infection | S. Filipe |
| R36AP | R36A lysogenic for phage SV1 | Expression of LytA and Svl | R36A infected with SV1 |
| R36AP ΔlytA | R36A ΔlytA lysogenic for phage SV1; Eryr | Expression of Svl, absence of LytA expression | R36A ΔlytA infected with SV1 |
| R36AP Δsvl | R36A lysogenic for phage SV1 Δsvl::cat; Cmr | Expression of LytA, absence of Svl expression | R36A infected with SV1 with svl deleted |
| R36AP ΔlytA Δsvl | R36A ΔlytA lysogenic for phage SV1 Δsvl::cat; Cmr Eryr | Absence of LytA and Svl expression | R36A ΔlytA infected with SV1 with svl deleted |
| E. coli | |||
| DH5α | lacZΔM15 | Recipient for pZ1-pZ3 | Invitrogen |
| JM109 | F′ lacIqZΔM15 | Recipient for pZ4-pZ6 | |
| Plasmids | |||
| pGEM-3Z | lacZα Ampr | Cloning vector | |
| pZ1 | pGEM-3Z::lytAup; Ampr | Cloning vector | lytAup (AFLYTA, ARLYTA) |
| pZ2 | pZ1::lytAdw; Ampr | Cloning vector | lytAdw (BFLYTA, BRLYTA) |
| pZ3 | pZ2::erm(B); Ampr Eryr | Template for PCR product aLTA | |
| pZ4 | pGEM-3Z::svlup; Ampr | Cloning vector | svlup (AFLYS1, ARLYS1) |
| pZ5 | pZ4::svldw; Ampr | Cloning vector | svldw (BFLYS1, BRLYS1) |
| pZ6 | pZ5::cat; Ampr Cmr | Template for PCR product aLS1 | CAT (BM18H, PEVP3-1R) |
| pEVP3 | Cmr | Source of cat gene; template for PCR product CAT | 5 |
| pJDC9 | Eryr | Source of erm(B) gene | 4 |
| Amplicons | |||
| aLTA | lytAup::erm(B)::lytAdw | For insertion-deletion replacement for lytA | aLTA (AFLYTA, BRLYTA) |
| aLS1 | svlup::cat::svldw | For insertion-deletion replacement for svl | aLS1 (AFLYS1, BRLYS1) |
Eryr, erythromycin resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Novr, novobiocin resistance.
Strain construction by a cross carried out by using transformation is indicated as recipient × DNA donor. Primers used to amplify DNA fragments by PCR are indicated in parentheses.
Antibiotic susceptibility.
Chloramphenicol and erythromycin MICs were determined by Etest following the manufacturer's guidelines (AB Biodisk, Solna, Sweden). Susceptibility to novobiocin was tested with impregnated paper discs (Oxoid, Hampshire, England). PCR to detect the cat gene, which amplifies a 338-bp fragment internal to the gene, in the SVMC28 strain was performed using the primer pair CATd and CATr (24) (Table 2).
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′→3′)a | Recognition site |
|---|---|---|
| CATd | TTAGGYTATTGGGATAAGTTA | |
| CATr | CATGRTAACCATCACAWACCAG | |
| AFLYTA | AGCGAATTCGGCAGGATATAAGGGTGTTATC | EcoRI |
| ARLYTA | ATAGGATCCATTCTACTCCTTATCAATTAAAAC | BamHI |
| BFLYTA | GCAGTCGACTAATGGAATGTCTTTCAAATC | XmiI |
| BRLYTA | CAATAGCATGCGATATTCCTTTCACCTTTTTCC | PaeI |
| DINF-D4 | GCAAAAGATCCTTCCTCTAGTTTC | |
| ORF1-R3 | CTTCACCATCAGCTCCCAAC | |
| AFLYS1 | AGCGAATTCAGGGGTTCTCTTACTGATGATC | EcoRI |
| ARLYS1 | ATAGGATCCTCCCTATCGTCCTTTCCATGC | BamHI |
| BFLYS1 | GCAGTCGACTGAAGACAGGCTGGGTCAAGTAC | XmiI |
| BRLYS1 | CAATAGCATGCGCTATTTCCCAAGGTGCTGG | PaeI |
| BM18H | ATAGGATCCGGGTTCCGAGGCTCAACGTCAA | BamHI |
| PEVP3-1R | CGAGGTCGACGGTATCGATAAGCT | XmiI |
| 28HA37-X1 | TCAGGTTACTTGAAAAGGCAATAG | |
| 28HA37-R5 | CAACGTCGCCGTTCTGTTGAATC |
Recognition sites are underlined.
DNA techniques.
All routine DNA manipulations were performed according to standard methods (40). The PCR primers are listed in Table 2. Chromosomal DNA from S. pneumoniae and phage DNA were isolated similarly to previously described procedures (1, 27, 46). Plasmids were prepared using either the High Pure plasmid isolation system or the Genopure plasmid Midi system (Roche, Mannheim, Germany), and PCR products and endonuclease digests were purified using the High Pure PCR product purification system (Roche, Mannheim, Germany). The enzymes used in the manipulation of DNA were purchased from MBI Fermentas (Vilnius, Lithuania). All oligonucleotides were obtained from the Invitrogen Co. (Paisley, Scotland). Nucleotide sequences were analyzed using VECTOR NTI Deluxe (Invitrogen, Barcelona, Spain) software.
Construction of pneumococcal mutants by insertional deletion of lytA and svl.
The mutant strain SVMC28 ΔlytA, in which the lytA gene was deleted and replaced by the erm(B) gene, was constructed essentially as described previously (9). First, lytAup (529 bp), the sequence encoding the upstream fragment of lytA, was amplified with the primers AFLYTA and ARLYTA from SVMC28 chromosomal DNA. The PCR product was digested with EcoRI and BamHI and inserted into the plasmid pGEM-3Z to generate pZ1. Next, lytAdw (509 bp), the sequence encoding the downstream fragment of lytA, was amplified from chromosomal DNA of SVMC28 using the primers BFLYTA and BRLYTA, and the PCR product was inserted as an XmiI-PaeI fragment into pZ1, generating pZ2. Lastly, the BamHI/ClaI fragment (2,051 bp) from pJDC9, which contained the Ermr cassette [erm(B) gene] (4), was cloned into the BamHI/XmiI-digested plasmid pZ2, yielding pZ3. Plasmid pZ3 contained the erm(B) marker flanked by the upstream and downstream regions of lytA and was used as a template for PCR with AFLYTA and BRLYTA to produce an aLTA fragment. After transformation of SVMC28 with aLTA, the deletion of lytA in the erythromycin-resistant SVMC28 ΔlytA mutant was confirmed by PCR amplification and subsequent sequencing with primers DINF-D4 and ORF1-R3, external to AFLYTA and BRLYTA. An identical strategy was used to construct the mutant SVMC28 Δsvl, in which svl was replaced by the cat gene. svlup (583 bp) and svldw (477 bp) were PCR amplified from SV1 phage DNA using the primer pairs AFLYS1-ARLYS1 and BFLYS1-BRLYS1. svldw contained 185 bp of svl. A CAT fragment (1,053 bp), containing the Cmr marker (cat), was amplified from pEVP3 with BM18H and PEVP3-1R and cloned into pZ5 as a BamHI-XmiI fragment. aLS1 was produced with AFLYS1 and BRLYS1 from pZ6, and the resulting PCR product was used to transform the pneumococcal strain SVMC28. The deletion of svl in the chloramphenicol-resistant SVMC28 Δsvl mutant was confirmed by PCR amplification and sequencing with primers 28HA37-R5 and 28HA37-X1, external to AFLYS1 and BRLYS1. To construct the double-deletion mutant SVMC28 Δsvl ΔlytA, the PCR fragment aLTA was used to transform SVMC28 Δsvl. The chloramphenicol- and erythromycin-resistant strain SVMC28 Δsvl ΔlytA was confirmed to carry deletions of the lytA and svl genes by the procedures described above. For all PCRs, except those used as templates in sequencing reactions, the High Fidelity PCR Enzyme Mix kit (MBI Fermentas, Vilnius, Lithuania) was used. The amplicons (DNA products amplified by PCR) are described in Table 1.
