Abstract
The acquisition of β-lactamases in members of the Enterobacteriaceae family poses a challenge to antimicrobial susceptibility testing in the clinical laboratory. We correlated the distribution of the MICs for Klebsiella spp. and Escherichia coli with the presence of extended-spectrum β-lactamase (ESBL) and plasmid-mediated AmpC β-lactamase (pAmpC) genes. A total of 264 isolates were subjected to cefazolin, ceftriaxone, cefotaxime, ceftazidime, cefepime, and aztreonam agar dilution MIC determination; ESBL screening and confirmatory testing by the methods of the Clinical and Laboratory Standards Institute (CLSI); and for isolates for which the MICs of extended-spectrum cephalosporins were ≥1 μg/ml or the MICs of cefpodoxime were >4 μg/ml, PCR amplification and sequencing of the ESBL and pAmpC genes. PCR was positive for 73/81 isolates (45 isolates with an ESBL gene alone, 24 isolates with a pAmpC gene alone, with 4 isolates with both genes). Compared to PCR, confirmatory testing by the CLSI method yielded a sensitivity and a specificity of 98.0 and 96.3%, respectively; there were six false-positive results and one false-negative result. No distinction in the MIC distribution was apparent between isolates with the ESBL gene and isolates with the pAmpC gene. A substantial percentage of the isolates with PCR-confirmed ESBL and/or pAmpC genes fell within the current CLSI susceptible category. For a ceftazidime, ceftriaxone, or cefotaxime MIC of ≥2 μg/ml, a dichotomy existed between isolates with and without ESBL and pAmpC genes in most cases. This suggests that the presence of the ESBL and the pAmpC enzymes may yield similar MICs of extended-spectrum cephalosporins, many of which fall within the current nonresistant categories. Lowering of the current CLSI breakpoints for cephalosporins appears to be warranted.
Resistance to cephalosporins in members of the Enterobacteriaceae family is of concern, as the prevalence of horizontally acquired resistance mechanisms has increased significantly in the past 20 years. Isolates of the Enterobacteriaceae family have historically recruited narrow-spectrum Ambler class A β-lactamases of the TEM and SHV lineages, which were associated with nosocomial outbreaks in the 1980s (24). Continued evolution under strong selective pressure led to a broadening of the hydrolytic spectrum, as exemplified by the global spread of TEM and SHV derivatives (10). More recently, CTX-M class A enzymes have appeared globally and are of particular concern in Europe (14). In the United States, ESBL production occurred in 2 to 3% of clinical Escherichia coli isolates associated with intra-abdominal infections between 2002 and 2005 (2), and the prevalence of Klebsiella pneumoniae isolates that produce ESBLs has shown a slow increase, from 6% in 2002 to 8% in 2005 (2, 5, 19, 27). During the same period, a worldwide increase in the prevalence of ESBL producers among E. coli and K. pneumoniae isolates was observed: from 7 to 12% and from 13 to 18%, respectively (2, 5, 19, 27). In addition, chromosomal Ambler class C AmpC genes have been mobilized and are now being disseminated on plasmids, reminiscent of the early dissemination and evolution of ESBLs (21). Increasingly, reports document the detection of plasmid-mediated AmpC β-lactamase (pAmpC) resistance in the Enterobacteriaceae (7, 9, 22). Data from the SENTRY Antimicrobial Surveillance Program for North America show that 19 of 65 ESBL screen-positive E. coli isolates harbored pAmpC (8). The ESBL and pAmpC enzymes now constitute a diversified threat to the activities of the penicillins, cephalosporins, monobactams, and carbapenems. ESBL- and pAmpC-producing strains are no longer confined to the hospital environment, from which they were first reported, but of late, they are being isolated from the community at increasing frequencies (22, 23). These newer strains, which harbor multiple resistance mechanisms, warrant continued surveillance studies, as well as a reappraisal of laboratory detection strategies.
