Abstract
Streptococcus gordonii and Veillonella atypica, two early-colonizing members of the dental plaque biofilm, participate in a relationship that results in increased transcription of the S. gordonii gene amyB, encoding an α-amylase. We show that the transcription factor CcpA is required for this interspecies interaction.
Human dental plaque is a well-recognized example of a multispecies bacterial community and a site of interspecies cell-cell interactions. These interactions include physical contacts mediated by molecules on the surfaces of bacterial cells, known as coaggregation, as well as metabolic interactions that have an effect on growth and gene expression (8, 9). Two early-colonizing oral bacteria, Streptococcus gordonii and Veillonella atypica, undergo such interactions. Fermentation of sugar by S. gordonii produces lactic acid, a substrate for energy generation by V. atypica. During coculture growth of S. gordonii and V. atypica, a signaling event that results in increased transcription of the S. gordonii gene amyB, encoding an α-amylase, occurs (4). We have proposed that AmyB functions intracellularly to hydrolyze stored glycogen, resulting in the availability of sugar for S. gordonii and, upon fermentation, lactic acid for V. atypica (4). Consequently, V. atypica-induced expression of the S. gordonii amylase may contribute to cross-feeding between these organisms and may be advantageous to one or both species involved in the symbiosis.
We have previously shown that the signaling between V. atypica and S. gordonii involves a diffusible signal that has not been identified (4). In other systems, signaling between species of oral bacteria includes the use of autoinducer-2 (8, 9, 14). However, we have been unable to detect autoinducer-2 in V. atypica culture supernatants (4). Here we begin to investigate the mechanism by which V. atypica causes changes in gene expression in S. gordonii by examining the role of the S. gordonii transcription factor CcpA in V. atypica-induced amylase expression.
The S. gordonii amylase encoded by amyB was initially identified as a gene that is induced by coculture of S. gordonii with V. atypica (4). To gain an understanding of the mechanism by which amyB transcription is regulated by V. atypica, we sought to identify factors in S. gordonii that control amyB expression. The amyB promoter region contains a 7-bp inverted repeat matching the catabolite response element consensus (5) at 11 of 14 positions (4). In other systems, the catabolite response element consensus site is bound by a member of the CcpA family of transcriptional regulators (5). The S. gordonii CcpA homolog, previously called RegG, has been previously described (1, 2, 15).
Mutagenesis of S. gordonii V288 ccpA.
To see if CcpA also plays a role in the expression of AmyB during symbiosis with V. atypica, we constructed a ccpA mutant of S. gordonii strain V288 using PCR ligation mutagenesis (10). Two segments of ccpA (GenBank accession no. NC_009785), corresponding to bases −9 to 483 and 515 to 964 relative to the ATG start codon, were PCR amplified from S. gordonii V288 genomic DNA using primers 5′-ccpA (5′-GGAAACAATATGAACACAGACG-3′) and ccpA-XhoI (5′-CGCTCGAGTAACACTTGGAAGTTGGTGC-3′) (for the segment from bases −9 to 483) and primers EcoRI-ccpA (5′-GGAATTCCTATCACATTCCTTGCTAAG-3′) and 3′-ccpA (5′-GAACAACTTCACGTTCTTCC-3′) (for the segment from bases 515 to 964), where underlining indicates the restriction enzyme sites. The kanamycin resistance gene aphAIII was PCR amplified from plasmid pDL276 (3) using primers XhoI-aphAIII (5′-CGCTCGAGTGTGGTTTCAAAATCGGCTC-3′) and aphAIII-EcoRI (5′-GGAATTCCATCTAAATCTAGGTACTAAAAC-3′). The PCR products were digested with EcoRI and XhoI and ligated together. The product of the ligation was used as the template for a PCR with the 5′-most primer, 5′-ccpA, and the 3′-most primer, 3′-ccpA. The resulting PCR product was purified and transformed into S. gordonii V288 (11). The transformation mixture was plated on Todd-Hewitt agar (Becton Dickinson, Sparks, MD) containing 500 μg/ml kanamycin to select for isolates that had undergone double recombination. PCR analysis of genomic DNA prepared from one isolate, designated AC001, was performed, and the insertion of the kanamycin resistance gene in ccpA was verified.
