Abstract
We compared the results of typing methicillin-resistant Staphylococcus aureus (MRSA) isolates using the DiversiLab system (DL) to the results obtained using pulsed-field gel electrophoresis (PFGE). One hundred five MRSA isolates of PFGE types USA100 to USA1100 and the Brazilian clone, from the Centers for Disease Control and Prevention (CDC) and Project ICARE strain collections, were typed using DL. In addition, four unique sets of MRSA isolates from purported MRSA outbreaks that had been previously typed by DL, each consisting of six isolates (where five isolates were classified as indistinguishable by DL and one was an unrelated DL type) were typed by PFGE. DL separated the 105 MRSA isolates of known USA types into 11 clusters and six unique banding patterns. DL grouped most of the USA100, USA200, and USA1100 isolates into unique clusters. Multilocus sequence type 8 isolates (i.e., USA300 and USA500) often clustered together at >95% similarity in DL dendrograms. Nevertheless, USA300 and USA500 DL patterns could be distinguished using the pattern overlay function of the DL software. Among the hospital outbreak clusters, PFGE and DL identified the same “unrelated” organism in three of four sets. However, PFGE showed more pattern diversity than did DL, suggesting that two of the sets were less likely to represent true outbreaks. In summary, DL is useful for screening MRSA isolates to rule out potential outbreaks of MRSA in hospitals, but PFGE provides better discrimination of potential outbreak strains and is more useful for confirming strain relatedness and specific USA types.
Although pulsed-field gel electrophoresis (PFGE) is often considered the gold standard for typing methicillin-resistant Staphylococcus aureus (MRSA) isolates for epidemiologic studies (8, 12, 13), PFGE requires several days to complete and the results are often difficult for inexperienced users to interpret. On the other hand, DNA sequence-based methods, such as spa typing, which has also been shown to be useful for epidemiologic studies of MRSA (3), are not practical for many clinical laboratories in the United States, which lack access to DNA sequencing facilities. An alternative strain typing method, which is available commercially, is the DiversiLab typing system (DL) (bioMérieux, Inc., Durham, NC), which uses the presence of DNA repetitive elements present in the organism's genome to determine the genetic relatedness of bacterial and fungal isolates (4-6, 9, 18). DL has been used successfully in several MRSA typing studies to distinguish sporadic from outbreak-related isolates and is noted to be more rapid to perform and easier to learn than PFGE (14, 15). Agreement between DL clusters of organisms and USA PFGE types, as defined by McDougal et al. (12), was reported for five well-defined U.S. outbreaks, although specific data were not shown (14). However, a recent study of representative MRSA strains from the Harmony collection in Europe concluded that while DL, PFGE, and multilocus sequence typing (MLST) provided concordant classification of strains, PFGE showed a higher level of strain discrimination than either DL or MLST (17). Thus, whether DL can differentiate accurately among USA types remains an open question.
The goal of this study was to use DL to characterize a series of MRSA isolates of known PFGE types from U.S. hospitals to determine whether DL could (i) differentiate among PFGE types USA100 through USA1100, (ii) identify DL banding patterns that correlated with specific USA types, and (iii) differentiate contemporary outbreak-related MRSA isolates from sporadic isolates collected from U.S. hospitals.
MATERIALS AND METHODS
Bacterial isolates.