An initial concern over ESBLs was that of possible cross-resistance to drugs of the same class. This resulted in the issuance of recommendations in the 1990s that expressed the need for caution, with guidelines suggesting that ESBLs may show cross-resistance to all penicillins, cephalosporins, and monobactams (12, 13, 18). Nevertheless, studies have shown that cephalosporins displaying lower MICs may in fact still prove to be clinically useful (4, 24, 25, 29).
The emergence of novel enzymes with varied and specific hydrolysis spectra is placing increased pressure on clinical laboratories to accurately and efficiently screen for and identify those isolates suspected of harboring ESBL genes, with previous studies calling for a reevaluation of breakpoints (1, 11, 15). The Clinical and Laboratory Standards Institute (CLSI) recommends screening of E. coli, K. pneumoniae, Klebsiella oxytoca, and Proteus mirabilis isolates for ESBL production by the use of cefpodoxime, ceftazidime, aztreonam, cefotaxime, or ceftriaxone, followed by phenotypic confirmation with clavulanate (6). However, multiple resistance mechanisms may obscure the ability to detect ESBLs, as the new classes of non-ESBL enzymes that are emerging may result in resistance patterns that overlap those caused by the ESBLs, leading to the failure of conventional clavulanate-based ESBL detection. In addition, commercial susceptibility testing systems may be challenged by the coexistence of multiple resistance determinants (3, 26). No recommendations are available on methods for the screening and confirmation of the presence of other prevalent β-lactamases, in particular, pAmpC genes, the MICs of which often overlap those of ESBLs. Finally, an alternative to specific screening for ESBLs is to reexamine the appropriate MIC breakpoints for cephalosporins.
The aim of the study described here was to characterize a large collection of contemporary strains of E. coli and Klebsiella species to correlate the MIC distribution with the molecular characteristics of the acquired secondary β-lactamases.
(This work was presented in part at the 109th General Meeting of the American Society for Microbiology, Philadelphia, PA, 17 to 21 May 2009.)
MATERIALS AND METHODS
Bacterial strains.
Two hundred sixty-four isolates were investigated in the present study, including 199 consecutive clinical isolates of E. coli (n = 180), K. pneumoniae (n = 8), and K. oxytoca (n = 11) received at or isolated by the Clinical Microbiology Laboratory, Mayo Clinic Rochester, as part of routine diagnostic activities. In addition, 25 E. coli isolates received as part of routine activities in 2008 were included on the basis of their reduced susceptibility to cefoxitin, suggestive of pAmpC production. Forty additional control strains comprising 12 ESBL-producing E. coli isolates, 27 ESBL-producing K. pneumoniae, and 1 ESBL-producing K. oxytoca isolate, graciously provided by Karen Bush (Johnson & Johnson Pharmaceutical Research & Development LLC, Raritan, NJ) or David Paterson (University of Pittsburgh), were included.
Antimicrobial susceptibility testing.
Agar dilution susceptibility testing was performed with cefazolin, ceftriaxone, cefotaxime, ceftazidime, cefepime, and aztreonam according to the recommendations of the CLSI (6). In addition, cefpodoxime at 4 μg/ml was used for screening for ESBL production. A cefpodoxime MIC of >4 μg/ml prompted confirmatory testing for ESBL production by the CLSI-recommended clavulanate disk augmentation test with disks (BD Diagnostics, Sparks, MD) impregnated with ceftazidime (30 μg) with and without clavulanate (10 μg) and cefotaxime (30 μg) with and without clavulanate (10 μg). An increase in the zone size of ≥5 mm for either of the cephalosporins in the presence of clavulanate was considered a positive result for ESBL expression, in accordance with the recommendations of the CLSI (6). Isolates for which the MIC of any extended-spectrum cephalosporin was ≥1 μg/ml or the cefpodoxime MIC was >4 μg/ml were subjected to ESBL and pAmpC gene detection by PCR.
DNA preparation.
