Abstract
PCR detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA is the method recommended for use for the diagnosis of HIV-1 infection in infants in limited-resource settings. Currently, testing must be performed in central laboratories, which are usually located some distance from health care facilities. While the collection and transportation of samples, such as dried blood spots, has improved test accessibility, the results are often not returned for several weeks. To enable PCR to be performed at the point of care while the mothers wait, we have developed a vertical filtration method that uses a separation membrane and an absorbent pad to extract cellular DNA from whole blood in less than 2 min. Cells are trapped in the separation membrane as the specimen is collected, and then a lysis buffer is added. The membrane retains the DNA, while the buffer washes away PCR inhibitors, which get wicked into the absorbent blotter pad. The membrane containing the entrapped DNA is then added to the PCR mixture without further purification. The method demonstrates a high degree of reproducibility and analytical sensitivity and allows the quantification of as few as 20 copies of HIV-1 proviral DNA from 100 μl of blood. In a blinded study with 182 longitudinal samples from infants (ages, 0 to 72 weeks) obtained from the Women and Infants Transmission Study, our assay demonstrated a sensitivity of 99% and a specificity of 100%.
Human immunodeficiency virus (HIV) infection is a major cause of death among infants in developing countries. In 2007, there were approximately 420,000 new infections in children worldwide (20). The mortality rate among HIV type 1 (HIV-1)-infected infants is as high as 45% by the first year of age and 59% by the second year (7). The initiation of anti-HIV treatment before 12 weeks of age could reduce rate of infant mortality by 75% (21). Thus, the early diagnosis of HIV infection with timely treatment initiation has the potential to drastically reduce the rate of infant mortality from HIV infection.
While antibody tests are commonly used to diagnose HIV infection in adults, the persistence of passively transferred maternal antibodies make rapid tests inaccurate for infants (2). World Health Organization guidelines recommend the use of proviral DNA PCR or RNA reverse transcription-PCR confirmatory tests for the diagnosis of a suspected HIV infection in infants less than 2 years of age (22). The most commonly used assay is the DNA PCR, which has nearly 100% sensitivity and 100% specificity with samples obtained from infants at 6 weeks of age (17). This also coincides with the time of the first infant immunization, thus increasing the chance of obtaining maximum infant participation in HIV testing if the tests are performed at the same time as the immunization.
In sub-Saharan Africa, the most widely used DNA PCR test is the Amplicor HIV-1 (version 1.5) assay (Roche Diagnostic Systems, Inc., Branchburg, NJ) (17, 23). Performance of the assay requires an extensive laboratory infrastructure and equipment typically located in urban central laboratories. To improve access to PCR testing for remote areas (18), dried blood spots (DBSs) are acquired at remote sites and shipped to central testing laboratories (5). DBS-based testing has dramatically increased the percentage of HIV-exposed infants who are tested in Botswana and South Africa (5). However, logistical problems associated with sample acquisition and transportation can cause substantial delays in returning the results to the DBS collection sites. According to the findings of a survey of seven testing sites in Uganda, 38 to 97% of the results are not returned within 30 days (K. Palamountain, personal communication), producing high rates of loss to follow-up.
Point-of-care testing could reduce the rates of loss to follow-up and significantly increase the number of patients who may receive a diagnosis by performing the test while the mother waits. While quantitative real-time PCR (qPCR) thermal cycling and detection have been performed with low-cost, handheld instruments such as the Bio-Seeq Plus instrument (Smiths Detection Group Ltd., Watford, Herts, United Kingdom), field-deployable, integrated systems which also prepare specimens for amplification are far more complex and expensive. Such systems include the GeneXpert system (Cepheid Inc., Sunnyvale, CA), Smiths Detection's Portable Veterinary Diagnostic Laboratory system, the Liat analyzer (IQuum Inc., Marlborough, MA), and the FL system (Enigma Diagnostics Ltd, San Francisco, CA). The greater expense associated with those assays is primarily a consequence of the use of DNA isolation methods that require multiple solutions and separation steps.