A total of 105 MRSA isolates, 103 of which were identified as PFGE types USA100 through USA1100 and 2 as isolates of the Brazilian clone, were obtained from the culture collections of Project ICARE (Intensive Care Antimicrobial Resistance Epidemiology) (2) and the Centers for Disease Control and Prevention (CDC) (Table 1). In addition, four sets of isolates consisting of six MRSA isolates each (where five isolates were indistinguishable by DL and one was an unrelated DL type), each from a unique purported MRSA outbreak, were solicited from among DL users in the United States. For outbreak A, isolates were collected over a 3-month period from surgical patients in a single hospital. The infections from which the MRSA isolates were obtained were classified by the infection control staff as health care related. Isolates from outbreak B were collected from the same hospital but spanned a different 2-month time period and were also classified by infection control staff as health care related. The patients' locations and additional epidemiologic data were not provided for either outbreak. The third set of isolates came from a different hospital that had just implemented MRSA surveillance. The isolates were obtained by infection control personnel over a single 2-day period. An epidemiologic link among the patients was suspected, but no further details were provided by the hospital. The final set of isolates, collected over a 6-month period, was sent by a third medical center; the isolates were classified as outbreak related, but no further epidemiologic data were provided by the hospital to validate this designation. The sixth isolate in each set was selected by the hospital laboratory from strains typed by DL that were presumed to be unrelated to the “outbreak strain.” The isolates were retested at the DL reference laboratory to ensure that the six DL patterns (five indistinguishable and one unrelated) were reproducible. No differences in DL patterns were noted among the indistinguishable isolates from each outbreak set when the pattern overlay function and similarity matrices were used. The isolates were coded and labeled so that the outlier was not identifiable, and they were sent blinded to the Project ICARE laboratory for PFGE typing. The organisms were confirmed as S. aureus by catalase and coagulase testing, and oxacillin resistance was confirmed by oxacillin or cefoxitin disk diffusion testing using the Clinical and Laboratory Standards Institute (CLSI) disk diffusion method (1) on Mueller-Hinton agar (BD Diagnostics, Sparks, MD). S. aureus ATCC 25923 was used for quality control on each day of susceptibility testing.
TABLE 1.
Comparison of DL and PFGE typing results for MRSA study isolatesa
| Cluster/singleton designationb | No. of isolates in cluster | USA type(s) (no. of isolates) included in DL cluster on dendrogramc |
|---|---|---|
| A | 8 | 800 (4) T, 700 (3) T, 400 (1) T |
| B | 6 | 800 (6) |
| C | 41 | 300 (18) T, 500 (18) T, 800 (2), Brazilian (3) T |
| D | 7 | 400 (7) |
| E | 3 | 1000 (2), 800 (1) |
| F | 3 | 300 (3) |
| G | 4 | 1100 (4) |
| H | 4 | 100 (4) |
| I | 16 | 100 (16) T |
| J | 5 | 200 (5) T |
| K | 2 | 600 (2) T |
| 1 | 1 | 1000 T |
| 2 | 1 | 500 |
| 3 | 1 | 100 |
| 4 | 1 | 1100 T |
| 5 | 1 | 600 |
| 6 | 1 | 100 |
Isolates were obtained from Project ICARE and CDC strain collections.
Capital letters indicate DL clusters, and numbers indicate DL singleton strain types.
“T” indicates that the cluster contains the type strain for the given USA PFGE type or that the singleton is the type strain for that USA type.
Bacterial typing methods.
PFGE was performed as previously described using restriction endonuclease SmaI (12), and the banding patterns were analyzed by using Bionumerics version 5.0 (Applied Maths, Austin, TX). Patterns were assigned to USA types or the Brazilian clone using >80% similarity as previously described by McDougal et al. (12). The band-based PFGE typing criteria defined by Tenover et al. for epidemiologic studies were used for PFGE to analyze the outbreak data (16).
The organisms were typed using DL as recommended by the manufacturer. DNA was extracted using a Mo Bio UltraClean microbial DNA isolation kit (Carlsbad, CA), repetitive-element PCR was performed using the DL Staphylococcus kit, and the amplification products were analyzed using DL with microfluidics chip technology. The DL data for each organism consisted of a dendrogram (which presents the results for each organism visually as a banding pattern), a graph of fluorescence intensity corresponding to the banding pattern, and a similarity matrix. DL strain relatedness was defined as a minimum of >95% similarity with a difference of up to one band in the dendrogram. The fluorescence patterns produced by repetitive-element PCR for each potential pair of related isolates were overlaid using the pattern overlay function of the DL software to look for subtle variations in the fluorescence profiles to confirm strain relatedness for both the Project ICARE and the outbreak isolates. Amplifications that yielded low-fluorescence-intensity profiles were repeated with freshly extracted DNA.
RESULTS
DL analysis of isolates of known PFGE type.