Four to five single colonies from an overnight growth plate were inoculated into in-house-prepared tubes containing glass beads and SETS neutralization buffer (Roche Diagnostics, Mannheim, Germany). The tubes were heated at 99°C with shaking at 1,400 rpm for 6 min with an Eppendorf thermomixer (Hamburg, Germany). The lysate was centrifuged, and the pellet was resuspended in 500 μl distilled water for amplification.
Amplification and detection.
PCR amplification of the TEM, SHV, and CTX-M ESBL genes was performed as described previously (16, 17, 28). Multiplex amplification of the pAmpC genes was performed with primers targeting MOX-, CMY-, LAT-, DHA-, ACC-, MIR-, and FOX-type β-lactamases, as described by Perez-Perez and Hanson (20) (Table 1). Amplification was performed as described by Pai et al. (16), Park et al. (17), Saladin et al. (28), and Perez-Perez and Hanson (20) with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) in an ABI 9600 thermocycler (Applied Biosystems, Foster City, CA). The amplified DNA was detected by electrophoresis with a 2% E-gel 96 gel (Invitrogen), and the sizes of the bands were estimated by comparison to the bands on a 100-bp ladder (Invitrogen) (Table 1).
TABLE 1.
PCR primers employed
| Target | β-Lactamase(s) | Primers | Amplification product size (bp) | Reference |
|---|---|---|---|---|
| ESBL | TEM | blaTEMF, blaTEMR | 1,079 | 17 |
| SHV | blaSHVR, blaSHVR | 868 | ||
| CTX-M1 group | CTX-1F, CTX-1R | 864 | 28 | |
| CTX-M2 group | CTX-2F, CTX-2R | 866 | ||
| CTX-M9 group | CTX-9F, CTX-9R | 785 | 16 | |
| pAmpC | MOX-1, MOX-2, CMY-1, CMY-8 to CMY-11 | MOXMF, MOXMR | 520 | 20 |
| LAT-1, CMY-2 to CMY-7 | CITMF, CITMR | 462 | ||
| DHA-1, DHA-2 | DHAMF, DHAMR | 405 | ||
| ACC | ACCMR, ACCMF | 346 | ||
| MIR-1T, ACT-1 | EBCMF, EBCMR | 302 | ||
| FOX-1 to FOX-5b | FOXMF, FOXMR | 190 |
DNA sequencing and analysis.
The amplification reaction mixtures were prepared for sequencing by removal of the remaining primers and deoxynucleoside triphosphates by digestion with exonuclease I and shrimp alkaline phosphatase (ExoSap-IT; USB Corporation, Cleveland, OH), according to the manufacturer's recommendations. The purified amplified DNA was subjected to DNA sequencing with an ABI Prism BigDye Terminator (version 1.1) cycle sequencing ready reaction kit with AmpliTaq DNA polymerase and was analyzed with ABI Prism 3730 XL DNA analyzers (Perkin-Elmer Applied Biosystems, Foster City, CA) in the Molecular Biology Core Facility, Mayo Clinic Rochester. TEM and SHV DNA sequence data were compared to the sequence data for known types (from the reference database of ESBL type-specific amino acid mutations; G. Jacoby and K. Bush, Lahey Clinic; accessible through http://www.lahey.org/studies/) by employing the online ESBL Genotyping Tool hosted by the Institute of Infection Medicine, University Medical Center Schleswig-Holstein Campus, Kiel, Germany (accessible through http://www.informatik.uni-kiel.de/∼amino/). The CTX-M and pAmpC gene sequences were compared to published sequences by use of the NCBI Basic Local Alignment Search Tool (BLAST).
RESULTS
ESBL screening and molecular detection.
