Abstract
We present the first documented human case of Salmonella enterica serovar Apapa infection, isolated concurrently from a hospital inpatient and a pet lizard. The isolates were identical by biochemical profiling and pulsed-field gel electrophoresis. This rare serotype is known to be associated with reptiles. The current practice for avoiding reptile-associated infections is reviewed.
CASE REPORT
A 67-year-old Caucasian female presented with a 1-day history of sudden-onset left iliac fossa pain associated with nausea, vomiting, and a productive cough. Her extensive past medical history included primary biliary cirrhosis, esophageal varices, small intestinal mucosa-associated lymphoid tissue lymphoma, a benign ovarian cyst, and osteoporosis. On examination, she was unwell, with a fever (37.8°C), tachycardia (heart rate, 110), and hypotension (70/40 mmHg). She had signs of peritonism in the left iliac fossa and respiratory signs consistent with right mid-zone consolidation. The differential diagnosis included diverticulitis and bowel perforation with accompanying pneumonia. In view of her persistent hypotension and hypoxia, she was transferred to the intensive care unit. She required respiratory ventilation and was commenced on intravenous antibiotics according to hospital policy, including ciprofloxacin. Her abdominal symptoms subsided without surgical intervention. She remained in the intensive care unit for a total of 18 days until her respiratory function improved. She made a good recovery and was discharged home after a further 12 days.
Tracheal aspirate samples collected the day after admission grew Klebsiella pneumoniae, which was thought to represent upper airway colonization. Three sets of blood cultures were negative. A stool sample submitted the day after admission yielded a non-lactose-fermenting coliform, consistent with a Salmonella sp. This was later identified as Salmonella enterica serovar Apapa at the Health Protection Agency Laboratory of Enteric Pathogens (Table 1). Repeat stool cultures from the patient at day 12 and at week 16 were negative.
TABLE 1.
Characteristics of the three isolates of Salmonella Apapa described in this report
| Isolate (source) | Date of isolation | API 20E profile | Serotype | Antibiogram | MLST | PFGE |
|---|---|---|---|---|---|---|
| H073120204 (human) | 7/25/07 | 6704552 | I45:m,t:− | Sensitive to ampicillin, chloramphenicol, ciprofloxacin, and trimethoprim | ST647 | Bands identical to those of isolate H073120204 |
| H073400283 (lizard) | 8/10/07 | 6704552 | I45:m,t:− | Sensitive to ampicillin, chloramphenicol, ciprofloxacin, and trimethoprim | ST647 | Bands identical to those of isolate H073400283 |
| H073520339 (tank water) | 8/22/07 | 6704552 | IV44:Z4,Z23:− | Sensitive to ampicillin, chloramphenicol, ciprofloxacin, and trimethoprim | ST433 | 12 bands different (<20% similarity) |
Further questioning revealed that the patient's son had recently moved back to live with her, along with his two bearded dragon lizards. He kept the lizards in a tank, and the son was the only household member who handled the lizards and cleaned out the tank; the patient herself reported no direct contact with the creatures. Samples of lizard feces and tank water were obtained, and these also yielded a Salmonella sp. These isolates were identified and further characterized by the reference laboratory by pulsed-field gel electrophoresis (PFGE) for fingerprinting of the organisms isolated from the different sources (Table 1 and Fig. 1). The reference laboratory performed PFGE according to the PulseNet protocol, with restriction enzyme XbaI and S. Braenderup H9812 as the reference marker strain (10). The isolates were subjected to multilocus sequence typing (MLST) by determination of the sequences of seven housekeeping genes, aroC, dnaN, hemD, hisD, purE, sucA, and thrA (6). Comparison was made with the Salmonella MLST database hosted at University College Cork, Ireland (http://mlst.ucc.ie/mlst/dbs/Senterica).
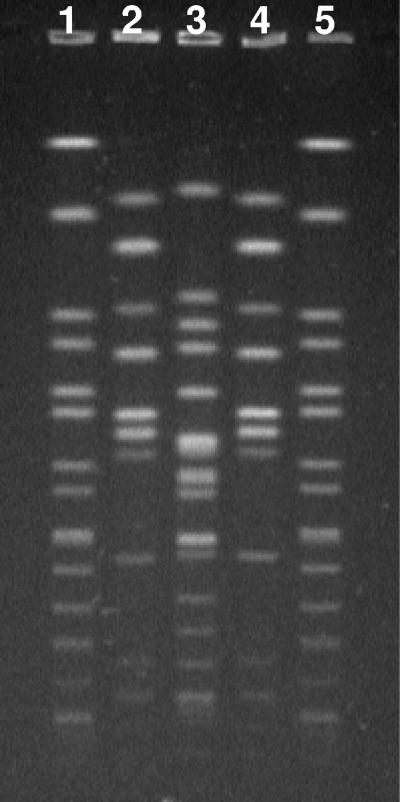
FIG. 1.