In this study, we demonstrate a novel DNA extraction method, filtration isolation of nucleic acids (FINA), which can be performed by clinic personnel in less than 2 min with specimens collected from heel or finger sticks. It is applicable to testing in central laboratories as well as point-of-care testing. In combination with qPCR, FINA detects proviral DNA with high degrees of sensitivity and specificity.
MATERIALS AND METHODS
Reference strains.
8E5-LAV cells (10) harboring a single copy of the HIV-1 provirus were obtained from the Virology Quality Assurance Laboratory (Rush Presbyterian/St. Luke's Medical Center, Chicago, IL) as frozen cell pellets of 4,000 cells/μl. The cell pellets were thawed, the cell counts were confirmed with a hemocytometer; and standard panel dilutions were made and spiked into fresh anticoagulated whole blood from an HIV-negative volunteer over a concentration range of 0.2 to 400 HIV-1 copies/μl. One hundred microliters of the samples was introduced into the DNA purification protocol.
Clinical samples.
Samples were prepared from 182 frozen infant peripheral blood mononuclear cell (PBMC) pellets collected as part of the Women and Infants Transmission Study (WITS) between 1991 and 1995 (15) and obtained from Clinical Trials and Surveys Corp. (C-TASC), Baltimore, MD. Specimens were drawn at 1, 4, 8, 16, 24, 36, 48, 72, and 96 weeks after birth. Cell counts were obtained with a hemocytometer to assess the quality of the thawed samples. The pellets were reconstituted in HIV-negative, leukocyte-depleted blood (Evanston Hospital, Evanston, IL). Cellular DNA was extracted from 100-μl samples and was performed in duplicate. Samples were considered positive if a threshold cycle (CT) value was obtained for at least one of the replicates. All samples were run with a standard curve prepared with 8E5-LAV cells, and the number of HIV-1 proviral DNA copies per 106 PBMCs was determined. Upon completion of the study, the sample's HIV status was compared to the previously determined plasma HIV-1 RNA levels assayed by the Roche Amplicor HIV-1 Monitor reverse transcription-PCR (15); these data were provided to us by the WITS sample repository (C-TASC) for calculation of the assay's sensitivity and specificity.
DNA isolation.
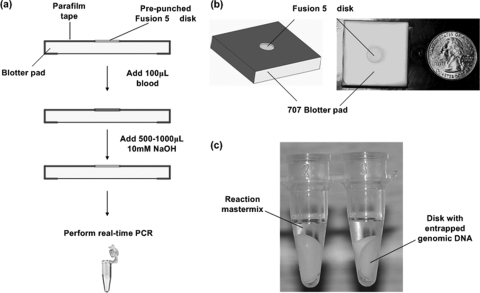
Cellular DNA was extracted from fresh whole blood by our novel FINA method. FINA modules were prepared by sandwiching a Fusion 5 membrane disk (diameter, 7.14 mm; Whatman Inc., Florham Park, NJ) between a square 707 blotter pad (25 by 25 mm; VWR International, West Chester, PA) and a thin sheet of Parafilm with a 5.1-mm-diameter hole in the center such that the disk and the hole on the Parafilm sheet aligned with each other. Pressure was applied on the sandwich by pressing on the surface of the Parafilm to ensure sufficient contact between the disk and the blotter pad. The disk has a particle retention capability of 2.3 μm. Blood was added to the disk, followed by a single wash with 500 to 1,000 μl of 10 mM NaOH until there was the visible clearance of hemoglobin from the disk. A watertight seal between the Parafilm and the Fusion 5 membrane ensured that the blood flowed through the membrane, and good contact between the Fusion 5 membrane and blotter pad assembly ensured rapid wicking of the sample. Preliminary studies demonstrated that the residual NaOH on the disks does not inhibit the subsequent PCRs (unpublished observations). The 10 mM NaOH was added dropwise for most of this study. However, the bulk addition of 600 μl 10 mM NaOH to the FINA module did not adversely affect the extraction process, since the solution formed a droplet on the hydrophobic Parafilm layer and wicked through the membrane in approximately 20 s. After the disk was washed, the disk was separated from the blotter pad with forceps and placed in the PCR tube. Occasionally, the application of excess pressure during the preparation of the FINA modules resulted in partitioning of the membrane layers during separation from the blotter pad. These disks were discarded and the samples were retested with fresh FINA modules.