The cefpodoxime MIC was >4 μg/ml or the MIC of an extended-spectrum cephalosporin was ≥1 μg/ml for 81 of the 264 isolates. PCR amplification of these 81 isolates yielded positive results for an ESBL gene alone for 45 isolates, a pAmpC gene alone for 24 isolates, and both ESBL and pAmpC genes for 4 isolates. The ESBL genes detected included 19 TEM genes (other than TEM-1 and TEM-2), 22 SHV genes (other than SHV-1), and 19 CTX-M genes. The TEM genes detected were those for TEM-1 (n = 25), TEM-3 (n = 1), TEM-6 (n = 3), TEM-10 (n = 3), TEM-12 (n = 1), TEM-26 (n = 5), TEM-43 (n = 1), and TEM-104 (n = 1). Four TEM gene sequences were unidentifiable due to short sequence reads (although on the basis of the available sequence data, the possibility that they were TEM-1 and TEM-2 was excluded). The SHV genes detected were those for SHV-1 (n = 7), SHV-2 (n = 2), SHV-11 (n = 2), SHV-12 (n = 3), and SHV5/55 (n = 7). Ten SHV gene sequences were unidentifiable due to short sequence reads (although on the basis of the available sequence data, the possibility that they were SHV-1 was excluded). The CTX-M genes detected were those for CTX-M-1 (n = 3), CTX-M-9 (n = 3), CTX-M-15 (n = 2), CTX-M-24 (n = 2), and CTX-M-28 (n = 3). For six CTX-M-1-like sequences, further type assignment was not possible due to short sequence reads. The pAmpC genes detected were those for CMY-2 (n = 21), CMY-8 (n = 1), and FOX-5 (n = 2). Four pAmpC gene sequences were not sufficient for type assignment.
ESBL detection methods.
Screening and confirmatory testing for ESBL production yielded a sensitivity of 98.0% and a specificity of 96.3% compared to the results of PCR amplification (Table 2). There were six false-positive results by the disk method of detection of ESBL production; and these were for isolates containing TEM-1 and SHV-1, SHV-1, TEM-1 and FOX-5, TEM-1 and CMY-2, CMY-2, and no secondary β-lactamase, respectively. There was one false-negative result by the disk method, and that was for an E. coli isolate harboring TEM-104.
TABLE 2.
Comparison of results of phenotypic ESBL confirmatory testing and PCR for ESBL genesa
| PCR result | No. of strains with the following result by disk augmentation:
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 48 | 1 | 49 |
| Negative | 6 | 26 | 32 |
| Total | 54 | 27 | 81 |
Only ESBL screen-positive isolates were subjected to disk augmentation and PCR. Screening and confirmatory testing for ESBL production yielded a sensitivity of 98.0% and a specificity of 96.3%.
MIC distribution.
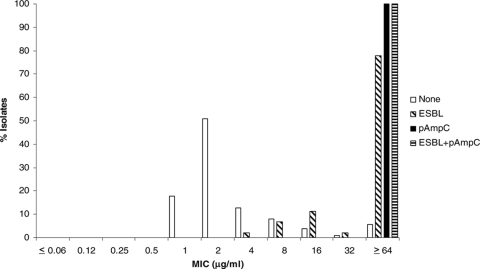
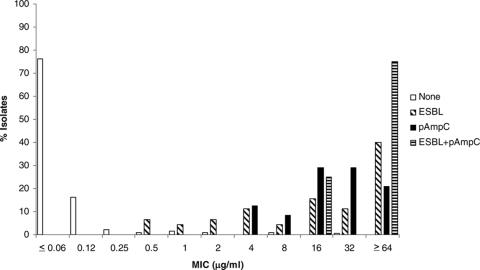