To verify that insertion of the kanamycin resistance gene in ccpA did not result in polar effects on the gene downstream of ccpA, reverse transcriptase PCR was performed on the downstream open reading frame (GenBank accession no. NC_009785). RNA was extracted from wild-type and ccpA mutant cells grown under the test conditions using the Qiagen RNeasy minikit (Valencia, CA). The RNA was treated with DNase I from New England Biolabs (Ipswich, MA) and used as a template for reverse transcriptase PCR with primers 5′GT (5′-CCTCTTACATGCCGCCAAGAG-3′) and 3′GT (5′-CAATCTGGCATCCTTCTAGCC-3′). DNA fragments of the expected size, 390 bp, were amplified from cDNA of both the wild-type and ccpA mutant strains. Controls in which the RNA templates were used for conventional PCR with no reverse transcriptase did not yield PCR products (data not shown).
The growth rates of the wild type and ccpA mutant in tryptone-yeast extract medium (TYE; 1% tryptone, 0.5% yeast extract, 0.3% K2HPO4) containing either 0.2% or 2.0% glucose or maltose were tested. Under all four conditions tested, the wild type and ccpA mutant had doubling times averaging 50 to 60 min, with no statistical difference between results for the two strains.
CcpA is required for V. atypica-induced expression of S. gordonii amyB.
To examine the possibility that CcpA is involved in the ability of S. gordonii to regulate amyB expression in response to V. atypica, we measured the amylase activities of the wild-type and ccpA strains grown as monocultures and as cocultures with V. atypica PK1885 (7) in medium containing 0.2% glucose. Amylase activity was measured using variations on a previously described method (12). Cell extracts were prepared from 1.0 ml of logarithmic cultures or cocultures. Cells were harvested by centrifugation, washed and resuspended in 50 mM Tris-HCl (pH 6.8), and lysed by bead beating with 0.1-mm glass beads. To compare the amylase activity in cell extracts of S. gordonii cells grown in coculture with V. atypica to those of cells grown as monocultures, it was necessary to express amylase activity relative to the number of S. gordonii cells rather than the amount of protein in cell extracts. Therefore, to normalize the amylase activity to the number of S. gordonii cells in the culture, samples were taken prior to harvesting, serially diluted, and plated to determine the number of S. gordonii cells per sample. Wild-type S. gordonii grown in the presence of V. atypica induced amylase activity threefold over the level in S. gordonii cells grown alone (P = 0.007) (Fig. 1). However, amylase activity in the ccpA mutant remained at basal levels and was unaffected by growth in coculture with V. atypica (Fig. 1).
FIG. 1.
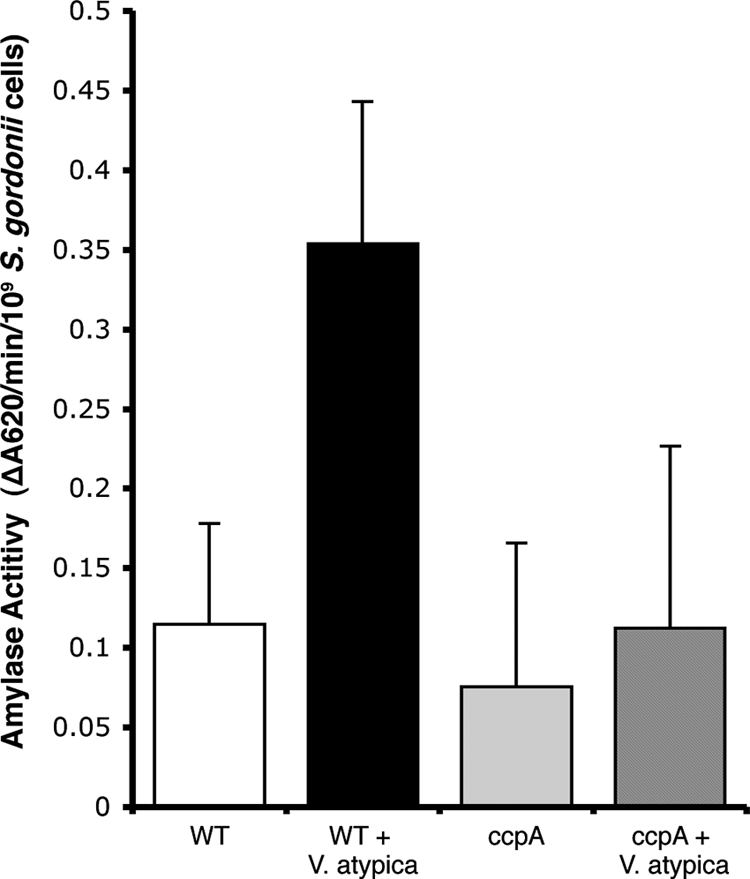
Amylase activities from wild-type (WT) and ccpA mutant S. gordonii cells grown alone and in coculture with V. atypica. To normalize the activities between monocultures and cocultures, amylase activity is expressed as ΔA620/min/109 S. gordonii cells. The results presented are the averages from four independent trials. Error bars represent standard deviations.