PFGE of Salmonella Apapa isolates. Human isolate H073400283 (lane 2) and lizard feces isolate H073120204 (lane 4) show identical band patterns following digestion of their genomic DNAs with XbaI. Tank water isolate H073520339 (lane 3) is different from these two isolates. The control, S. Braenderup H9812, is in lanes 1 and 5.
Reference laboratory results demonstrated that the isolate from lizard feces was also Salmonella Apapa. This was identical to the patient isolate by resistance typing, serotyping, MLST, and PFGE, thus demonstrating an epidemiological link. In contrast, the isolate from the culture plate of the tank water was antigenically different from the other two isolates. The patient and lizard isolates produced identical sequences by MLST, belonging to a previously unrecognized sequence type (ST), ST647, while the tank water isolate was ST433.
Discussion.
This is the first published case of human infection with Salmonella Apapa. This rare serotype is named after the port of Apapa in Lagos, Nigeria. Its antigenic profile is I45:m,t:− by the Kauffman-White scheme (9). A medical literature search identified no publications, but the Google search engine highlighted several veterinary publications (4, 5, 8) and hence alerted us to the association with reptiles.
The patient's family provided samples of lizard feces and tank water, which were also culture positive for a Salmonella sp. Serological identification supplemented by PFGE subtyping confirmed that the isolates from the patient and lizard feces were indistinguishable. It was not surprising that the tank water isolate was antigenically different from the patient and lizard isolates. It is likely that the tank water harbored many serotypes of Salmonella, and the one that was sent to the reference laboratory for further characterization was not serovar Apapa. It is well recognized that a single reptile or its environment may harbor more than one serotype. Thus, it has been suggested that several suspected Salmonella colonies from primary detection media should be selected for further identification and serotyping. Alternatively, use of a more intensive method with an additional preenrichment stage, such as used for food analysis, may help detect the different Salmonella serotypes (12).
The patient's initial clinical presentation was atypical in that there was no history of diarrhea. She had acute abdominal symptoms, but they resolved without surgical intervention. She might have been a carrier of Salmonella Apapa, and this organism could have been an incidental finding that was not causing her clinical symptoms. Stool cultures on day 12 were negative, and it was assumed that the 10-day course of ciprofloxacin for her community-acquired pneumonia had cleared the salmonellae. Follow-up stool cultures at 4 months did not detect salmonellae.
The health protection nurses and environmental health teams were closely involved with this case, and the patient's family completed a questionnaire. The patient stated that she did not directly touch the lizards or clean out the lizard tank. It is likely that she acquired the infection feco-orally, through poor hygiene in food preparation or by accidentally touching the fecally contaminated hands of other family members. Alternatively, contamination of the environment, either by the hands of other family members or through splashing or aerosols from the lizard tank water, may have been important. Of note, no other family members reported diarrhea or other gastrointestinal symptoms.
Apapa is a rare serotype, with only 18 isolations from cases of human infection identified by the United Kingdom reference laboratory between 1 January 2000 and 31 August 2007, including the present isolate. Of these, one was from a patient with a history of foreign travel; in the remaining 17 cases, there was no indication of foreign travel but at least five patients were known to have had contact with reptiles. Additionally, an isolate of Salmonella Apapa from a reptile unrelated to the present cases was identified in May 2007, confirming the association of this serotype with reptiles.
The number of cases of reptile-associated salmonella infections in Europe is not known, although a review of the current situation has recently been published (3). In Germany in 2006, S. Apapa was isolated from three 5-month-old triplets also associated with two bearded dragon lizards, as was our patient. Of interest, the reptiles also harbored three other serotypes, which were also pathogenic for humans. The same serotype that was isolated from our tank water was reported from four other people who had had contact with a snake (two cases), a bearded dragon lizard (one case), and a snake or a bearded dragon lizard (one case) (3). There were regular reports from the United States of reptile-associated Salmonella infections between 1994 and 2002 (1-3). Of the estimated 1.4 million human cases of Salmonella infections that occur annually in the United States, approximately 74,000 are thought to be due to exposure to reptiles and amphibians (7).