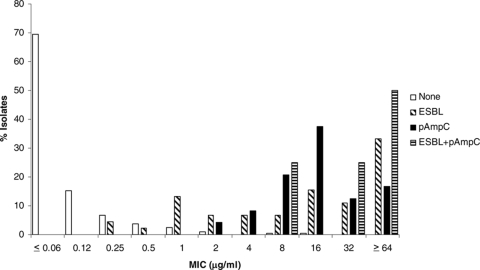
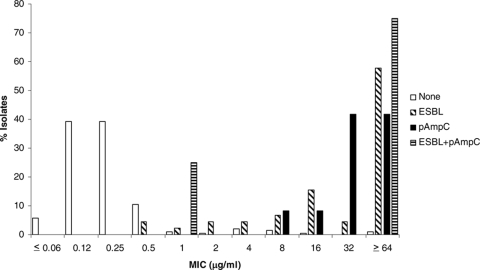
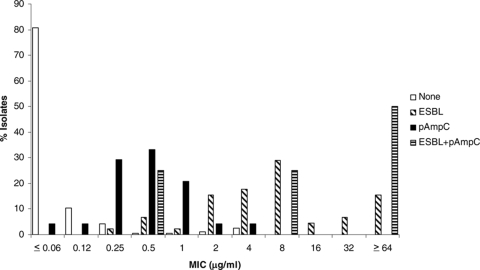
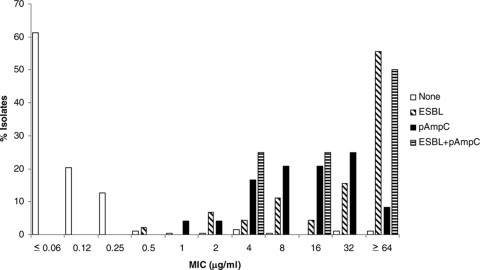
The MICs for isolates with and without ESBL and pAmpC genes are shown in Fig. 1 through 6 and Table 3. All isolates for which the cefpodoxime MIC was ≥8 μg/ml or the MIC of an extended-spectrum cephalosporin was ≥1 μg/ml were subjected to ESBL- and pAmpC-directed PCRs. In Fig. 1 to 6, the MIC distributions are presented as the percentage of isolates with or without ESBL and/or pAmpC genes. Table 3 and Fig. 1 to 6 show a dichotomy between the results for isolates with and without secondary β-lactamases. Notably, many isolates harboring ESBL or pAmpC genes did not fall within the current CLSI resistance category (Table 4).
FIG. 1.
Cumulative cefazolin MIC distribution.
FIG. 2.
Cumulative ceftriaxone MIC distribution.
FIG. 3.
Cumulative cefotaxime MIC distribution.
FIG. 4.
Cumulative ceftazidime MIC distribution.
FIG. 5.
Cumulative cefepime MIC distribution.
FIG. 6.
Cumulative aztreonam MIC distribution.
TABLE 3.
Distribution of MICs for study isolates
| Antimicrobial agent | β-Lactamasea | No. of isolates with the following MIC (μg/ml)b:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.004 | 0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥ 64 | ||
| Cefazolin | None | 34 | 98 | 24 | 15 | 7 | 2 | 11 | ||||||||
| ESBL | 1 | 3 | 5 | 1 | 35 | |||||||||||
| pAmpC | 24 | |||||||||||||||
| ESBL + pAmpC | 4 | |||||||||||||||
| Ceftriaxone | None | 1 | 6 | 61 | 78 | 31 | 4 | 2 | 3 | 2 | 2 | 1 | ||||
| ESBL | 3 | 2 | 3 | 5 | 2 | 7 | 5 | 18 | ||||||||
| pAmpC | 3 | 2 | 7 | 7 | 5 | |||||||||||
| ESBL + pAmpC | 1 | 3 | ||||||||||||||
| Cefotaxime | None | 1 | 3 | 25 | 104 | 29 | 13 | 7 | 5 | 2 | 1 | 1 | ||||
| ESBL | 2 | 1 | 6 | 3 | 3 | 3 | 7 | 5 | 15 | |||||||
| pAmpC | 1 | 2 | 5 | 9 | 3 | 4 | ||||||||||
| ESBL + pAmpC | 1 | 1 | 2 | |||||||||||||
| Ceftazidime | None | 1 | 1 | 9 | 75 | 75 | 22 | 2 | 1 | 4 | 3 | 1 | 2 | |||
| ESBL | 2 | 1 | 2 | 2 | 3 | 7 | 2 | 26 | ||||||||
| pAmpC | 2 | 2 | 10 | 10 | ||||||||||||
| ESBL + pAmpC | 1 | 3 | ||||||||||||||
| Cefepime | None | 1 | 5 | 79 | 69 | 20 | 8 | 5 | 1 | 2 | 1 | |||||
| ESBL | 1 | 3 | 1 | 7 | 8 | 13 | 2 | 3 | 7 | |||||||
| pAmpC | 1 | 1 | 7 | 8 | 5 | 1 | 1 | |||||||||
| ESBL + pAmpC | 1 | 1 | 2 | |||||||||||||
| Aztreonam | None | 1 | 4 | 23 | 89 | 39 | 24 | 2 | 1 | 1 | 3 | 1 | 2 | 2 | ||
| ESBL | 1 | 3 | 2 | 5 | 2 | 7 | 25 | |||||||||
| pAmpC | 1 | 1 | 4 | 5 | 5 | 6 | 2 | |||||||||
| ESBL + pAmpC | 1 | 1 | 2 | |||||||||||||
The β-lactamase detected by ESBL- and/or pAmpC-directed PCR amplification.
