Abstract
A multiplex ligation-dependent probe amplification assay for simultaneous detection of six virus species was developed and tested on clinical cerebrospinal fluid (CSF) samples. The assay, termed MeningoFinder, showed an accordance of 97%, concordance of 96%, interlaboratory sensitivity of 90%, and interlaboratory specificity of 94% compared to PCRs.
The diagnosis of central nervous system (CNS) infections is nowadays mostly performed by molecular methods such as (real-time) PCR (13). However, simultaneous PCR detection of agents is limited by the use of multiple primers, as this increases the risk of primer-dimer formation and constrains suitable reaction conditions (9). An amplification technique that allows for simultaneous detection of targets without the use of multiple primer sets is the multiplex ligation-dependent probe amplification (MLPA) technology. MLPA technology was shown to allow amplification of up to 40 different targets simultaneously through the use of one universal primer set in the final amplification (12). Recently, the MLPA technology was applied for the first time for detection of infectious agents causing respiratory tract infections (11). The goal of the current study was to design and evaluate the MeningoFinder assay (Fig. 1), a novel MLPA assay for simultaneous detection of six clinically relevant agents causing CNS infections, i.e., enterovirus, cytomegalovirus (CMV), varicella-zoster virus (VZV), Epstein-Barr virus (EBV), and herpes simplex virus type 1 and 2 (HSV-1 and HSV-2). The assay was tested independently in two different laboratories and compared to conventional PCR-based diagnostics.
FIG. 1.
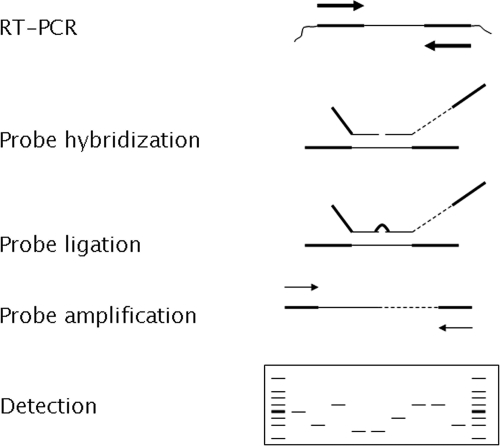
Schematic representation of the MeningoFinder MLPA assay setup. A one-step reverse transcription-PCR (RT-PCR) is performed using specific primers for all seven targets. Subsequently, the MLPA reaction is performed, including a probe hybridization step, a ligation step, and a amplification step. The amplification is performed with one universal primer set that can amplify all ligated probes. After amplification, the MLPA reaction is analyzed by electrophoresis. Each MLPA probe can be discerned due to its specific length. Recently, the protocol has been further optimized by combining the probe ligation and probe amplification in one reaction.
Sample panels containing quantified viral loads of all targets were obtained from Quality Control for Molecular Diagnostics (Glasgow, United Kingdom). Clinical cerebrospinal fluid samples (n = 50) were collected by the Laboratory for Pathology and Medical Microbiology, Eindhoven, The Netherlands (PAMM). Samples were divided in multiple parts for randomized double-blind analysis by the different laboratories. The MeningoFinder assay was compared to routine in-house PCRs. The PCRs for enterovirus, HSV-1, HSV-2, and VZV were performed by PAMM (1, 6, 15), and the PCRs for CMV and EBV were performed at the Laboratory of Medical Microbiology at the Maastricht University Medical Center (MUMC) (10, 16). Viral RNA/DNA from clinical samples (200-μl cerebrospinal fluid specimens) for PCR or MLPA was isolated using either the Nuclisense easyMAG (bioMerieux, Boxtel, The Netherlands) or the MagnaPure LC nucleic acid system (Roche Diagnostics) according to the manufacturers' instructions. Before starting the extraction, a competitive internal control (IAC) was added; IAC was constructed as described previously (11). The MeningoFinder probes were designed against conserved viral genomic regions, i.e., the CMV UL89 gene (gb DQ48009.1, positions 1767 to 1824), the EBV nuclear antigen 1 gene (gb AY825078.1, positions 280 to 346), the enterovirus polyprotein gene (emb AJ295206.1 HEC295206, positions 469 to 525), the HSV-1 and HSV-2 US6 genes (gi 30961596 gb AY240830.1, positions 89 to 155, and gb EU018091.1, positions 154 to 220) and the VZV immediate-early 62 gene (gb AY253719.1, positions 1996 to 2062). The primers were used for both specific reverse transcription as well as preamplification (Table 1). As shown in Fig. 2, the MeningoFinder setup produces seven specific PCR products. The MeningoFinder MLPA was performed by PAMM and Pathofinder B.V. (PF). Preamplification and MLPA were performed as previously described (11), except for the primer concentration during preamplification, which was raised to 4 μM for each primer (Biolegio, Malden, The Netherlands). Amplified MLPA products were analyzed either by agarose gel electrophoresis (Cambrex, Verviers, Belgium) or by acrylamide gel electrophoresis using a LI-COR 4300 DNA analyzer. For the multicenter study, the interlaboratory sensitivity was defined as the percentage of positive samples giving a correct positive signal (as determined by PCR). The interlaboratory specificity was defined as the percentage of negative samples giving a correct negative signal (European Committee for Standardization, 2002) (4). The accordance (repeatability) and concordance (reproducibility) were defined as the percentages of finding the same result (positive or negative) from two samples analyzed in either the same laboratory or different laboratories under standard conditions (4).