DNA was extracted from equal volumes of fresh whole blood by use of a QIAamp DNA microkit (Qiagen Inc., Valencia, CA), according to the manufacturer's recommendations. The DNA was eluted from the QIAamp column in 30 μl of DNase- and RNase-free H2O, and 25 μl was introduced into the qPCR mixture to add the maximum amount of sample possible to the qPCR mixture.
qPCR amplification and detection.
PCR detection was performed by the Abbott HIV-1 RealTime assay (Abbott Molecular Inc., Des Plaines, IL) with a 50-μl reaction volume, but the reverse transcription step was omitted, as described previously (19). The sequences of the forward and reverse primers, hybridization probe, and quenching oligonucleotide (QO14) were described previously (19). For FINA, the disk containing the entrapped DNA was added as the PCR template, and for the assay with the QIAamp microkit, 25 μl of eluate was added. All PCRs were run in a Stratagene Mx3005p instrument by using the 6-carboxyfluorescein channel for data collection. Standard curves of the average CT value versus log(copy number) were plotted, and the slope was used to calculate the efficiency of the PCR (13). The limit of detection (LOD) of the FINA method was compared to that of the commercially available QIAamp DNA microkit (Qiagen Inc.) by performing the qPCR with the same PCR mixture but with DNA that had been extracted according to the manufacturer's protocol. The LODs of the extraction methods were determined by testing multiple replicates of 0.05 to 4 copies/μl blood samples. To assess the reproducibility of the FINA method, blood samples containing 40 and 4 HIV-1 copies/μl were tested on two different days, and the intra-assay and interassay reproducibilities were assessed.
Statistical analysis.
CT values were plotted against log(copy number) by using Stratagene's MxPro software to obtain standard curves. Slope parameters were estimated by linear regression of CT values versus log(copy number). For the slope estimates, 95% confidence intervals were obtained by using Prism (version 4.0) software (GraphPad Software, La Jolla, CA). The value of the slope was used to calculate the efficiency of the PCR (13). To assess the variability of the method when it was performed on two different days, a t test was performed with the CT values obtained on both days for blood samples containing 40 and 4 8E5-LAV cells/μl. GraphPad Prism (version 4.0) software was used for the statistical analyses. A two-tailed P value was used to assess the difference between the CT values obtained on the two different days.
RESULTS
Performance of FINA for extraction of HIV proviral DNA.
To extract leukocyte DNA from whole blood at the point of care for PCR, a novel method, FINA, has been developed (Fig. 1a). A cell separation membrane disk is placed in direct contact with an absorbent pad, which drives fluid flow by capillary pressure (Fig. 1b). When 100 μl of whole blood was added on the disk, leukocytes and erythrocytes were trapped in the cell separation membrane, while plasma flowed through into the absorbent pad. Subsequently, the membrane-entrapped cells were lysed by the addition of 500 to 1,000 μl of 10 mM NaOH, which released the cellular DNA. Cell debris, hemoglobin, and other PCR inhibitors were wicked into the absorbent pad, along with excess lysis buffer. The disk with the entrapped DNA was then added directly to a qPCR assay mixture as the template (Fig. 1c).
FIG. 1.
(a) Schematic of DNA extraction by FINA; (b) the in-house FINA module; (c) the disk transferred to a tube for PCR.