Underlined numbers indicate susceptible category by current CLSI recommendations (≤8 μg/ml). Isolates for which cefpodoxime MICs were ≥8 μg/ml or the MICs of extended-spectrum cephalosporins were ≥1 μg/ml were subjected to PCR for detection of ESBL and pAmpC genes.
TABLE 4.
Categorization of susceptibilities of isolates containing ESBL and pAmpC genes by use of current CLSI breakpoints
| Antimicrobial agent | % Isolates producing the following gene and having the indicated susceptibility categorization:
|
|||||
|---|---|---|---|---|---|---|
| ESBL (n = 45)
|
pAmpC (n = 24)
|
ESBL plus pAmpC (n = 4)
|
||||
| Susceptible | Intermediate | Susceptible | Intermediate | Susceptible | Intermediate | |
| Cefazolin | 9 | 11 | 0 | 0 | 0 | 0 |
| Ceftriaxone | 33 | 27 | 21 | 58 | 0 | 25 |
| Cefotaxime | 40 | 27 | 33 | 50 | 25 | 25 |
| Ceftazidime | 22 | 16 | 8 | 8 | 25 | 0 |
| Cefepime | 73 | 4 | 100 | 0 | 50 | 0 |
| Aztreonam | 24 | 4 | 45 | 21 | 25 | 25 |
DISCUSSION
Thirty percent (81/264) of the isolates tested had a positive screening result with cefpodoxime. Of these, 45/81 (55.6%) tested positive for ESBL genes, 24/81 (29.6%) tested positive for pAmpC genes, and 4/81 (4.9%) tested positive for both ESBL and pAmpC genes. No secondary β-lactamases were detected by PCR in 8/81 (9.9%) screening-positive isolates. The TEM, SHV, and CTX-M ESBLs were represented in the collection, as were the CMY-2 and FOX-5 pAmpC genes. Disk augmentation with cefotaxime and ceftazidime with and without clavulanate detected ESBLs with a high degree of sensitivity and specificity compared to the results obtained by PCR (98.0% and 96.3%, respectively). Isolates with ESBL or pAmpC genes yielded higher MICs than isolates free of secondary β-lactamases. It is of interest that pAmpC-containing isolates yielded a smaller distribution around higher MICs than ESBL-containing isolates for cefazolin, ceftriaxone, cefotaxime, and ceftazidime, possibly due to the greater variety of ESBL genes than pAmpC genes represented. Cefepime and aztreonam had slightly greater activities against pAmpC-containing isolates than ESBL isolates, consistent with the hydrolysis spectra, although an overlap in activity was seen. Of note, a large number of isolates with secondary β-lactamases fell within the current susceptible and intermediate categories, and this was equally so for both the ESBL-producing and the pAmpC-producing groups.