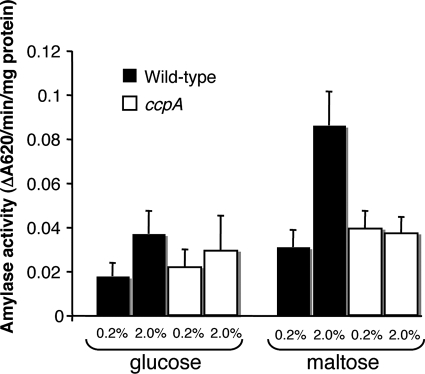
Maltose induces CcpA-dependent expression of amyB, independently of coculture with V. atypica.
Characterization of amyB homologs from other streptococci has shown induction of enzyme activity by growth in the presence of various carbohydrates (16). The possible role for CcpA in the expression of amyB in response to carbohydrates was tested by measuring the amylase activities of both wild-type and ccpA mutant S. gordonii cell extracts grown in TYE with glucose or maltose. When grown on either 0.2% or 2.0% glucose, the amylase activities of the wild type and mutant were very similar (Fig. 2). In contrast, amylase activity from wild-type S. gordonii grown as a monoculture was induced nearly threefold by the growth with 2% maltose (Fig. 2), mirroring the threefold induction by coculture with V. atypica. This induction in response to high concentrations of maltose was absent in the ccpA mutant. This result suggests that induction of amyB by coculture with V. atypica and by maltose may involve a common regulatory pathway.
FIG. 2.
Amylase activities from wild-type and ccpA mutant S. gordonii cells grown with either 0.2% or 2% maltose or glucose. The results presented are the averages from four independent trials. Error bars represent standard deviations.
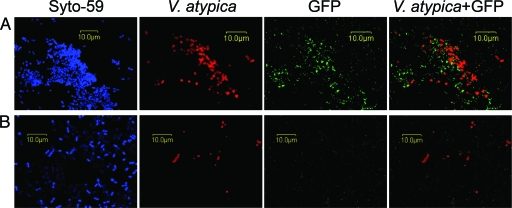
CcpA is required for transcription of amyB in coculture biofilms with V. atypica.
The possibility that CcpA is required for increased transcription from the amyB promoter during the growth of S. gordonii and V. atypica PK1910 (7) in a mixed-species biofilm community was investigated using techniques for biofilm growth and imaging described previously (4, 13). A plasmid containing the amyB promoter region fused to a promoterless gfp, pPamy-′gfp (4), was used to assess transcription from the amyB promoter in the wild-type and ccpA mutant S. gordonii strains. Coculture biofilms of either wild-type or ccpA mutant S. gordonii containing pPamy-′gfp and V. atypica were grown for 4 hours with saliva as the sole nutrient source. Biofilms were stained for all cells present with the DNA stain Syto-59 (Invitrogen, Carlsbad, CA) and for V. atypica cells by primary immunofluorescence with Alexa Fluor 546-conjugated anti-V. atypica antibodies (6). While wild-type S. gordonii cells expressed green fluorescent protein (GFP) when growing in mixed-species microcolonies with V. atypica, the ccpA mutant did not (Fig. 3A and B). Neither the wild type nor the ccpA mutant induced amyB expression in the absence of V. atypica (not shown). These results further demonstrate that pathways involving CcpA are required for signaling between these organisms in mixed-species biofilms.