TABLE 1.
Preamplification primers used in the MeningoFinder assay
| Target | Sequence | Amplicon size (bp) |
|---|---|---|
| HSV-1 | 5′-GAAGACCCCGAGGATTCG-3′ | 204 |
| 5′-CCAGTACACAATTCCGCAAA-3′ | ||
| HSV-2 | 5′-CATGGGGCGTTTGACCTC-3′ | 249 |
| 5′-TACACAGTGATCGGGATGCT-3′ | ||
| VZV | 5′-ATGCATTAGCTCTTGTCGAGGAGGCTTCTG-3′ | 302 |
| 5′-ATGCATGGAATGGCTATGAGCCGTCGATAC-3′ | ||
| Enterovirus | 5′-ATGCACTAGCTCCTCCGGCCCCTGAATGCGGCTAA-3′ | 181 |
| 5′-ATGCATGGACCTTTCAATTGTCACCATAAGCAGCCA-3′ | ||
| EBV | 5′-CGCAGATGACCCAGGAGA-3′ | 408 |
| 5′-CCCTCAGCAAATATATGAGTTTGTA-3′ | ||
| CMV | 5′-ATGCACTAGCCATCGAACAGCCCTTCTACC-3′ | 252 |
| 5′-ATGCATGGACAGATACGATTCTGGCGCTTG-3′ | ||
| IAC | 5′-ACATGTAACCGCCCCCATT-3′ | 118 |
| 5′-TCCACGCACGCACTACTATG-3′ |
FIG. 2.
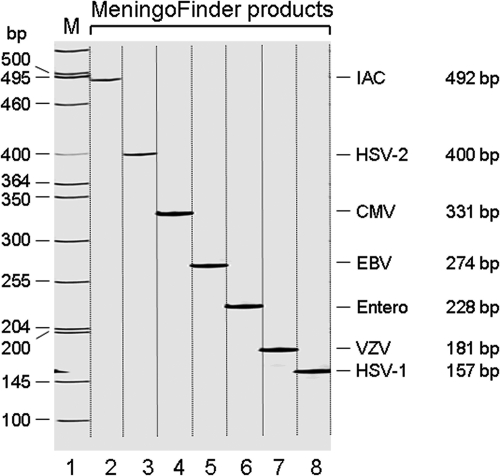
Gel electrophoresis results of the MeningoFinder assay showing the different sized bands representing detection of the respective viruses or IAC. Lane M contains molecular size markers, and lanes 1 to 7 show the different MLPA products. Entero, enterovirus.
MeningoFinder detection limit and specificity.
The analytical detection limit of the MeningoFinder MLPA assay was evaluated using external quality control samples (Quality Control for Molecular Diagnostics) and was found to be 4 copies/MLPA assay for CMV, 50 copies/MLPA assay for EBV, 15 copies/MLPA assay for enterovirus, 28 copies/MLPA assay for HSV-1, 180 copies/MLPA assay for HSV-2, and 154 copies/MLPA assay for VZV. Similar detection limits were previously found for a multiplex MLPA assay detecting respiratory viruses (11), as well as for multiplex real-time PCR assays and microarray tests detecting viruses causing CNS infections (2)(8, 14). The specificity of the MeningoFinder assay was demonstrated by the lack of cross-reactivity between any of the targets in the panel (data not shown).