Transformation of S. pneumoniae and selection of transformants.
Transformation of pneumococcal cells was carried out as described previously (9). Transformants were selected on tryptic soy agar supplemented with 5% (vol/vol) sheep blood and chloramphenicol (4 μg/ml) and/or erythromycin (2 μg/ml).
Analysis of phage excision.
The pulsed-field gel electrophoresis (PFGE) procedure for the visualization of extrachromosomal phage DNA was adapted from the method of Ramirez et al. (34), except that cells were harvested after a 2-h period of mitomycin C (MitC) (Sigma, Steinheim, Germany) treatment. The electrophoresis conditions were 6 V/cm, ramping of the pulse between 1 and 2 s, and a total running time of 16 h. The buffer was maintained at 14°C during the run.
Construction of lysogenized strains.
The lysogenized strains were constructed using the laboratory strain R36A. SV1, obtained from wild-type SVMC28, was used to lysogenize strains R36A and R36A ΔlytA, resulting in strains R36AP and R36AP ΔlytA (Table 1). To obtain lysogenic strains defective in the phage lysin, R36A and R36A ΔlytA were infected with SV1 Δsvl extract from SVMC28 Δsvl. The resulting lysogens were named R36AP Δsvl and R36AP ΔlytA Δsvl, respectively (Table 1). Phage infection was performed by phage plaque assays (see below). Agar was picked at the edges of plaques. The presence of both wild-type and mutant phages was tested for by PCR with primers for SV1 (AFLYS1 and ARLYS1 [Table 2]). MitC-induced lysis was performed on selected PCR-positive colonies. We considered that response to MitC was indicative of phage excision and consequently of successful prior lysogeny. The released phages were able to infect and lyse cells of the wild-type R36A strain, producing phage plaques and confirming that the strains were indeed lysogenic and that the phages were fully functional.
Lysis assays.
Overnight cultures of wild-type, mutant, and lysogenized strains in C+Y supplemented with the appropriate antibiotics were diluted 1:100 in fresh medium (without antibiotics), and the cultures were grown until the optical density at 600 nm (OD600) reached approximately 0.2 to 0.25. MitC was then added to a final concentration of 0.1 μg/ml to induce the lytic cycle (32). Incubation was continued, and growth was monitored by the OD. Cultures treated with deoxycholate (DOC) (0.04% [wt/vol]) and nisin (1 μg/ml) (Sigma, Steinheim, Germany) were grown to an OD600 of 0.4 and 0.2 to 0.25, respectively. All assays were carried out at least in duplicate. Lysis was expressed directly as the OD600 drop or as the percentage decrease in the OD600 relative to its maximal value (the OD value prior to lysis). The degree of lysis at each time point was calculated from the following equation: lysis extent (percentage) = 100 − (percentage of maximal OD).
Phage plaque assays.
Plaque assays were performed as described elsewhere (22) with the following modifications: C+Y medium with 170 U catalase/ml agar was used, top agar was not added, and phage were applied in 10-μl aliquots directly on the soft agar (0.35%) with the indicator strain. Incubation was performed at 30°C. Lysogenic phages were induced with MitC. At specific times (between 40 and 180 min) after MitC treatment, cultures were filtered through a 0.45-μm-pore-size membrane, and the supernatant was stored at 4°C for a maximum of 24 h until it was used. To lysogenize the strains, cultures were filtered after total lysis (180 min). To eliminate the possibility that lysis of the indicator strain was caused by the bacterial products (e.g., bacteriocins) and not caused by phage infection, a fractionation of the culture medium was performed. Cultures treated for 180 min with MitC were prefiltered through a 0.45-μm-pore-size membrane, followed by filtering them with a 100-kDa mass cutoff polyethersulfone membrane (Vivaspin 20 concentrator; Sartorius Stedim Biotech, Goettingen, Germany), which retained the SV1 phage but not proteins that could cause bacterial lysis, such as LytA, Svl, holins, and bacteriocins (mass < 100 kDa). Both the filtrate containing proteins of <100 kDa and that containing the phage particles were used. Phage plaques were observed with a Leica MZ7.5 high-performance stereo microscope (Leica Microsystems, Germany).
Viability assays.
Flow cytometry analysis of cultures treated with MitC was performed. As a control for cell death, the cultures were treated with nisin and DOC. All compounds used were filtered through a 0.22-μm-pore-size membrane before flow cytometry measurement. In these assays, selective overnight cultures of wild-type SVMC28 and the derived mutants strains were diluted 1:100 in fresh 0.22-μm-filtered C+Y and grown to the appropriate OD600. Cells were collected immediately after exposure and at 20-min intervals during a 2-h period and then diluted in sterile-filtered 0.85% NaCl to a concentration of ∼1 × 106 cells/ml. Cell viability was assessed by using the Live/Dead BacLight bacterial viability kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 1.5 μl of Syto 9 (green fluorescent nucleic acid stain; 3.34 mM) at a final concentration of 5 μM and 1.5 μl of propidium iodide (PI) (red fluorescent nucleic acid stain; 2 mM) at a final concentration of 3 μM were added to each 1-ml diluted sample of cells. PI stock solution was diluted to 2 mM in sterile-filtered distilled H2O immediately prior to staining. The samples were then incubated at room temperature in the dark for 20 min and analyzed on a Partec CyFlow space flow cytometer (Partec GmbH, Münster, Germany) with 488-nm excitation from a blue solid-state laser at 50 mW. Forward scatter (FSC), side scatter (SSC), and two fluorescence signals were measured. Green fluorescence, indicating the population of live cells (nonpermeabilized cytoplasmic membranes) was detected in the FL1 channel, and red fluorescence, indicating the population of dead cells (permeabilized cytoplasmic membranes), was detected in the FL3 channel. Optical filters were set up so that FL1 measured at 520 nm and FL3 measured above 610 nm. The sample analysis rate was kept below 1,000 events/s. The trigger was set for the FSC channel, and the combination of FSC and SSC was used to discriminate bacteria from the background. Twelve thousand events were collected for each sample taken. Data were collected and analyzed using FloMax software (Partec GmbH, Münster, Germany). Assays were carried out at least in duplicate.
Cell viability was also assessed by fluorescence microscopy to confirm staining by the different strains after MitC treatment. Syto 9/PI-labeled cell suspensions were microscopically analyzed 40, 80, and 120 min after MitC addition with a Zeiss Axiovert 200 M microscope (Carl Zeiss, Germany) equipped with a 100-W halogen lamp, the appropriate excitation and emission filters for Syto 9 and PI (excitation wavelengths, 450 to 490 nm [Syto 9] and 540 to 552 nm [PI]; emission wavelengths, 515 to 565 nm [Syto 9] and >590 nm [PI]), a Plan Apochromat 63X/1.4 objective lens, and a CoolSnap HQ charge-coupled device camera (Roper Scientific Photometrics, Tucson, AZ). Fluorescence photographs were acquired with Metamorph software (version 6.1r0).
Statistical analysis.
Differences in the mean values of the lysis extent between strains were analyzed by a Student t test. For all comparisons, a P value of <0.05 was considered to represent statistical significance; 95% confidence intervals for the average of the OD measurements from different experiments were calculated based on the Student t distribution.