DL typing separated the 105 MRSA isolates (which formed 11 distinct clusters by PFGE, representing the 10 USA types and the Brazilian clone), into 11 different clusters (clusters A through K, defined as having >95% similarity) and six singleton patterns (singletons 1 through 6) (Table 1). Eight of 11 DL clusters contained isolates of a single USA type, three of which contained the appropriate USA “type strain.” The remaining three DL clusters contained more than a single USA type. Cluster A contained isolates of USA400, USA700, and USA800, including the type strains for each of those PFGE types. Cluster C was the largest and most diverse of the DL clusters, containing isolates of USA300, USA500, USA800, and the two Brazilian clone isolates. Cluster C also included the type strains for USA300 and USA500. The third cluster (cluster E) contained USA800 and USA1000 isolates.
Most of the USA100 isolates appeared in two closely linked clusters (clusters H and I), which showed an overall similarity of approximately 93%. Two additional USA100 isolates showed unique DL patterns, one of which was similar to a sequence associated with cluster H. Most isolates of PFGE types USA200, USA400, and USA1100 formed separate clusters on the DL dendrogram, although only the USA200 cluster (cluster J) contained its type strain. The USA1100 type strain, although a singleton, was linked to the USA1100 cluster at approximately 85% relatedness. The USA400 type strain was not linked to the other USA400 isolates in cluster D.
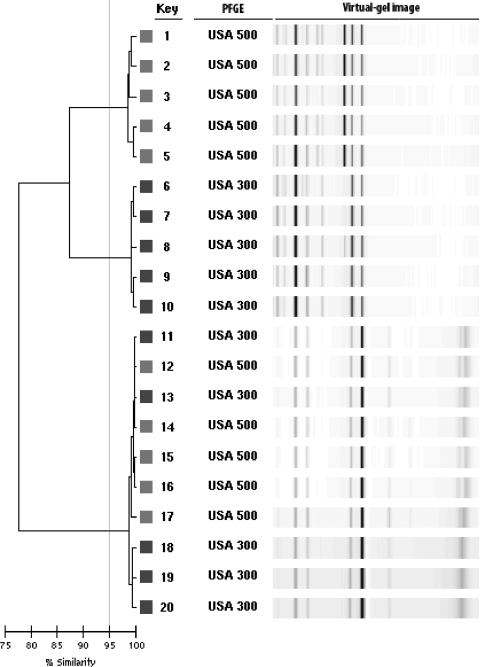
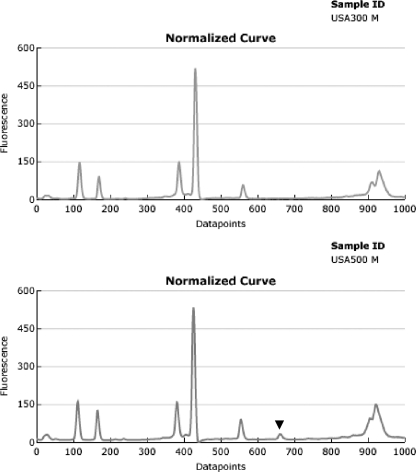
Most of the isolates with PFGE types that are typically in MLST clonal complex 8, including USA300 and USA500, clustered together on the dendrogram at >95% similarity in cluster C. A subset of the isolates in this large cluster is shown in Fig. 1. Isolates 11 through 14 demonstrate the tight clustering of these two USA types. However, the patterns of the USA300 and USA500 isolates could be differentiated by comparing them directly using the pattern overlay function of the DL software. USA500 isolates consistently produced a fluorescence peak at 660 that was not present in the traces of USA300 isolates (Fig. 2).
FIG. 1.
Dendrogram showing USA300 and USA500 isolates clustering together at >95% similarity.
FIG. 2.
Pattern overlay function of the fluorograms showing the unique peak of USA500 isolates at 660 data points that is not present in the patterns of USA300 isolates.