Evaluation of the MeningoFinder assay on clinical samples.
The accordance, concordance, and interlaboratory sensitivity and specificity were evaluated using a triple-center approach. The overall accordance and concordance of the assay were found to be 97% and 96%, respectively. All discrepancies were caused by failed detection in one of two runs at PAMM resulting from possible low loads of enterovirus or due to incidents of contamination. The values for accordance and concordance that were found in our study are higher or at least in the same range as those reported in other multicenter evaluations of molecular tests (3, 4).
To assess the interlaboratory sensitivity and specificity, the results from the MeningoFinder assay were also compared to results obtained by PCR. The MeningoFinder MLPA and PCR were tested on 50 clinical CSF samples (Table 2). Of the 26 samples positive by PCR, the MeningoFinder assay produced concordant results in 24 (PAMM) and 23 (PF) samples, resulting in an average overall interlaboratory sensitivity of 90%. Regarding the 24 samples that were found negative by PCR, the MeningoFinder assay generated concordant results in 22 and 23 results for PAMM and PF, respectively, resulting in an average overall interlaboratory specificity of 94%. Interlaboratory sensitivities and specificities were also calculated per individual causative agent. The interlaboratory specificities were found to lie between 98 and 100%, whereas the sensitivities were 100% for enterovirus, HSV-1, HSV-2, and VZV. The sensitivity for CMV could not be calculated due to the absence of any CMV-positive samples. The sensitivity for EBV was 0% due to a failure of the MeningoFinder assay to detect EBV in two samples. However, the EBV load in these samples was lower (<10 copies/PCR) than the detection limit of the MeningoFinder assay for EBV (50 copies/MLPA assay). Further development of the assay will be geared toward improving the detection limit for EBV, as well as for HSV-2 and VZV. The values for sensitivity and specificity of the current MeningoFinder assay are comparable to those for molecular methods used in other multicenter trials (4, 5, 7). The main limitation of the current study is that samples have been analyzed in a retrospective fashion. Future studies will have to be performed to evaluate the assay prospectively. In conclusion, the MeningoFinder assay is a flexible and convenient assay that does not require specialized treatment. It offers a promising alternative for simultaneous routine detection of multiple viruses causing CNS infections.
TABLE 2.
Comparison of the results of MeningoFinder MLPA and PCR on 50 clinical CSF samples
| No. of samples | PCR resulta | MeningoFinder MLPA result by:b
|
|
|---|---|---|---|
| PAMM | PF | ||
| 12 | Enterovirus | Enterovirus | Enterovirus |
| 2 | Enterovirus | Enterovirusc | Enterovirus |
| 1 | Enterovirus | Enterovirusc | HSV-2 + enterovirus |
| 1 | NEG | EBV | EBV |
| 4 | HSV-1 | HSV-1 | HSV-1 |
| 1 | HSV-2 | HSV-2 | HSV-2 |
| 2 | HSV-2 + EBV | HSV-2 | HSV-2 |
| 4 | VZV | VZV | VZV |
| 1 | NEG | VZV | NEG |
| 22 | NEG | NEG | NEG |
PCR analyses were performed by PAMM for HSV-1, HSV-2, enterovirus, and VZV. EBV and CMV real-time PCRs were performed by MUMC. NEG, negative.
All MeningoFinder assays were performed in duplicate, both at PAMM and PF. All results were identical in duplicate experiments unless indicated otherwise. NEG, negative.
Enterovirus was detected in one out of two experiments for these three enterovirus results.