Nucleotide sequence accession number.
The DNA sequence of the 1.719-kb SV1 lytic cassette has been assigned GenBank accession number FJ765451.
RESULTS
The prophage SV1 lytic cassette and construction of the lysin mutants.
Strain SVMC28, an S. pneumoniae clinical isolate lysogenic for the inducible phage SV1, was selected for study (34). The SV1 lytic cassette is localized downstream of the structural cluster and adjacent to the attP site in the SV1 genome, similarly to other pneumococcal lytic cassettes, and shows a high nucleotide sequence identity to the pneumococcal phage MM1 (31). In addition, it exhibits a typical three-component organization in which the two open reading frames encoding putative holins (Svh1 and Svh2) precede the endolysin gene, which encodes a putative amidase (Svl). Attempts to clone svh1 and svh2 open reading frames in E. coli resulted in loss of viability, strongly indicating that these proteins correspond to holins (data not shown).
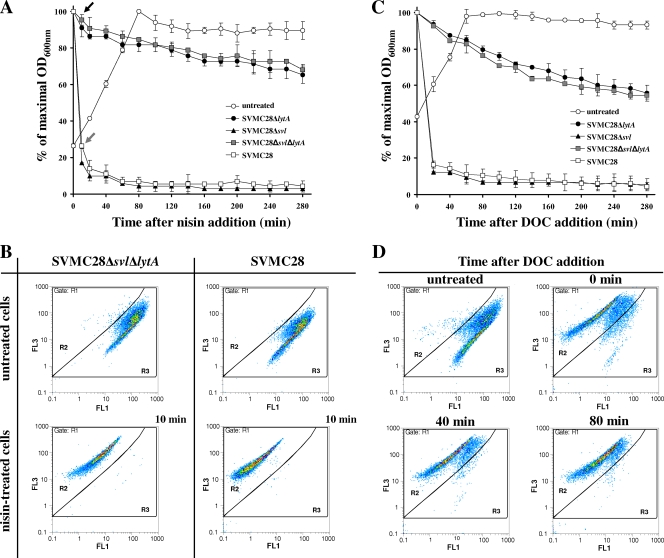
In order to construct S. pneumoniae mutants without bacterial and phage lytic activities, the lytA and svl genes were eliminated by insertion-deletion in the clinical isolate SVMC28 (Table 1). Elimination of the lytic genes did not alter the growth rates of the mutants. In fact, all strains displayed growth curves indistinguishable from those of the parental strain, indicating that the modifications introduced had no significant impact on pneumococcal physiology. As expected, the lytA-deficient strains SVMC28 ΔlytA and SVMC28 Δsvl ΔlytA were greatly resistant to autolysis in stationary phase (data not shown) and when treated with nisin or DOC (see Fig. 3). In these cases, only 30% to 45% lysis occurred 280 min after nisin or DOC addition, which was far less than that exhibited by the wild-type strain SVMC28 (close to 95%). The phage lysin played no role in the response to these stimuli, since SVMC28 ΔlytA and SVMC28 Δsvl ΔlytA showed similar lytic phenotypes. Thus, residual lysis was probably caused by other pneumococcal autolysins, such as LytC, which is also responsible for lysis in stationary phase but with a lower activity than LytA at 37°C (6, 12). The elimination of svl in the SVMC28 Δsvl mutant did not alter the lytic response upon nisin and DOC treatment, which was characterized by the same lysis rate as the wild-type strain (see Fig. 3), confirming that LytA is fully functional in this mutant strain.
FIG. 3.
Kinetics of nisin- and DOC-triggered lysis of wild-type S. pneumoniae SVMC28 and mutants and corresponding flow cytometry analysis. (A) Kinetics of nisin-triggered lysis. Nisin at a final concentration of 1 μg/ml was added to cultures at an OD600 of 0.2 to 0.25, and the OD was monitored. The arrows indicate the times at which nisin-treated cultures were harvested for flow cytometry analysis (10 min). (B) Flow cytometry analysis of wild-type SVMC28 and SVMC28 Δsvl ΔlytA after nisin treatment. Exponentially growing cells were treated with nisin or left untreated (panel A), stained with a mixture of Syto 9 and PI, and analyzed on a flow cytometer. Similar analysis patterns were obtained for SVMC28 ΔlytA and SVMC28 Δsvl (data not shown). Gates R2 and R3 differentiated between damaged and undamaged cell populations, respectively, and were designed over gate R1, which included the total stained population. The results are representative of a minimum of two independent experiments. (C) Kinetics of DOC-triggered lysis. DOC at a final concentration of 0.04% (wt/vol) was added at time zero to cultures in mid-exponential phase (OD600 = 0.4), and the turbidity was monitored. (D) Flow cytometry analysis of SVMC28 Δsvl ΔlytA exposed to DOC. Exponentially growing cells were treated with DOC at 0.04% (wt/vol). Culture samples were collected at 0, 40, and 80 min after DOC addition (panel C), stained with a mixture of Syto 9 and PI, and analyzed on a flow cytometer. As a control, the same cells were left untreated. Gate definitions were as for panel A. The results are representative of a minimum of two independent experiments. In panels A and C, the results presented for each strain correspond to the mean value of at least two independent assays, and 95% confidence intervals are indicated.
Phenotypic evaluation of phage-induced lysis.
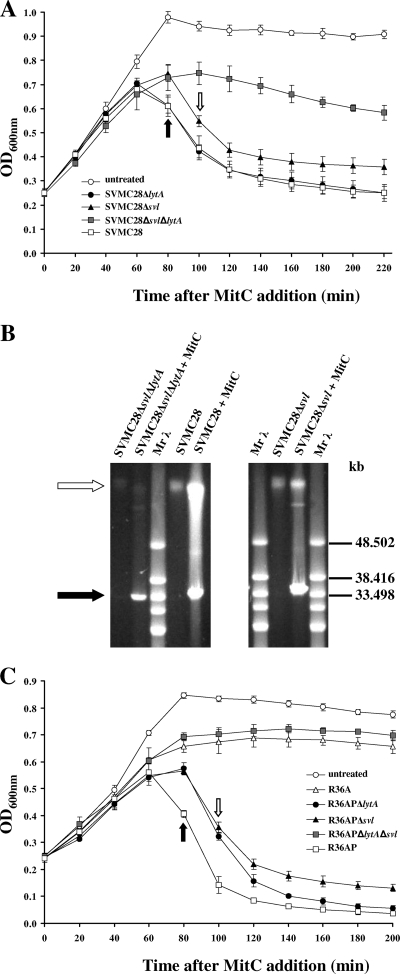
To investigate the possible role of the autolysin LytA in phage release in S. pneumoniae, we started by comparing the lytic phenotypes of the parental and mutant strains after induction of the SV1 lytic cycle with MitC. As indicated in Fig. 1A, SVMC28 ΔlytA cultures showed pronounced lysis 80 min after MitC addition, in contrast to the control untreated cultures. This is consistent with the known role of phage lysins in bacterial cell wall degradation to allow the release of new phage particles. Remarkably, in the mutant lacking endolysin (SVMC28 Δsvl), lysis was unequivocally detected after treatment with MitC, clearly demonstrating that LytA is activated after prophage induction.
FIG. 1.