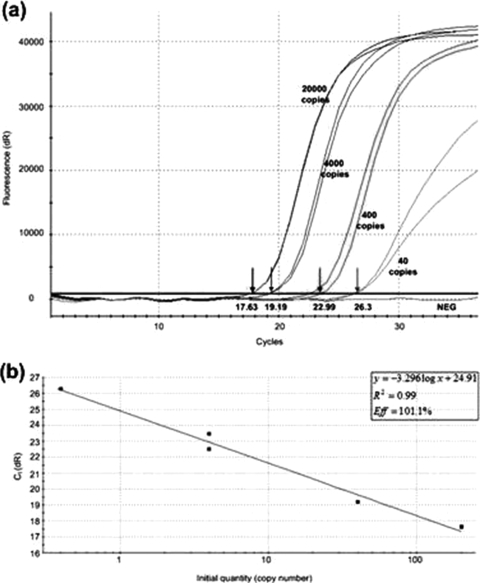
The efficiency of FINA was demonstrated by spiking HIV-negative blood samples with 8E5-LAV cells containing HIV-1 over a concentration range of 0.4 to 200 HIV-1 copies/μl blood. DNA was extracted by the FINA method and was quantified by the commercially available Abbott RealTime HIV-1 assay, which was developed through a combination of the selection of a highly conserved target region and a mismatch-tolerant probe design to equivalently quantify the HIV-1 group M subtypes A to H, group O, and group N isolates found in sub-Saharan Africa (19). The CT values obtained from the amplification plots (Fig. 2a) were used to plot a standard curve for determination of the efficiency of the PCR. The efficiency of the PCR was calculated to be 101.1% (slope, −3.296; 95% confidence interval, −2.941 to −3.653), indicating that no PCR inhibitors were carried over from blood (Fig. 2b).
FIG. 2.
(a) Amplification plots obtained with 100 μl HIV-negative blood spiked with 0.4 to 200 HIV-1 copies/μl blood after DNA extraction by FINA and qPCR. The arrows indicate the average CT values obtained with each of the concentrations. NEG, negative control (HIV-negative blood). (b) Standard curve of average CT value versus log(copy number) obtained with 100 μl HIV-negative blood spiked with 0.4 to 200 HIV-1 copies/μl blood after DNA extraction by FINA and qPCR. The slope of −3.296 indicates a PCR efficiency (Eff) of 101.1%.
FINA was compared to conventional genomic DNA purification with the QIAamp DNA microkit by performing qPCR with DNA extracted by both methods. Replicate whole-blood samples spiked with 0.05 to 4 8E5-LAV cells/μl were extracted and assayed by qPCR. By FINA, 0.2 HIV-1 copies/μl, or a total of 20 HIV-1 copies, could be detected 100% of the time, which is an LOD similar to the LOD obtained with the QIAamp purification kit (Table 1). The LOD of the Roche Amplicor HIV-1 DNA test (version 1.5) has been reported to be 33 copies per reaction (11). This corresponds to 0.3 copies/μl when 100 μl whole blood is used. The LOD of the FINA PCR was therefore equivalent to that of the Roche Amplicor HIV-1 DNA PCR test (version 1.5).
TABLE 1.
Determination of LOD of FINA and QIAamp extraction methodsa
| No. of HIV-1 copies/μl blood | Total no. of HIV-1 copies before extraction | % Detectionb
|
|
|---|---|---|---|
| FINAc | QIAamp assayd | ||
| 4 | 400 | 100 | 100 |
| 2 | 200 | 100 | 100 |
| 0.4 | 40 | 100 | 75 |
| 0.2 | 20 | 100 | 75 |
| 0.1 | 10 | 50 | Not tested |
| 0.05 | 5 | 33.3 | Not tested |
The LOD of the FINA-PCR is equivalent to that of the Roche Amplicor HIV-1 DNA PCR test (version 1.5), which has an LOD of 33 copies in one reaction.
Percent detection = 100 × (number of samples detected/number of samples tested).
Six replicates were tested for each concentration.
Four replicates were tested for each concentration.
We assessed the reproducibility of the FINA method by testing 10 blood samples containing 40 and 4 HIV-1 copies/μl on two different days (Table 2). A t test indicated that there was no significant difference (P = 0.53) between the runs on the two different days.
TABLE 2.