Acknowledgments
The research described here has been facilitated by a Triple-In TenderFonds grant provided by the Province of Limburg, The Netherlands.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Aurelius, E., B. Johansson, B. Skoldenberg, A. Staland, and M. Forsgren. 1991. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet 337189-192. [DOI] [PubMed] [Google Scholar]
- 2.Boriskin, Y. S., P. S. Rice, R. A. Stabler, J. Hinds, H. Al-Ghusein, K. Vass, and P. D. Butcher. 2004. DNA microarrays for virus detection in cases of central nervous system infection. J. Clin. Microbiol. 425811-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claes, F., S. Deborggraeve, D. Verloo, P. Mertens, J. R. Crowther, T. Leclipteux, and P. Buscher. 2007. Validation of a PCR-oligochromatography test for detection of Trypanozoon parasites in a multicenter collaborative trial. J. Clin. Microbiol. 453785-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Josefsen, M. H., N. Cook, M. D'Agostino, F. Hansen, M. Wagner, K. Demnerova, A. E. Heuvelink, P. T. Tassios, H. Lindmark, V. Kmet, M. Barbanera, P. Fach, S. Loncarevic, and J. Hoorfar. 2004. Validation of a PCR-based method for detection of food-borne thermotolerant campylobacters in a multicenter collaborative trial. Appl. Environ. Microbiol. 704379-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornegay, J. R., M. Roger, P. O. Davies, A. P. Shepard, N. A. Guerrero, B. Lloveras, D. Evans, and F. Coutlee. 2003. International proficiency study of a consensus L1 PCR assay for the detection and typing of human papillomavirus DNA: evaluation of accuracy and intralaboratory and interlaboratory agreement. J. Clin. Microbiol. 411080-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonnrot, M., M. Sjoroos, K. Salminen, M. Maaronen, T. Hyypia, and H. Hyoty. 1999. Diagnosis of enterovirus and rhinovirus infections by RT-PCR and time-resolved fluorometry with lanthanide chelate labeled probes. J. Med. Virol. 59378-384. [PubMed] [Google Scholar]
- 7.Malorny, B., N. Cook, M. D'Agostino, D. De Medici, L. Croci, A. Abdulmawjood, P. Fach, R. Karpiskova, T. Aymerich, K. Kwaitek, and J. Hoorfar. 2004. Multicenter validation of PCR-based method for detection of Salmonella in chicken and pig samples. J. AOAC Int. 87861-866. [PubMed] [Google Scholar]
- 8.Markoulatos, P., A. Georgopoulou, N. Siafakas, E. Plakokefalos, G. Tzanakaki, and J. Kourea-Kremastinou. 2001. Laboratory diagnosis of common herpesvirus infections of the central nervous system by a multiplex PCR assay. J. Clin. Microbiol. 394426-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markoulatos, P., N. Siafakas, and M. Moncany. 2002. Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 1647-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niesters, H. G., J. van Esser, E. Fries, K. C. Wolthers, J. Cornelissen, and A. D. Osterhaus. 2000. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J. Clin. Microbiol. 38712-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reijans, M., G. Dingemans, C. H. Klaassen, J. F. Meis, J. Keijdener, B. Mulders, K. Eadie, W. van Leeuwen, A. van Belkum, A. M. Horrevorts, and G. Simons. 2008. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J. Clin. Microbiol. 461232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schouten, J. P., C. J. McElgunn, R. Waaijer, D. Zwijnenburg, F. Diepvens, and G. Pals. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solbrig, M. V., A. N. Hasso, and C. A. Jay. 2008. CNS viruses—diagnostic approach. Neuroimaging Clin. N. Am. 181-18. [DOI] [PubMed] [Google Scholar]
- 14.Tafreshi, N. K., M. Sadeghizadeh, S. Amini-Bavil-Olyaee, A. M. Ahadi, I. Jahanzad, and M. H. Roostaee. 2005. Development of a multiplex nested consensus PCR for detection and identification of major human herpesviruses in CNS infections. J. Clin. Virol. 32318-324. [DOI] [PubMed] [Google Scholar]
- 15.van Beek, J., A. zur Hausen, E. K. Kranenbarg, R. J. Warring, E. Bloemena, M. E. Craanen, C. J. van de Velde, J. M. Middeldorp, C. J. Meijer, and A. J. van den Brule. 2002. A rapid and reliable enzyme immunoassay PCR-based screening method to identify EBV-carrying gastric carcinomas. Mod. Pathol. 15870-877. [DOI] [PubMed] [Google Scholar]
- 16.van Doornum, G. J., J. Guldemeester, A. D. Osterhaus, and H. G. Niesters. 2003. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J. Clin. Microbiol. 41576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]