Three isolates of USA600 (cluster K plus a singleton) and single isolates of USA100, USA300, and USA500 that had DL patterns reflecting adequate fluorescence intensity levels were retested because their initial DL patterns clustered with unrelated PFGE types. The repeat DL patterns of all six isolates were consistent, and their groupings did not change. However, repeat testing of two isolates that initially had low-intensity DL patterns (one USA100 and one USA800) resulted in changes in their overall DL patterns when higher fluorescence intensity levels were achieved. This result was primarily due to the appearance of small peaks that were not recognizable in the original pattern. The changes were sufficient to alter the clustering of the two isolates on the dendrogram (moving the USA100 to cluster I and the USA800 to cluster E) (data not shown).
PFGE analysis of “outbreak” isolates.
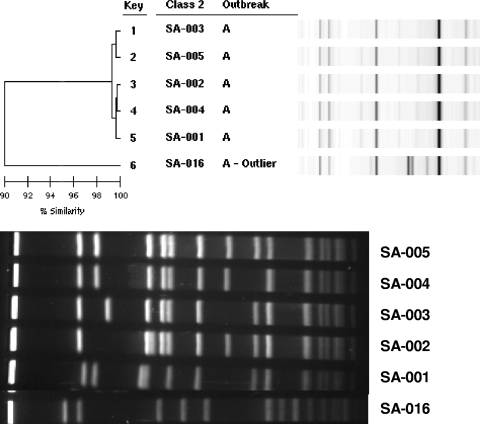
MRSA isolates, previously typed by DL users and designated outbreak related, were typed by PFGE. In the first set of isolates (outbreak A), both DL and PFGE identified the same isolate as having a typing pattern unrelated to those of the others (i.e., the outlier pattern, which for PFGE was USA200). The other five patterns were indistinguishable from one another by DL but showed greater variation by PFGE (Fig. 3). In fact, no two PFGE patterns were alike, even though all were categorized as USA100 (PFGE relatedness of >80%). This degree of PFGE pattern diversity is unusually high for a hospital outbreak, even one that lasted over a 3-month period. Among the isolates from outbreak B, DL again showed five indistinguishable patterns and one outlier pattern. The DL pattern of the five outbreak B isolates was different from the pattern of the outbreak A isolates, even though they were from the same hospital. Once again, all five PFGE patterns were different from each other and distinct from those in outbreak A, even though all of 10 isolates were PFGE type USA100 (data not shown). The outlier PFGE type was a non-USA type pattern. Outbreak C isolates were collected within a 2-day period during a hospital MRSA surveillance study and were all classified by the hospital epidemiologist as “community-associated MRSA.” There was complete concordance between the results of DL and PFGE for this group of isolates. The related isolates all showed indistinguishable USA300 patterns, while the outlier was categorized as USA100. Finally, although the patterns of the five outbreak D isolates were indistinguishable by DL and were different from that of the DL outlier isolate, the PFGE patterns of the all six isolates, including the outlier, were USA100 and the one pattern that showed a subtle 2-band difference was not the isolate designated as the outlier by DL.
FIG. 3.
Comparison of DL results and PFGE patterns for isolates from outbreak A, showing the greater diversity of the PFGE profiles.
DISCUSSION
Strain typing is an important tool for infection control investigations in hospitals, particularly to confirm suspected outbreaks of disease. However, many clinical laboratories do not have the ability to perform strain typing studies on site, often because they lack either the specialized equipment or the technical expertise. PFGE is often considered the gold standard for epidemiologic studies of MRSA because of its high discriminatory power and the strong correlation between PFGE strain types and epidemiologic data (7, 8, 13). Unfortunately, PFGE typing is performed in few clinical laboratories in the United States and sending isolates to a state health department or other reference laboratory can be time consuming and, in the latter case, expensive. The DL system provides a commercial alternative to PFGE for typing both MRSA and methicillin-susceptible Staphylococcus aureus (14, 15) that is simple enough to be performed in most clinical microbiology laboratories.