CT values obtained by the FINA extraction method for concentrations of 40 and 4 HIV-1 copies/μl blood on two different days
| No. of HIV-1 copies/μl blooda |
CT value (avg ± SD)
|
|
|---|---|---|
| Day 1 | Day 2 | |
| 40 | 20.35 ± 0.29 | 20.69 ± 0.25 |
| 4 | 23.67 ± 0.41 | 23.66 ± 0.28 |
Ten replicates were tested for each concentration.
Evaluation of FINA performance with clinical infant samples.
To determine the sensitivity and the specificity of the FINA extraction method, 182 blinded samples were obtained from WITS (15) between 1991 and 1995. Samples were obtained longitudinally from infants at times ranging from 1 week through 24 months after birth, and PBMC pellets were stored at −80°C until this study. The frozen PBMC pellets were reconstituted with HIV-negative, leukocyte-depleted blood to yield reconstituted whole-blood samples; the cellular DNA was extracted and amplified as described above. Upon completion of the study, the results were compared to the plasma viral load data provided by WITS researchers (15). A total of 86 true-positive results, 95 true-negative results, and 1 false-negative result were obtained; no false-positive results were obtained. The HIV proviral DNA concentration in the frozen pellets ranged from 19 to 55,000/106 PBMCs. The sensitivity was calculated to be 98.8% (95% confidence interval, 92.9 to 99.9%), and the specificity was calculated to be 100% (95% confidence interval, 95.2 to 100%). In order to achieve this high rate of sensitivity, the samples were tested in duplicate because many of the samples tested had very low PBMC counts. Infants typically have approximately 5,000 PBMCs/μl (1, 16); however, a number of the samples tested in this study had less than 200 PBMCs/μl. Five samples that had small amounts of PBMCs had discordant duplicate results and were designated positive on the basis of the single positive PCR result. It is not clear what caused the one duplicate false-negative result in this study. We retested this sample in duplicate, and the results of both tests were again negative. The corresponding plasma viral load for this time point was 77,900 copies/ml, and specimens from this patient were positive at both an earlier time point and a later time point, indicating that the assay could detect the patient's HIV subtype. The sample did have a low PBMC count (106 cells/μl), so it may be that the number of cells harboring HIV DNA was below our assay's level of detection.
Stability of cellular DNA on FINA disks.
The stability of the DNA entrapped in the FINA modules was investigated to determine the effect of delays in PCR testing. HIV-positive 8E5-LAV cells (0.8 and 8 HIV-1 copies/μl blood) were spiked into whole blood, and 100 μl was added to the FINA module and dried. The modules were stored at 37°C and assayed at 0, 1, 2, 3, 4, and 5 weeks after a single wash with 10 mM NaOH was performed. The temperature was chosen to match the high ambient temperatures in sub-Saharan Africa (4). HIV was detected in all of the samples with 0.8 and 8 HIV-1 copies/μl of blood at all the time points (Fig. 3). With the exception of samples tested at week 2, the copy numbers in all samples were quantified to ±0.5 log unit of the original copy number. It was observed that for samples amplified at week 2, the saturation fluorescence was much lower than that for samples tested at the other time points, indicating possible inhibition of the PCR. These results indicate that DNA is at most only mildly affected by storage in FINA modules for up to 5 weeks.
FIG. 3.
Plots showing the effect of storage on HIV-1 proviral DNA. The graphs show the mean log(copy number) versus storage time (in weeks) when the starting concentration was 8 copies/μl (a total of 800 copies before extraction by FINA) (a) and 0.8 copies/μl (a total of 80 copies before extraction by FINA) (b). The bars indicate ±1 standard deviation.