Participation of bacterial and phage lysins in pneumococcal-phage-induced lysis. (A) Lysis profiles of SVMC28 strains. Wild-type SVMC28 and the derived mutants were grown to an OD600 of 0.2 to 0.25, and 0.1 μg/ml of MitC was added to induce phage excision (0 min). (B) PFGE analysis of extrachromosomal phage DNA induced with MitC. Total DNA was isolated from cultures of SVMC28, SVMC28 Δsvl, and SVMC28 Δsvl ΔlytA treated with MitC or left untreated (control). The preparations were separated by PFGE. The white arrow indicates the bacterial chromosome (about 2.2.Mb), while the black arrow indicates phage DNA. Similar PFGE profiles were obtained for SVMC28 ΔlytA (data not shown). Mrλ, lambda ladder PFGE marker (New England Biolabs, Beverly, MA). (C) Lysis profiles of lysogenized R36A strains. R36AP, R36AP ΔlytA, R36AP Δsvl, and R36AP ΔlytA Δsvl were grown to an OD600 of 0.2 to 0.25, and 0.1 μg/ml of MitC was added to induce phage excision (0 min). In panels A and C, the arrows represent the times at which lysis started: 80 min after MitC addition for SVMC28, SVMC28 ΔlytA, and R36AP (black arrows) and 100 min for SVMC28 Δsvl, R36AP Δsvl, and R36AP ΔlytA (white arrows). The untreated SVMC28 and R36AP cultures are representative of the growth curves of all untreated strains. The results are averages of a minimum of four independent experiments, and 95% confidence intervals are indicated.
Analysis of the MitC-treated double mutant SVMC28 Δsvl ΔlytA, which lacks LytA and Svl, revealed negligible lysis with a maximum of 35% lysis at 180 min (Fig. 1A), which may reflect the activity of LytC (12). A PFGE analysis of the total DNA showed no considerable changes in prophage excision compared to the wild type in SVMC28 Δsvl and SVMC28 Δsvl ΔlytA mutant strains, with all MitC-treated cultures showing free phage DNA (∼33 kb), which was not detected in the untreated control cultures (Fig. 1B). This was also observed for the lytA-deficient mutant SVMC28 ΔlytA (data not shown). The extra fragments visualized below the chromosome are concatemers of the phage genome, as already demonstrated for SV1 (34). These observations clearly demonstrate that, similarly to the wild type, in all mutant strains, the phage successfully initiated the lytic cycle with DNA excision and replication, indicating that the observed lysis after MitC addition was phage mediated.
To test whether this newly discovered role of LytA was independent of the bacterial genetic context, we characterized the lysis of similar mutants generated in the R36A background by lysogenizing the laboratory strains R36A and R36A ΔlytA with phages SV1 and SV1 with svl deleted (SV1 Δsvl) (Table 1). As depicted in Fig. 1C, MitC induction of phage excision in the endolysin-deficient R36AP Δsvl strain resulted in pronounced lysis, confirming the activation of LytA. Thus, independently of the host genetic background, LytA mediates phage-induced lysis of the host cell in the absence of the endolysin.
It is noteworthy that the lysis rates of the Δsvl mutant were similar to those of the ΔlytA mutant in both SVMC28 and R36A backgrounds (Fig. 1A and C), indicating that the exclusive presence of the autolysin LytA in the absence of any phage lytic enzyme is sufficient for an accentuated decrease in OD at the end of the lytic cycle. However, the bacterial autolysin appears not to influence the lysis rate or the lysis extent promoted by Svl, since these parameters were similar between the SVMC28 and R36AP strains (containing both enzymes) and the respective ΔlytA mutants, in which just the Svl activity is present (Fig. 1A and C). Nevertheless, for R36A in the absence of LytA (strain R36AP ΔlytA), the lysis timing was delayed. Thus, in contrast to the experiment performed in the SVMC28 genetic background, in the R36A strain, LytA is essential to control the exact timing of lysis.
In both genetic backgrounds, exclusive reliance on LytA for lysis resulted in a 20-min delay relative to when both phage and host lysins were present (Fig. 1A and C). This delay was not a reflection of a difference in growth rates, as the doubling time of the Δsvl mutants was comparable to those of the SVMC28 and R36AP strains. In addition, a substantial reduction of the total lysis percentage in the Δsvl mutants relative to the strains carrying both bacterial and phage lysins was also observed (at 180 min, SVMC28 Δsvl and SVMC28, P < 10−4 [Fig. 1A], and R36AP Δsvl and R36AP, P < 10−4 [Fig. 1C]).
Collectively, these results suggest that in the absence of phage endolysin, although LytA is able to mediate bacterial lysis, already assembled phage particles may be retained inside the bacterial host for a longer time, and that relying exclusively on the host autolysin could have an important impact on the quantity of phage particles released.
Phenotypic assessment of phage release.
Although the different lytic phenotypes provide clues about the changes in the phage particles released, the measurement of phage production was essential to determine if a significant difference in the release of phage progeny was observed. A phage plaque assay, using strain R36A as an indicator, was therefore performed using the supernatant of the MitC-induced strains SVMC28 and R36AP and the corresponding mutants at different time points. When the R36A lysogens (with phage already adapted to infect R36A) were used, the plating efficiency was improved relative to the phage obtained directly from SVMC28, providing excellent conditions to explore the differences attributed to LytA in virion release.
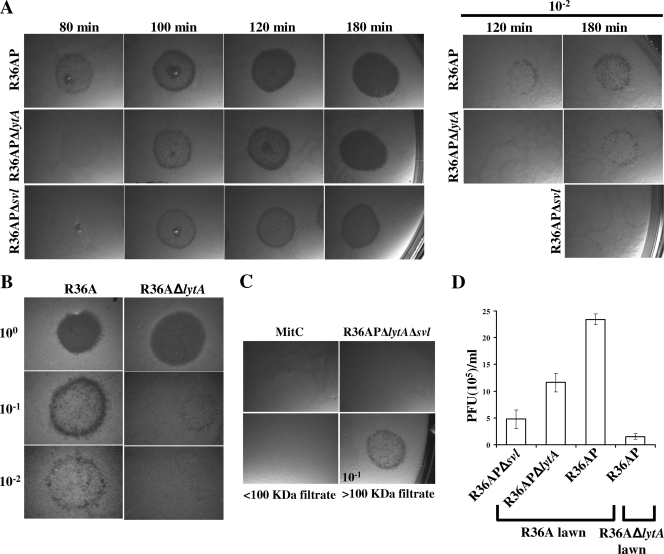
In the absence of endolysin, phage plaques were clearly observed, indicating that LytA by itself allows the release of functional phages capable of completing their life cycle (Fig. 2A). However, in accord with the delayed lysis timing, phage plaques obtained for R36AP Δsvl were observed only from 100 min onward, whereas for R36AP, phage plaques were already visible 80 min after MitC addition, when culture lysis was detected (Fig. 2A). In agreement with the reduced lysis mediated exclusively by LytA relative to that observed in the presence of both host and phage lysins, the number of phage released when only LytA was present was also significantly diminished (at 180 min, P < 10−5) (Fig. 2D). Thus, these results support previous observations and confirm that lysis strictly dependent on LytA may severely impair phage fitness by reducing phage progeny release and delaying its timing.
FIG. 2.