FIG. 3.
Confocal scanning laser microscopy analysis of dual-species biofilms of the wild type (A) and ccpA mutant (B) with V. atypica. The four images in each row are maximum projections of a single field of view showing fluorescence from Syto-59 (blue; all cells), Alexa Fluor 546-conjugated anti-V. atypica antibodies (red), and GFP (green; S. gordonii expressing amyB). The three fluorescence channels are shown separately in the left three panels and, in the right panel, as an overlay of GFP with V. atypica.
In this study, CcpA was found to be required for induction of the S. gordonii amylase gene, amyB, in response to growth with V. atypica. The fact that induction can occur based on the presence of maltose, as well as by coculture with V. atypica, suggests that the signal supplied by V. atypica could be maltose or a related sugar from the lipopolysaccharide in the V. atypica outer membrane. Alternatively, independent signaling pathways may converge, with CcpA responding to an unrelated signal from V. atypica. This information provides an understanding of the requirements for the transcription of amyB and the signaling pathways that are involved. Further analysis of the activators of CcpA activity, the signal derived from V. atypica and protein machinery in S. gordonii, will be required to understand the mechanisms for changes in gene expression that result from this symbiosis.
Acknowledgments
This work was supported by NIH grant 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources.
We thank P. E. Kolenbrander for generously supplying the anti-V. atypica antibodies. We acknowledge the Western South Dakota DNA Core Facility (WestCore) at Black Hills State University for sequencing of PCR products. We thank Peg Ustad for her expert assistance with formatting the confocal images. We thank Tomoko Ise for expert technical assistance.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Bizzini, A., J. M. Entenza, and P. Moreillon. 2007. Loss of penicillin tolerance by inactivating the carbon catabolite repression determinant CcpA in Streptococcus gordonii. J. Antimicrob. Chemother. 59607-615. [DOI] [PubMed] [Google Scholar]
- 2.Dong, Y., Y.-Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 1862511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 571194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 10116917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res. Microbiol. 145503-518. [DOI] [PubMed] [Google Scholar]
- 6.Hughes, C. V., R. N. Andersen, and P. E. Kolenbrander. 1992. Characterization of Veillonella atypica PK1910 adhesin-mediated coaggregation with oral Streptococcus spp. Infect. Immun. 601178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes, C. V., C. A. Roseberry, and P. E. Kolenbrander. 1990. Isolation and characterization of coaggregation-defective mutants of Veillonella atypica. Arch. Oral Biol. 35123S-125S. [DOI] [PubMed] [Google Scholar]
- 8.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49193-205. [DOI] [PubMed] [Google Scholar]
- 11.Macrina, F. L., P. H. Wood, and K. R. Jones. 1980. Genetic transformation of Streptococcus sanguis (Challis) with cryptic plasmids from Streptococcus ferus. Infect. Immun. 28692-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson, W. L., and G. H. Chambliss. 1985. Isolation and characterization of a cis-acting mutation conferring catabolite repression resistance to α-amylase synthesis in Bacillus subtilis. J. Bacteriol. 161875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer, R. J., Jr., K. M. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 695794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 601446-1456. [DOI] [PubMed] [Google Scholar]
- 15.Rogers, J. D., and F. A. Scannapieco. 2001. RegG, a CcpA homolog, participates in regulation of amylase-binding protein A gene (abpA) expression in Streptococcus gordonii. J. Bacteriol. 1833521-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson, C. L., and R. R. B. Russell. 1998. Intracellular α-amylase of Streptococcus mutans. J. Bacteriol. 1804711-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]