Our study confirms that DL, although sometimes less discriminatory than PFGE for typing MRSA, can provide information that is useful for infection control investigations in hospitals. Typing of 15 isolates can be completed in as little as 4 hours, and isolates that are unrelated by DL are typically unrelated by PFGE. Thus, DL can be used effectively to rule out outbreaks of MRSA in hospitals. Similar data regarding the ability of DL to discriminate among MRSA strains have been reported by Ross et al. (14), Shutt et al. (15), and te Witt et al. (17). All three studies showed that DL is less discriminatory than PFGE.
A recent report by Limbago et al. (11) describing the PFGE types of MRSA isolates obtained during a population-based study of invasive MRSA infections in the United States in 2005 to 2006 confirms that USA100 and USA300 are the predominant MRSA PFGE types in the United States (8). In our study, USA100 isolates formed a single large cluster of ∼93% similarity, just below the >95% cutoff value defined by DL for “relatedness.” The USA100 type strain pattern from the DL MRSA library was present in the main USA100 cluster, suggesting that at least the USA100 PFGE type could be predicted based on high similarity to the type strain in the DL library. USA300 and USA500 isolates clustered together on the DL dendrogram, which is consistent with recent data suggesting that USA300 isolates are derived from an USA500 ancestor (10). These two PFGE types can be differentiated by using the similarity matrix and the pattern overlay function. Thus, by testing the appropriate USA100 and USA300 control strains and using the similarity matrix and pattern overlay functions, it may be possible to use DL to predict the likelihood that an isolate is USA100 or USA300. This would not be true of USA400, USA700, or USA800, however, since the type strains for these patterns clustered together.
The importance of using the similarity matrix and the pattern overlay functions to compare pairs of isolates that are suspected to have the same strain type is one aspect of the DL software that has not been emphasized in prior studies. In the current study, this aspect was particularly important for differentiating among USA300 and USA500 isolates, which often clustered together in DL dendrograms at >95% similarity. The PFGE patterns of USA300 and USA500 isolates are similar but <80% related using Dice coefficients and the unweighted pair group method for analysis (12), even though both types belong to MLST clonal complex 8. Microbiologists have grown accustomed to using dendrograms for displaying PFGE data (12) and often share dendrograms with infection control personnel as a simple way of conveying information about which isolates from a suspected outbreak of disease are related. While the dendrograms provided by the DL system are helpful for defining broad categories of relatedness among isolates, obtaining the greater detail needed for epidemiologic investigations to confirm strains that belong to the same type requires the use of both the similarity matrix and the pattern overlay function.
A unique feature of this study was the analysis of strains not previously typed by PFGE that were obtained from purported outbreaks of MRSA in hospitals. Among the MRSA outbreak isolates, the PFGE patterns of two of four sets reflected greater pattern diversity than did DL, more so than would be expected during a hospital outbreak lasting 2 or 3 months. The pattern overlay and matrix similarity functions also showed the isolates to be indistinguishable. Because the DL patterns were indistinguishable, it is likely that the strain typing data rather than the epidemiologic data were used to define the outbreaks. The diversity of the PFGE typing results using the interpretive criteria of Tenover et al. (16) argues against the likelihood that these isolates were from a true outbreak of disease. The variability of the PFGE patterns is more consistent with the variations seen in an endemic MRSA strain over time. The differences between the DL and PFGE data for outbreaks A and B emphasize the importance of analyzing the strain typing results in conjunction with epidemiologic information rather than relying solely on the typing data to define an outbreak. This approach poses a potential problem for reference laboratories that receive bacterial isolates for typing without any accompanying epidemiologic data. Thus, the results of typing studies conducted with DL should be reported with caution if no epidemiologic data are available. In most cases, a group of isolates with different DL patterns (i.e., well below the <95% similarity cutoff value) will not be clonal, i.e., are not likely to be part of an outbreak, and can be reported as unrelated with confidence. However, a group of isolates with indistinguishable patterns, as shown here, will not always represent the same USA type or clonal group and thus may not indicate an outbreak. If epidemiologic data are available, and those data provide a link among a group of isolates (e.g., a point source for the organisms has been identified), indistinguishable DL patterns of the isolates provide useful additional evidence that an outbreak exists.