Participation of bacterial and phage lysins in phage release. (A) Phage release patterns. The culture media of R36AP, R36AP Δsvl, R36AP ΔlytA, and R36AP ΔlytA Δsvl treated with MitC were filtered (0.45 μm) at 20-min intervals after the start of lysis in the R36AP strain, and the supernatants were used directly in phage plaque assays of the indicator strain R36A. The time after MitC addition is indicated. The results are representative of three independent experiments. (B) Phage plaque assay using indicator strains differing only in the presence or absence of LytA. The supernatant of the culture medium of R36AP was collected 180 min after MitC treatment and used on indicator strains R36A and R36A ΔlytA. The results are representative of three independent experiments. (C) Indicator lawn lysis is due to phage induction. As a control, R36A indicator lawns were exposed to C+Y medium with MitC at the same concentration used for phage induction. For R36AP ΔlytA Δsvl, the supernatant collected 180 min post-MitC addition produced no phage plaques. The supernatant of an R36AP culture treated for 180 min with MitC was filtered through a 100-kDa-cutoff membrane to retain phages while eliminating small proteins. The <100-kDa filtrate showed no phage plaques, unlike the retained fraction (>100-kDa filtrate; 10−1 dilution), demonstrating that the plaques were due to phage and were not due to the action of bacterial and phage lysins, holins, or bacteriocins (mass < 100 kDa), which can cause cell lysis. (D) Comparison of the numbers of PFU per ml detected upon phage induction. The numbers of PFU per ml were determined for strains R36AP, R36AP Δsvl, and R36AP ΔlytA after 180 min of MitC treatment on indicator lawns of strains R36A and R36A ΔlytA, as indicated. Averages and 95% confidence intervals are indicated. For R36A in the indicator lawn, the comparisons were between supernatants obtained from R36APΔ lytA and R36AP (P < 10−4), R36AP Δsvl and R36AP ΔlytA (P < 10−3), and R36AP Δsvl and R36AP (P < 10−5). When the R36AP supernatant was plated on indicator lawns of strains R36A and R36A ΔlytA, P was <10−9. Magnification (A to C), ×6.3 or ×8.0.
Surprisingly, the bacterial-lawn clearance due to infection with supernatant from R36AP ΔlytA was different from that due to R36AP (Fig. 2A). The lysis delay between R36AP ΔlytA and R36AP prevented a direct comparison of the phage plaques obtained at most of the time points studied. Nevertheless, at 180 min, when the two lytic processes reached indistinguishable and approximately stable lysis extents (Fig. 1C), the bacterial lawn clearance due to infection with R36AP ΔlytA supernatant was much less pronounced than that due to infection with R36AP supernatant (Fig. 2A), resulting from fewer phage being released (P < 10−4) (Fig. 2D). Similarly, for the SVMC28 genetic background, in the absence of LytA, the bacterial clearance due to phage infection was always less marked at any given time than that observed for the wild type (data not shown). This was also unexpected, since the lysis of SVMC28 ΔlytA and the wild type was characterized by the same overall extent, timing, and rate (Fig. 1A). Thus, one would expect that the numbers of phage released would be equivalent at any given point of the lytic process for those strains.
Thus, these data reveal a negative impact on phage release in the absence of LytA, indicating that the bacterial autolysin, together with the phage endolysin, maximizes progeny release, contributing substantially to this process. In fact, another set of experiments further established this role. When the same phage preparation obtained after total lysis of MitC-treated R36AP was used to infect strains R36A and R36A ΔlytA as indicators (differing only in the presence of a functional LytA), fewer phage plaques were obtained for R36A ΔlytA (P < 10−9) (Fig. 2B and D), demonstrating the importance of the presence of the host lysin for plaque formation and supporting its role in phage release.
Membrane permeabilization and cell lysis.
It has been shown for some bacteria that holins strictly control the lysis timing by disrupting the bacterial cytoplasmic membrane, which results in the triggering of the endolysin activity (13, 39, 42, 51, 52). In S. pneumoniae, phage endolysins depend on holin activity for efficient peptidoglycan degradation (8, 28), but whether the timing of lysis, and thus the endolysin activity, is controlled by holins has yet to be determined. Given the observation that LytA is able to mediate phage-induced lysis, we tested if the permeabilization and consequent depolarization of the cytoplasmic membrane caused by the holins (13, 43) is responsible for the activation of both lysins.
To address this issue, a real-time flow cytometry analysis of bacterial viability based on membrane integrity was performed after phage induction. In this assay, using a Live/Dead BacLight bacterial viability kit (Invitrogen, Carlsbad, CA), cells with damaged membranes (the dead population) allow the uptake of PI, fluorescing red (FL3 channel), whereas undamaged cells (the live population) internalize only the Syto 9 dye, fluorescing green (FL1 channel). The first step consisted of setting gates that differentiated between the two populations. As a control for cell death, we used the antimicrobial agent nisin, which, by inserting into the cytoplasmic membrane, causes irreparable membrane damage and triggers pneumococcal LytA activity (Fig. 3A) (45). After 10 min, before complete lysis occurred in the wild-type culture, the untreated and the nisin-treated cells formed two well-defined and distinct populations (Fig. 3B). Therefore, the gates representing damaged (R2) and undamaged (R3) cells were constructed using a 1:1 mixture of untreated and nisin-treated cells. Gating with R2 and R3 revealed that 99% of the untreated cells remained intact compared with less than 0.2% of the nisin-treated cells (Fig. 3B). To assess the robustness of the constructed gates, we used the detergent DOC, a well-known trigger of LytA activity (33, 47), which is assumed to have membrane-permeabilizing properties. In this experiment, the LytA-lacking mutant SVMC28 Δsvl ΔlytA was used to avoid immediate lysis (Fig. 3C and D). The untreated cells fit entirely into gate R3, and immediately after DOC addition, the cells were distributed almost exclusively in gate R2 (Fig. 3D). Similar results were obtained with strain SVMC28 ΔlytA (data not shown). Therefore, the chosen gates allowed complete differentiation of damaged cell populations due to other membrane perturbations, in addition to those induced by nisin.
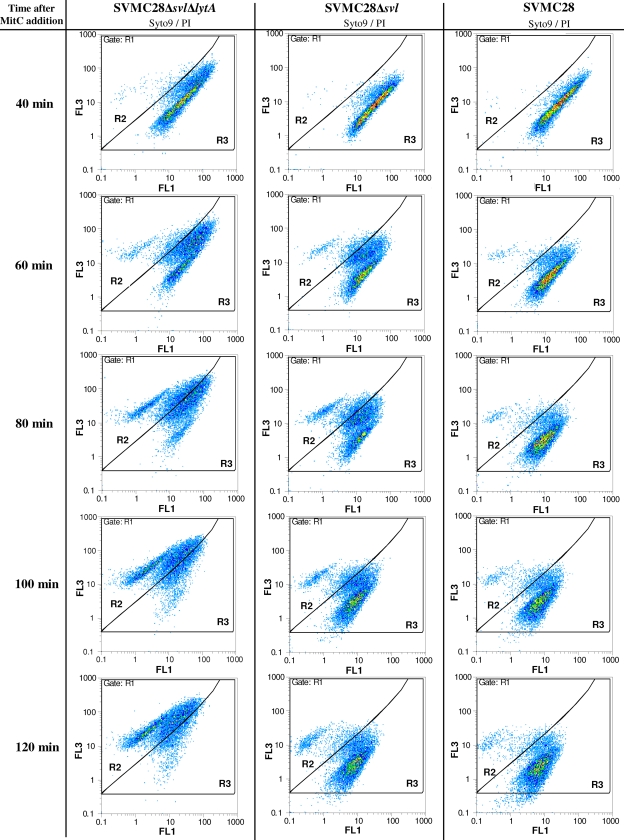
The validated gates were then applied to determine the proportions of viable and permeabilized populations in wild-type and mutant cultures treated with MitC. Depending on the strain analyzed, different fluorescence patterns emerged. For the double mutant SVMC28 Δsvl ΔlytA, a shift of the bacterial population toward increased red fluorescence intensity (R3 to R2) was observed, indicating that the cells were becoming permeabilized with increasing time after phage induction (Fig. 4). In fact, after 120 min of phage induction, almost all cells were within the R2 gate. This feature was not observed for the wild-type strain (Fig. 4) or the SVMC28 ΔlytA strain (data not shown). At 80 min, membrane integrity was already compromised in a significant fraction of the SVMC28 Δsvl ΔlytA bacterial population, precisely when lysis was evident in the wild type (Fig. 1A). In contrast, in both the SVMC28 ΔlytA and wild-type strains, the majority of the population detected was viable, with the holin-damaged fraction corresponding to about 7% (Fig. 4). This can be attributed to rapid lysis upon holin membrane permeabilization. The compartment of damaged cells that had not yet lysed was therefore very sparsely populated due to the extremely fast lysis triggered by holin activity. These data provide definite evidence that holin-induced membrane lesions trigger the lytic activity of the phage endolysin.