DISCUSSION
This report describes a rapid and effective technique for the isolation of cellular DNA from whole blood which could be performed by health care workers at the point of care. It utilizes a sandwich filtration module and capillary pressure to produce qPCR-ready DNA in less than 2 min without the use of precision pipetting or laboratory equipment. In addition to the Fusion 5 membrane described here, other cell separation membranes, such as VF1, MF1, and LF1 (Whatman Inc.), have successfully been used; and the FINA method is also compatible with all commonly used blood collection anticoagulants (S. R. Jangam and D. H. Yamada, unpublished data). The template from the membrane disks was successfully amplified by using different real-time PCR instruments, including the Stratagene Mx3005p instrument (Stratagene, La Jolla, CA), the SmartCycler instrument (Cepheid Inc.), and the ABI StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA). This highly flexible method can also be used to amplify any genetic targets found in blood. Smaller volumes of blood can be processed for the amplification of abundant targets, and larger volumes can be processed for the amplification of less abundant targets. The blood samples are stable during storage on the FINA modules and can be collected under field conditions for later shipment to central laboratories for PCR analysis, making this method ideal for use in epidemiological studies.
We are developing the FINA module to be integrated into a point-of-care qPCR device for the diagnosis of HIV infection in infants. Because maternal antibodies persist in infant blood for up to 18 months after birth, the detection of HIV in infants requires direct virological testing, such as testing of nucleic acid for proviral DNA or viral RNA (5). RNA testing has been shown to have a higher sensitivity with samples collected at between 0 and 6 weeks of age, while both RNA and DNA testing have ∼100% sensitivity with samples collected at 6 to 7 weeks of age (6). In the absence of breast-feeding, the mother-to-child transmission of HIV takes place perinatally, and these infections can be detected by 4 to 6 weeks of age by either RNA or DNA testing. Testing at this time point is ideal, because it also corresponds to the time when the child will be entering the health care system for immunizations (5). If the infant is breast-fed, testing should take place 4 to 6 weeks after the child is weaned. FINA purification plus qPCR will provide a simple and affordable approach for the early diagnosis of HIV-1 infection in infants.
Combined with the Abbott RealTime HIV-1 assay, DNA extracted by the FINA method had an LOD of 20 copies/100 μl of whole blood, which is equivalent to the LOD of more complex extraction procedures (9). The proviral DNA loads in most HIV-1-infected infants is in excess of 200 copies/106 PBMCs (1), which would readily be detected by our assay. However, because several of the WITS samples had very low PBMC counts (<200 cells/μl), it was necessary to perform the study in duplicate to achieve the reported 99% sensitivity level. Five of the samples with low PBMC counts showed discordant results by duplicate tests, indicating that the probability of detecting HIV-1 in positive samples with <200 cells/μl was 50%.
Another potential limitation to the study was the limit of quantification (400 viral copies/ml of plasma) of the Roche Amplicor HIV-1 Monitor test used by the WITS researchers (15). Although the comparison would have been more rigorous with recently developed viral load assays with limits of quantification as low as 40 to 50 copies/ml, such as the Abbott HIV-1 RealTime assay or the ultrasensitive Roche Amplicor HIV-1 Monitor test (version 1.5), sample volume limitations prevented determination of the viral loads by those assays. However, analysis of the data reveals that this potential detection gap would have affected only two of the longitudinally evaluated subjects: one infant had an undetectable viral load at birth but had a detectable viral load (11,100 copies/ml) at 4 weeks, and the second infant had an undetectable viral load at birth and at 4 weeks of age but had a detectable viral load (34,029 copies/ml) at the 16-week time point. It is possible that these three samples could have had viral loads of between 40 and 400 copies/ml and thus would have been false negative if virus had been detected by the more sensitive reference viral load assay. Classification of the results for these samples as false negative would decrease the sensitivity to 95.4%.
An additional limitation of our WITS samples was that the previously frozen PBMC pellets were spiked into leukocyte-depleted blood to model fresh infant blood. Infant blood has a cellular composition different from that of adult blood (8), which could have the potential to affect the results of the FINA purification. A prospective study with fresh infant blood samples will be needed to establish the performance of the FINA PCR with clinical samples. Because the rate of mother-to-child transmission of HIV-1 is very low in the United States (79 infants were diagnosed with HIV-1 infection in 2007), these tests will need to be performed in a country with a higher prevalence of HIV infection among infants (3). With the specimens available to us, the findings of our current studies indicate that the performance of the FINA method is equivalent to that of conventional methods for DNA purification from blood, but the time and complexity of DNA extraction are significantly reduced. This eliminates the need for a fully equipped laboratory and may enable testing to be moved closer to the patients.