In conclusion, DL is useful for screening isolates of MRSA for potential outbreaks, especially when the analysis includes the use of both the similarity matrix and pattern overlay function to differentiate USA300 isolates from USA500 isolates. Isolates that are unrelated by DL are likely to be unrelated by PFGE, based on our data and those from previous studies (14, 15, 17). However, additional typing may be necessary to confirm strain relatedness or to identify a specific USA strain type or other MRSA type. Thus, PFGE remains superior for discriminating among outbreak strains, confirming strain relatedness, and identifying specific USA types.
Acknowledgments
We thank Nimalie Stone for helpful discussions.
Phase 5 of Project ICARE was supported in part by unrestricted research grants to the Rollins School of Public Health of Emory University by Astra-Zeneca Pharmaceuticals, Wilmington, DE; Elan Pharmaceuticals, San Diego, CA; Johnson & Johnson Pharmaceutical Research & Development, LLC, Raritan, NJ; Pfizer Incorporated, New York, NY; and 3M Health Care Products, St. Paul, MN.
The findings and conclusions in this publication are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests, 9th ed. Approved standard M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.Fridkin, S. K., C. D. Steward, J. R. Edwards, E. R. Pryor, J. E. McGowan, Jr., L. K. Archibald, R. P. Gaynes, F. C. Tenover, and Project Intensive Care Antimicrobial Resistance Epidemiology (ICARE) Hospitals. 1999. Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: Project ICARE phase 2. Clin. Infect. Dis. 29245-252. [DOI] [PubMed] [Google Scholar]
- 3.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 415442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Healy, M., K. Reece, D. Walton, J. Huong, S. Frye, I. I. Raad, and D. P. Kontoyiannis. 2005. Use of the DiversiLab system for species and strain differentiation of Fusarium species isolates. J. Clin. Microbiol. 435278-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Healy, M., K. Reece, D. Walton, J. Huong, K. Shah, and D. P. Kontoyiannis. 2004. Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 424016-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352468-475. [DOI] [PubMed] [Google Scholar]
- 8.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 9.Kontoyiannis, D. P., M. S. Lionakis, R. E. Lewis, G. Chamilos, M. Healy, C. Perego, A. Safdar, H. Kantarjian, R. Champlin, T. J. Walsh, and I. I. Raad. 2005. Zygomycosis in a tertiary-care cancer center in the era of Aspergillus-active antifungal therapy: a case-control observational study of 27 recent cases. J. Infect. Dis. 1911350-1360. [DOI] [PubMed] [Google Scholar]
- 10.Li, M., B. A. Diep, A. E. Villaruz, K. R. Braughton, X. Jiang, F. R. DeLeo, H. F. Chambers, Y. Lu, and M. Otto. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1065883-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limbago, B., G. E. Fosheim, V. Schoonover, C. E. Crane, J. Nadle, S. Petit, D. Heltzel, S. M. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, W. Schaffner, Y. Mu, and S. K. Fridkin for the Active Bacterial Core surveillance (ABCs) MRSA Investigators. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: A population-based analysis. J. Clin. Microbiol. 471344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355666-674. [DOI] [PubMed] [Google Scholar]
- 14.Ross, T. L., W. G. Merz, M. Farkosh, and K. C. Carroll. 2005. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shutt, C. K., J. I. Pounder, S. R. Page, B. J. Schaecher, and G. L. Woods. 2005. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J. Clin. Microbiol. 431187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.te Witt, R., V. Kanhai, and W. B. van Leeuwen. 2009. Comparison of the DiversiLab system, pulsed-field gel electrophoresis and multi-locus sequence typing for the characterization of epidemic reference MRSA strains. J. Microbiol. Methods 77130-133. [DOI] [PubMed] [Google Scholar]
- 18.Wise, M. G., M. Healy, K. Reece, R. Smith, D. Walton, W. Dutch, A. Renwick, J. Huong, S. Young, J. Tarrand, and D. P. Kontoyiannis. 2007. Species identification and strain differentiation of clinical Candida isolates using the DiversiLab system of automated repetitive sequence-based PCR. J. Med. Microbiol. 56778-787. [DOI] [PubMed] [Google Scholar]