These data suggest that with the current breakpoints and guidelines, the ESBL and pAmpC enzymes have similar patterns of resistance to extended-spectrum cephalosporins, most of which fall within current nonresistant categories. This raises the question of the clinical usefulness of laboratory procedures aimed at identifying ESBLs only and editing of the susceptibility interpretations on the basis of a positive clavulanate augmentation result. Lowering of the breakpoints for ceftazidime, ceftriaxone, and cefotaxime is an excellent strategy for the accurate reporting of resistance associated with ESBL and pAmpC genes without the need for any special screening.
Acknowledgments
We are grateful to Karen Bush and David Paterson for providing ESBL-producing isolates. We thank the laboratory technologists in the Bacteriology Laboratory, Mayo Clinic Clinical Microbiology Laboratory, for assistance.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Andes, D., and W. A. Craig. 2005. Treatment of infections with ESBL-producing organisms: pharmacokinetic and pharmacodynamic considerations. Clin. Microbiol. Infect. 11(Suppl. 6)10-17. [DOI] [PubMed] [Google Scholar]
- 2.Baquero, F., P.-R. Hsueh, D. L. Paterson, F. Rossi, G. V. Bochicchio, G. Gallagher, K. Lantz, J. B. Villasenor, K. McCarroll, M. A. Abramson, and J. W. Chow. 2009. In vitro susceptibilities of aerobic and facultatively anaerobic gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: 2005 results from Study for Monitoring Antimicrobial Resistance Trends (SMART). Surg. Infect. 1099-104. [DOI] [PubMed] [Google Scholar]
- 3.Bell, J. M., M. Chitsaz, J. D. Turnidge, M. Barton, L. J. Walters, and R. N. Jones. 2007. Prevalence and significance of a negative extended-spectrum beta-lactamase (ESBL) confirmation test result after a positive ESBL screening test result for isolates of Escherichia coli and Klebsiella pneumoniae: results from the SENTRY Asia-Pacific Surveillance Program. J. Clin. Microbiol. 451478-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun-Buisson, C., A. Philippon, M. Ansquer, P. Legrand, F. Montravers, and J. Duval. 1987. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet 330302-306. [DOI] [PubMed] [Google Scholar]
- 5.Chow, J. W., V. Satishchandran, T. A. Snyder, C. M. Harvey, I. R. Friedland, and M. J. Dinubile. 2005. In vitro susceptibilities of aerobic and facultative gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: the 2002 Study for Monitoring Antimicrobial Resistance Trends (SMART). Surg. Infect. 6439-448. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement, CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Coudron, P. E., E. S. Moland, and K. S. Thomson. 2000. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J. Clin. Microbiol. 381791-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence of plasmidic AmpC type β-lactamase-mediated resistance in Escherichia coli: report from the SENTRY antimicrobial surveillance program (North America, 2004). Int. J. Antimicrob. Agents 28578-581. [DOI] [PubMed] [Google Scholar]
- 9.Ding, H., Y. Yang, Q. Lu, Y. Wang, Y. Chen, L. Deng, A. Wang, Q. Deng, H. Zhang, C. Wang, L. Liu, X. Xu, L. Wang, and X. Shen. 2008. The prevalence of plasmid-mediated AmpC beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae from five children's hospitals in China. Eur. J. Clin. Microbiol. Infect. Dis. 27915-921. [DOI] [PubMed] [Google Scholar]
- 10.Heritage, J., F. H. M'Zali, D. Gascoyne-Binzi, and P. M. Hawkey. 1999. Evolution and spread of SHV extended-spectrum β-lactamases in gram-negative bacteria. J. Antimicrob. Chemother. 44309-318. [DOI] [PubMed] [Google Scholar]
- 11.Jones, R. N., W. A. Craig, P. G. Ambrose, M. N. Dudley, and S. Pottumarthy. 2005. Reevaluation of Enterobacteriaceae MIC/disk diffusion zone diameter regression scattergrams for 9 β-lactams: adjustments of breakpoints for strains producing extended spectrum β-lactamases. Diagn. Microbiol. Infect. Dis. 52235-246. [DOI] [PubMed] [Google Scholar]