FIG. 4.
Effect of phage holin activity on S. pneumoniae cell membrane permeabilization. Cultures of SVMC28, SVMC28 Δsvl ΔlytA, and SVMC28 Δsvl were treated with MitC and tested for membrane permeabilization at various times by flow cytometry analysis using a mixture of Syto 9 and PI staining (Live/Dead BacLight bacterial viability kit; Invitrogen, Carlsbad, CA). Experimentally defined gates R2 and R3 were used to differentiate between damaged and undamaged cell populations and were designed over gate R1, which included the total stained population (Fig. 3). The left column shows a shift in the Syto 9/PI staining pattern through time after phage induction of strain SVMC28 Δsvl ΔlytA, which lacks both the phage endolysin and the S. pneumoniae autolysin, LytA. In the presence of lytic enzymes (middle and right columns), a different scenario was observed, with only a few damaged cells detected. The data are from a representative experiment of a minimum of three independent experiments.
We then went on to analyze by flow cytometry the mutant SVMC28 Δsvl after MitC treatment to test the hypothesis that LytA may also be activated by the same holin lesions that induce Svl endolysin activation. In line with the observed lysis caused by LytA, the SVMC28 Δsvl pattern resembled that of the wild-type strain, with a large population of intact cells (gate R3) and a smaller fraction of damaged cells (gate R2) at every time point after MitC phage induction (Fig. 4). Contrary to the large reservoir of dead bacteria observed in SVMC28 Δsvl ΔlytA, the fraction of damaged cells was substantially smaller in SVMC28 Δsvl, as the population driven into the R2 gate by the holins underwent lysis, becoming undetectable by flow cytometry. Thus, membrane permeabilization caused by the holins is responsible for triggering LytA activity. However, in SVMC28 Δsvl, a larger population of membrane-damaged cells (gate R2) was clearly visible from 80 min onward, in contrast to the residual fraction observed in the wild type (Fig. 4). This observation is in agreement with the previously observed lysis delay mediated by LytA in the absence of endolysin. In this case, as the time between holin action and LytA-induced lysis was more prolonged, a greater percentage of holin-permeabilized but still unlysed cells was detected than with the wild type. Besides the well-defined population of membrane-permeabilized cells, exclusively distributed in the R2 gate, there was also a population located between the R2 and R3 gates. This mixed population was also observed in SVMC28 Δsvl ΔlytA, and since it was increasingly found in R2, it may correspond to chains containing both damaged and undamaged cells, where the number of damaged cells increased with time. This was indeed confirmed by fluorescence microscopy. As shown in Fig. 5, the number of PI-stained cells within the chains increased with time after SV1 induction. For instance, at 80 min, when the total population was almost evenly distributed between R2 and R3 (Fig. 4), approximately half of most chains in fact consisted of damaged, PI-stained cells (Fig. 5C). In addition, these observations also confirm the increased permeabilization perceived from flow cytometry analysis. In line with these data, in the SVMC28 Δsvl flow cytometry profile, more chains were observed with a mixture of damaged and undamaged cells than in the wild type (Fig. 4). This persistence of chains, which are not promptly dispersed by cell lysis, is caused by the delayed LytA-induced lysis of holin-damaged cells.
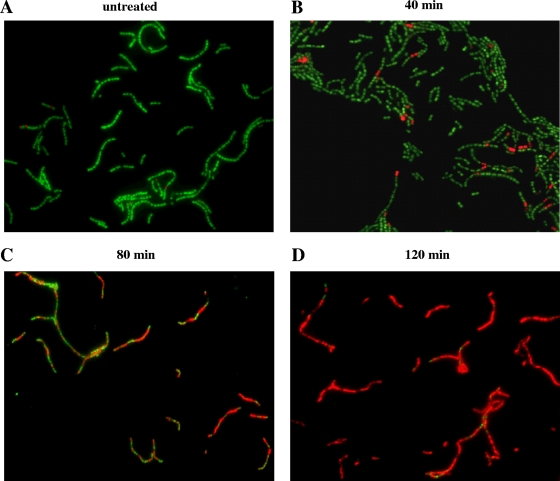
FIG. 5.
Fluorescence microscopy analysis of Syto 9/PI-stained strain SVMC28 Δsvl ΔlytA after MitC phage induction. SVMC28 Δsvl ΔlytA culture samples were collected at 40, 80, and 120 min after the addition of 0.1 μg/ml MitC, stained with a mixture of Syto 9 and PI, and visualized on a fluorescence microscope (magnification, ×630). As a control, the same cells were not treated with MitC. Different fluorescence patterns were clearly detected. (A) The untreated control corresponds mostly to bacteria exclusively stained with Syto 9. (B) After 40 min of phage induction, PI stained a few cells (dead cells), although the majority of cells stained only with Syto 9, indicative of intact membranes. (C) Eighty minutes after phage induction, almost half of the cells were stained with PI, with chains containing a mixture of PI- and Syto 9-stained cells. (D) After 120 min of phage induction, PI stained almost every cell. The chains showed few bacteria stained only with Syto 9. Each panel is from a representative experiment of four independent assays.
DISCUSSION
The “holin-lysin” strategy to release phage progeny through host lysis is the most widespread system in nature (53) and appears to be present in every S. pneumoniae phage (8, 28, 31). In this system, phage endolysins ultimately destroy the host envelope, allowing the escape of fully assembled virions, and therefore, this phage-encoded function is essential (8, 28, 54). However, the presence in S. pneumoniae of the powerful autolytic amidase LytA, which both structurally and functionally closely resembles pneumococcal-phage endolysins (10, 23, 34-36), raises the possibility that it could play an important role in phage-mediated lysis. Since holins, the other protein components of phage “holin-lysin” systems, form lesions in the host membrane (8, 13, 43) and membrane depolarization leads to autolysis in B. subtilis (16), it is tempting to hypothesize that cell wall-resident LytA could be activated by the holin-induced lesions. Although it has been suggested that the pneumococcal autolytic enzyme LytA contributes to phage release, convincing evidence has never been provided (11, 38). In the earlier reports, some experimental conditions used to analyze the role of LytA (e.g., culture transfer after Dp-1 infection to medium containing ethanolamine instead of choline) inhibit autolysin activity, as indicated by the authors (38), but also inhibit the activity of phage lysins, which depend on choline for proper function in the majority of phages, including Dp-1. Thus, the inhibition of phage release could be attributed to inhibition of the Dp-1 endolysin activity. On the other hand, LytA was essential for lysis of strain R6 at a low multiplicity of infection (<1) of the virulent phage Dp-1, since no lytic phenotype was observed in the derived lytA-deficient strain, regardless of an evident role of the endolysin at a high multiplicity of infection (>1) in the strain lacking LytA (11, 38). Given those inconsistencies, we set out to clarify the role of the bacterial autolysin in host lysis and release of newly assembled phage particles, using SV1, a lysogenic pneumococcal phage that carries a typical lytic cassette encoding putative holin (Svh) and lysin (Svl) activities.