Other human infections with specific Salmonella serotypes (such as Arizonae, Javiana, Poona, and Paratyphi B variant Java) are known to be associated with reptiles, including lizards (11). Most reptiles carry salmonellae in the intestinal tracts and may shed bacteria in their feces intermittently or continuously. Salmonella infection is usually asymptomatic in reptiles but may cause serious illness in humans. Administration of antibiotics to eliminate the bacteria from reptiles has been unsuccessful and may result in the emergence of antimicrobial resistance in their salmonellae. Routine precautions to prevent human infection include hand washing after handling reptiles, cages, equipment, and reptile feces. The Centers for Disease Control and Prevention, Atlanta, GA, recommends that children less than 5 years of age avoid contact with reptiles and that households with children less than 1 year of age or in which a baby is expected not own reptiles (3). People at increased risk of infection or serious complications of salmonellosis (e.g., immunocompromised persons) should also avoid contact with reptiles. Other recommendations include that pet reptiles not be allowed to roam freely throughout the home or living area and be kept out of the kitchen or food preparation area to prevent contamination. Kitchen sinks should not be used to bathe reptiles or to wash their dishes, cages, or aquariums. If baths are to be used for this purpose, they should be cleaned thoroughly and disinfected with bleach after use. Pet store owners, veterinarians, and others should inform owners and potential purchasers about the risk of acquiring salmonellosis from reptiles (3).
It is important that pet shops and owners and their families be aware of the dangers of handling exotic pets such as lizards and other reptiles. Basic hygiene measures, such as hand washing, should be applied after handling such animals or becoming contaminated with tank water from their environment.
Acknowledgments
We thank Audrey Pepperman (Health Protection Nurse) for her involvement with the case and Craig Corton for performing the MLST.
We sought no financial support for this study and have no conflicts of interest to declare.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Centers for Disease Control and Prevention. 1995. Reptile-associated salmonellosis—selected states, 1994-1995. MMWR Morb. Mortal. Wkly. Rep. 44347-350. [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1999. Reptile-associated salmonellosis—selected states, 1996-1998. MMWR Morb. Mortal. Wkly. Rep. 481009-1013. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2003. Reptile-associated salmonellosis—selected states, 1998-2002. MMWR Morb. Mortal. Wkly. Rep. 521206-1209. [PubMed] [Google Scholar]
- 4.Ebani, V. V., D. Cerri, F. Fratini, N. Meille, P. Valentini, and E. Andreani. 2005. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res. Vet. Sci. 78117-121. [DOI] [PubMed] [Google Scholar]
- 5.Intorre, L., M. Vanni, V. V. Ebani, D. Cerri, F. Fratini, G. Cardini, R. Tognetti, and G. Soldani. 2005. Antimicrobial susceptibility of animal strains of Salmonella enterica isolated in Italy from 2001 to 2003. J. Vet. Pharmacol. Ther. 28121-125. [DOI] [PubMed] [Google Scholar]
- 6.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 239-45. [DOI] [PubMed] [Google Scholar]
- 7.Mermin, J., L. Hutwagner, D. Vugia, S. Shallow, P. Daily, J. Bender, J. Koehler, R. Marcus, and F. J. Angulo. 2004. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin. Infect. Dis. 38(Suppl. 3)S253-S261. [DOI] [PubMed] [Google Scholar]
- 8.Pasmans, F., A. Martel, F. Boyen, D. Vandekerchove, I. Wybo, F. V. Immerseel, M. Heyndrickx, J. M. Collard, R. Ducatelle, and F. Haesebrouck. 2005. Characterization of Salmonella isolates from captive lizards. Vet. Microbiol. 110285-291. [DOI] [PubMed] [Google Scholar]
- 9.Popoff, M. Y., J. Bockemuhl, and L. L. Gheesling. 2004. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155568-570. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warwick, C., A. J. Lambiris, D. Westwood, and C. Steedman. 2001. Reptile-related salmonellosis. J. R. Soc. Med. 94124-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis, C., T. Wilson, M. Greenwood, and L. Ward. 2002. Pet reptiles associated with a case of salmonellosis in an infant were carrying multiple strains of Salmonella. J. Clin. Microbiol. 404802-4803. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]