- 12.Karas, J. A., D. G. Pillay, D. Muckart, and A. W. Sturm. 1996. Treatment failure due to extended spectrum β-lactamase. J. Antimicrob. Chemother. 37203-204. [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y.-K., H. Pai, H.-J. Lee, S.-E. Park, E.-H. Choi, J. Kim, J.-H. Kim, and E.-C. Kim. 2002. Bloodstream infections by extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in children: epidemiology and clinical outcome. Antimicrob. Agents Chemother. 461481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56451-454. [DOI] [PubMed] [Google Scholar]
- 15.MacGowan, A. Breakpoints for extended-spectrum β-lactamase-producing Enterobacteriaceae: pharmacokinetic/pharmacodynamic considerations. Clin. Microbiol. Infect. 14(Suppl. 1)166-168. [DOI] [PubMed]
- 16.Pai, H., E.-H. Choi, H.-J. Lee, J. Y. Hong, and G. A. Jacoby. 2001. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 393747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, Y.-J., S. Y. Park, E.-J. Oh, J.-J. Park, K.-Y. Lee, G.-J. Woo, and K. Lee. 2005. Occurrence of extended-spectrum beta-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 51265-269. [DOI] [PubMed] [Google Scholar]
- 18.Paterson, D. L., W.-C. Ko, A. Von Gottberg, J. M. Casellas, L. Mulazimoglu, K. P. Klugman, R. A. Bonomo, L. B. Rice, J. G. McCormack, and V. L. Yu. 2001. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. J. Clin. Microbiol. 392206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson, D. L., F. Rossi, F. Baquero, P.-R. Hsueh, G. L. Woods, V. Satishchandran, T. A. Snyder, C. M. Harvey, H. Teppler, M. J. DiNubile, and J. W. Chow. 2005. In vitro susceptibilities of aerobic and facultative gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: the 2003 Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 55965-973. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Perez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 402153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 461-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout, J. D., D. B. Gregson, D. L. Church, and K. B. Laupland. 2007. Population-based laboratory surveillance for AmpC beta-lactamase-producing Escherichia coli, Calgary. Emerg. Infect. Dis. 13443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitout, J. D. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 5652-59. [DOI] [PubMed] [Google Scholar]
- 24.Quinn, J. P., D. Miyashiro, D. Sahm, R. Flamm, and K. Bush. 1989. Novel plasmid-mediated beta-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 331451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, L. B., S. H. Willey, G. A. Papanicolaou, A. A. Medeiros, G. M. Eliopoulos, R. C. Moellering, Jr., and G. A. Jacoby. 1990. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamases at a Massachusetts chronic-care facility. Antimicrob. Agents Chemother. 342193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robberts, F. J. L., P. C. Kohner, and R. Patel. 2008. Unreliable ESBL detection in the presence of plasmid-mediated AmpC production in Escherichia coli clinical isolates. J. Clin. Microbiol. 47358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi, F., F. Baquero, P.-R. Hsueh, D. L. Paterson, G. V. Bochicchio, T. A. Snyder, V. Satishchandran, K. McCarroll, M. J. DiNubile, and J. W. Chow. 2006. In vitro susceptibilities of aerobic and facultatively anaerobic gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: 2004 results from SMART (Study for Monitoring Antimicrobial Resistance Trends). J. Antimicrob. Chemother. 58205-210. [DOI] [PubMed] [Google Scholar]
- 28.Saladin, M., V. T. B. Cao, T. Lambert, J.-L. Donay, J.-L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209161-168. [DOI] [PubMed] [Google Scholar]
- 29.Smith, C. E., B. S. Tillman, A. W. Howell, R. N. Longfield, and J. H. Jorgensen. 1990. Failure of ceftazidime-amikacin therapy for bacteremia and meningitis due to Klebsiella pneumoniae producing an extended-spectrum beta-lactamase. Antimicrob. Agents Chemother. 341290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]