The data presented here reveal unambiguously that LytA is activated during pneumococcal-phage-mediated lysis. In the absence of endolysin, this extremely powerful autolysin is able to mediate extensive host lysis that actually results in the release of a large number of fully functional phage capable of infecting other hosts. Thus, pneumococcal phages are able to use the bacterial autolysin LytA to exit from the host cell, completing their life cycle, in contrast to all other phages relying on a “holin-lysin” system. In the overwhelming majority of phages studied so far, mutants in the genes encoding endolysins are absolutely incapable of host lysis, trapping the phage progeny within the host cell (53). In T7 and T4 E. coli phages, artificial deletion of the endolysin did not prevent host lysis (15). However, in these unusual cases, the phages evolved to use another protein with muralytic activity encoded in their genomes, whose native function is to assist in the initial stages of infection to allow entry of the phage genome into the host cytoplasm (15). Dependence on lytic factors of cellular origin to disrupt the infected cell was indeed demonstrated only for phage PM2 of Pseudoalteromonas (19). Still, this phage does not encode an endolysin in its genome to autonomously achieve bacterial-host lysis and uses a novel system different from the typical “holin-lysin” strategy for progeny release (19).
If LytA is activated upon phage induction, leading to productive lysis in the absence of phage endolysins, what part does LytA play in the overall process under physiological conditions? From the lytic phenotypes, it seems that LytA activation does not contribute significantly to endolysin-mediated lysis (except perhaps for earlier lysis in the R36A bacterial background). Given these observations, we hypothesized that LytA activation is not crucial to accomplish efficient phage progeny release and is merely a side effect of the induction of the phage lytic system. However, the results from the phage plaque assays point to a different and more complex scenario. Phage release achieved by Svl endolysin is maximized by LytA, since the number of phage particles released is diminished in the absence of bacterial autolysin. This was observed both in lysogenic SVMC28 and lysogenized R36A strains. Interestingly, in the R36A genetic background, the absence of LytA (strain R36AP ΔlytA) also resulted in delayed lysis relative to that of the strain carrying both the phage and bacterial lysins. In conclusion, LytA activation after phage induction is not merely an inconsequential parallel process but seems to be essential for efficient phage progeny release.
Although LytA acts cooperatively with phage lysin to optimize phage progeny release, we observed that dependence solely on LytA might result in impaired phage fitness. In fact, host autolysin-induced lytic phenotypes showed a delay in lysis timing and a reduction in the proportion of total lysis. This corresponded to a delay in the release of phage particles and a significant reduction in the overall phage yield relative to what happens in the presence of both autolysin and endolysin. While retained in host cells, fully assembled phages lose the opportunity to infect naïve hosts, with a detrimental effect on phage propagation. Indeed, previous reports have shown an intimate relationship between lysis timing and phage fitness (15, 49). In addition, as the holins permeabilize the cells before lysis occurs, the LytA-induced delay in lysis traps new phages inside an already dead cell without biosynthetic capacity and thus incapable of further particle assembly. This LytA-mediated suboptimal phage release provides a provocative explanation for the crucial role of endolysins.
Another important conclusion from our data is that holin-induced lesions of the membrane not only activate phage endolysin, but also result in LytA activation. Thus, relying simply on holin function, phages elegantly accomplish the activation of the entire lytic arsenal at their disposal. Although holin-induced LytA activation could somehow be predictable, this is a significant finding, as dissipation of the membrane proton motive force does not always trigger the autolysins. Indeed, in B. subtilis, the major autolysin does not respond to proton motive force dissipation factors, despite other such enzymes being responsible for cell lysis following death from energy poisons (2, 25, 26). Thus, our data raise the possibility that the energy status of the membrane is important in LytA regulation. However, the underlying signaling mechanism induced by depolarization to trigger the activity of LytA is not understood. Cell depolarization may induce structural and spatial changes in the membrane (16, 18, 20) leading to LytA activation, probably by altering the inhibitory interactions between LytA and cell wall components, such as lipoteichoic acids (3, 7). It must be emphasized that LytA activity is not always indicative of bacterial lysis, since the enzyme has been implicated in other physiological processes, such as peptidoglycan synthesis and turnover and daughter cell separation (41, 47). Given its potentially lethal activity, however, LytA is tightly regulated to ensure the maintenance of cell integrity. We may therefore speculate that these physiological functions of LytA involve small and controlled local changes in membrane architecture activating LytA in a controllable fashion. In contrast, extensive depolarization, such as that imposed by holins, with major changes in the cell membrane and consequently in the cell wall architecture, may lead to massive and uncontrolled LytA activation, resulting in cell lysis. In fact, LytA-induced lysis upon the addition of β-lactams is related to the inhibition of peptidoglycan synthesis (48), which could also induce major changes in cell wall structure.
Since LytA resides in the cell wall, our observation that LytA is activated by holin lesions leads us to speculate on an alternative regulatory mechanism of phage endolysins. Pneumococcal-phage lysins are structurally and functionally similar to the bacterial cell wall hydrolase LytA (10, 23, 34, 36). Indeed, the constitutive expression of the pneumococcal-phage lysin HBL-3 or CPL-1 in S. pneumoniae M31, a mutant with the lytA gene deleted, restored the ability of the strain to undergo lysis in stationary phase and after exposure to DOC, two cellular responses that are dependent upon LytA activity (35). In spite of the absence of a canonical N-terminal sequence signal, LytA is translocated across the cytoplasmic membrane (7). Thus, pneumococcal-phage lysins could also be transported by the same unknown pathway that targets LytA to the cell wall and could be subjected to the same type of physiological control. Although the canonical model of “holin-lysin” systems indicates that holins provide access of cytoplasmic lysins to the cell wall through holes generated in the plasma membrane, in pneumococci, holins could function simply to activate these secreted endolysins through membrane depolarization, similarly to LytA activation, rather than allowing their egress.
Taken together, our data provide the first evidence of the involvement of bacterial lysins in the progeny release of endolysin-equipped phages. Pneumococcal-phage dependency on the host autolysin for optimal progeny release underscores the complex relationship between lysogenic phages and their bacterial hosts.
Acknowledgments
We thank Margarida Carrolo for her helpful assistance during the fluorescence microscopy assays, Elisabete Martins for support in the construction of the mutant strains, Teresa Figueiredo for providing the SV1 genome sequence data, and A. Tomasz and S. Filipe for providing strains.
M.J.F. was supported by grant SFRH/BD/38543/2007 from the Fundação para a Ciência e a Tecnologia, Portugal. This work was partly supported by Fundação para a Ciência e a Tecnologia (POCI/1999/BME/34418), Portugal.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith. 1999. Current protocols in molecular biology. Wiley-Interscience, New York, NY.
- 2.Blackman, S. A., T. J. Smith, and S. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 14657-64. [DOI] [PubMed] [Google Scholar]
- 3.Briese, T., and R. Hakenbeck. 1985. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur. J. Biochem. 146417-427. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64155-164. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164123-128. [DOI] [PubMed] [Google Scholar]
- 6.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 1844988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz, E., E. Garcia, C. Ascaso, E. Mendez, R. Lopez, and J. L. Garcia. 1989. Subcellular localization of the major pneumococcal autolysin: a peculiar mechanism of secretion in Escherichia coli. J. Biol. Chem. 2641238-1244. [PubMed] [Google Scholar]
- 8.Diaz, E., M. Munthali, H. Lunsdorf, J. V. Holtje, and K. N. Timmis. 1996. The two-step lysis system of pneumococcal bacteriophage EJ-1 is functional in gram-negative bacteria: triggering of the major pneumococcal autolysin in Escherichia coli. Mol. Microbiol. 19667-681. [DOI] [PubMed] [Google Scholar]
- 9.Filipe, S. R., E. Severina, and A. Tomasz. 2001. Functional analysis of Streptococcus pneumoniae MurM reveals the region responsible for its specificity in the synthesis of branched cell wall peptides. J. Biol. Chem. 27639618-39628. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, P., J. L. Garcia, E. Garcia, J. M. Sanchez-Puelles, and R. Lopez. 1990. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene 8681-88. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, P., R. Lopez, C. Ronda, E. Garcia, and A. Tomasz. 1983. Mechanism of phage-induced lysis in pneumococci. J. Gen. Microbiol. 129479-487. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, P., M. Paz Gonzalez, E. Garcia, J. L. Garcia, and R. Lopez. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33128-138. [DOI] [PubMed] [Google Scholar]
- 13.Grundling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 989348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundling, A., D. L. Smith, U. Blasi, and R. Young. 2000. Dimerization between the holin and holin inhibitor of phage lambda. J. Bacteriol. 1826075-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heineman, R. H., I. J. Molineux, and J. J. Bull. 2005. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J. Mol. Evol. 61181-191. [DOI] [PubMed] [Google Scholar]
- 16.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25753-763. [DOI] [PubMed] [Google Scholar]
- 17.Kemper, M. A., M. M. Urrutia, T. J. Beveridge, A. K. Koch, and R. J. Doyle. 1993. Proton motive force may regulate cell wall-associated enzymes of Bacillus subtilis. J. Bacteriol. 1755690-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komor, E., H. Weber, and W. Tanner. 1979. Greatly decrease susceptibility of nonmetabolizing cells towards detergents. Proc. Natl. Acad. Sci. USA 761814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupovic, M., R. Daugelavicius, and D. H. Bamford. 2007. A novel lysis system in PM2, a lipid-containing marine double-stranded DNA bacteriophage. Mol. Microbiol. 641635-1648. [DOI] [PubMed] [Google Scholar]
- 20.Labedan, B., and E. B. Goldberg. 1979. Requirement of membrane potential in injection of phage T4 DNA. Proc. Natl. Acad. Sci. USA 764669-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim. Byophys. Acta 39508-517. [DOI] [PubMed] [Google Scholar]
- 22.Loeffler, J. M., and V. A. Fischetti. 2006. Lysogeny of Streptococcus pneumoniae with MM1 phage: improved adherence and other phenotypic changes. Infect. Immun. 744486-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez, R., E. Garcia, P. Garcia, and J. L. Garcia. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3199-211. [DOI] [PubMed] [Google Scholar]
- 24.Marchese, A., M. Ramirez, G. C. Schito, and A. Tomasz. 1998. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J. Clin. Microbiol. 362944-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margot, P., C. Mauel, and D. Karamata. 1994. The gene of the N-acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase, is not involved in vegetative cell autolysis. Mol. Microbiol. 12535-545. [DOI] [PubMed] [Google Scholar]
- 26.Margot, P., M. Whalen, A. Gholamhoseinian, P. Piggot, and D. Karamata. 1998. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J. Bacteriol. 180749-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3208-218. [Google Scholar]
- 28.Martin, A. C., R. Lopez, and P. Garcia. 1998. Functional analysis of the two-gene lysis system of the pneumococcal phage Cp-1 in homologous and heterologous host cells. J. Bacteriol. 180210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Cuesta, M. C., J. Kok, E. Herranz, C. Pelaez, T. Raquena, and G. Buist. 2000. Requirement of autolytic activity for bacteriocin-induced lysis. Appl. Environ. Microbiol. 663174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obregon, V., J. L. Garcia, E. Garcia, R. Lopez, and P. Garcia. 2003. Genome organization and molecular analysis of the temperate bacteriophage MM1 of Streptococcus pneumoniae. J. Bacteriol. 1852362-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otsuji, N., M. Sekiguchi, T. Iijima, and Y. Takagi. 1959. Induction of phage formation in the lysogenic Escherichia coli K-12 by mitomycin C. Nature 184(Suppl. 14)1079-1080. [DOI] [PubMed] [Google Scholar]
- 33.Pozzi, G., M. R. Oggioni, and A. Tomasz. 1989. DNA probe for identification of Streptococcus pneumoniae. J. Clin. Microbiol. 27370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez, M., E. Severina, and A. Tomasz. 1999. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J. Bacteriol. 1813618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero, A., R. Lopez, and P. Garcia. 1993. Lytic action of cloned pneumococcal phage lysis genes in Streptococcus pneumoniae. FEMS Microbiol. Lett. 10887-92. [DOI] [PubMed] [Google Scholar]
- 36.Romero, A., R. Lopez, and P. Garcia. 1990. Sequence of the Streptococcus pneumoniae bacteriophage HB-3 amidase reveals high homology with the major host autolysin. J. Bacteriol. 1725064-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero, P., R. Lopez, and E. Garcia. 2004. Genomic organization and molecular analysis of the inducible prophage EJ-1, a mosaic myovirus from an atypical pneumococcus. Virology 322239-252. [DOI] [PubMed] [Google Scholar]
- 38.Ronda-Lain, C., R. Lopez, A. Tapia, and A. Tomasz. 1977. Role of the pneumococcal autolysin (murein hydrolase) in the release of progeny bacteriophage and in the bacteriophage-induced lysis of the host cells. J. Virol. 21366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, G. L., and A. D. Rutenberg. 2007. Clocking out: modeling phage-iduced lysis of Escherichia coli. J. Bacteriol. 1894749-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sanchez-Puelles, J. M., C. Ronda, J. L. Garcia, P. Garcia, R. Lopez, and E. Garcia. 1986. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158289-293. [DOI] [PubMed] [Google Scholar]
- 42.São-José, C., R. Parreira, G. Vieira, and M. A. Santos. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 1825823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savva, C. G., J. S. Dewey, J. Deaton, R. L. White, D. K. Struck, A. Holzenburg, and R. Young. 2008. The holin of bacteriophage lambda forms rings with large diameter. Mol. Microbiol. 69784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severina, E., M. Ramirez, and A. Tomasz. 1999. Prophage carriage as a molecular epidemiological marker in Streptococcus pneumoniae. J. Clin. Microbiol. 373308-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Severina, E., A. Severin, and A. Tomasz. 1998. Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J. Antimicrob. Chemother. 41341-347. [DOI] [PubMed] [Google Scholar]
- 46.Su, M. T., T. V. Venkatesh, and R. Bodmer. 1998. Large- and small-scale preparation of bacteriophage lambda lysate and DNA. BioTechniques 2544-46. [DOI] [PubMed] [Google Scholar]
- 47.Tomasz, A., P. Moreillon, and G. Pozzi. 1988. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J. Bacteriol. 1705931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasz, A., and S. Waks. 1975. Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc. Natl. Acad. Sci. USA 724162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, I.-N. 2006. Lysis timing and bacteriophage fitness. Genetics 17217-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, M., A. Arulandu, D. K. Struck, S. Swanson, J. C. Sacchettini, and R. Young. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307113-117. [DOI] [PubMed] [Google Scholar]
- 51.Xu, M., D. K. Struck, J. Deaton, I. N. Wang, and R. Young. 2004. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. USA 1016415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young, I., I. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8120-128. [DOI] [PubMed] [Google Scholar]
- 53.Young, R. 2005. Phage lysis, p. 92-127. In M. K. Waldor, D. I. Friedman, and S. L. Adhya (ed.), Phages, their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 54.Young, R., and U. Blasi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17191-205. [DOI] [PubMed] [Google Scholar]