Acknowledgments
This study was supported by the Bill and Melinda Gates Foundation Grand Challenges in Global Health grant 37774. Real-time PCR reagents and advice were provided by Abbott Molecular Inc., Des Plaines, IL. 8E5-LAV cells were provided by the Virology Quality Assurance Laboratory, Rush Presbyterian/St. Luke's Medical Center. HIV-negative blood was provided by Evanston Hospital. WITS samples were provided by C-TASC.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Beck, I. A., K. D. Drennan, A. J. Melvin, K. M. Mohan, A. M. Herz, J. Alarcon, J. Piscoya, C. Velazquez, and L. M. Frenkel. 2001. Simple, sensitive and specific detection of human immunodeficiency virus type 1 subtype B DNA in dried blood samples for diagnosis in infants in the field. J. Clin. Microbiol. 3929-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryson, Y. J., S. Pang, L. S. Wei, R. Dickover, A. Diagne, and I. S. Chen. 1995. Clearance of HIV infection in a perinatally infected infant. N. Engl. J. Med. 332833-838. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2007. HIV AIDS surveillance report—cases of HIV infection and AIDS in the United States and dependent areas, vol. 19. Centers for Disease Control and Prevention, Atlanta, GA.
- 4.Craig, M. H., R. W. Snow, and D. le Sueur. 1999. A climate-based distribution model of malaria transmission in sub-Saharan Africa. Parasitol. Today 15105-111. [DOI] [PubMed] [Google Scholar]
- 5.Creek, T. L., G. G. Sherman, J. Nkengasong, L. Lu, T. Finkbeiner, M. G. Fowler, E. Rivadeneira, and N. Shaffer. 2007. Infant human immunodeficiency virus diagnosis in resource-limited settings: issues, technologies, and country experiences. Am. J. Obstet. Gynecol. 197S64-S71. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, C., K. Charbonneau, T. Tina, K. Song, D. Patterson, T. Sullivan, T. Cummins, and B. Poiesz. 1999. Comparison of human immunodeficiency virus 1 DNA polymerase chain reaction and qualitative and quantitative RNA polymerase chain reaction in human immunodeficiency virus 1-exposed infants. Pediatr. Infect. Dis. J. 1830-35. [DOI] [PubMed] [Google Scholar]
- 7.Dabis, F., and E. R. Ekpini. 2002. HIV-1/AIDS and maternal and child health in Africa. Lancet 3592097-2104. [DOI] [PubMed] [Google Scholar]
- 8.Fischbach, F. 2003. A manual of laboratory and diagnostic tests. Lippincott Williams & Wilkins, Philadelphia, PA.
- 9.Fischer, A., C. Lejczak, C. Lambert, J. Servais, N. Makombe, J. Rusine, T. Staub, R. Hemmer, F. Schneider, J. C. Schmit, and V. Arendt. 2004. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J. Clin. Microbiol. 4216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folks, T. M., D. M. Powell, and M. A. Martin. June 1988. Cell line producing AIDS viral antigens without producing infectious virus particles. U.S. patent 4,752,565.
- 11.Germer, J. J., T. M. Gerads, J. N. Mandrekar, P. S. Mitchell, and J. D. Yao. 2006. Detection of HIV-1 proviral DNA with the AMPLICOR HIV-1 DNA test, version 1.5, following sample processing by the MagNA Pure LC instrument. J. Clin. Virol. 37195-198. [DOI] [PubMed] [Google Scholar]
- 12.Huang, S., J. Salituro, N. Tang, K. C. Luk, J. Hackett, P. Swanson, G. Cloherty, W. B. Mak, J. Robinson, and K. Abravaya. 2007. Thermodynamically modulated partially double-stranded linear DNA probe design for homogeneous real-time PCR. Nucleic Acids Res. 35e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutfalla, G., and G. Uze. 2006. Performing quantitative reverse-transcribed polymerase chain reaction experiments. DNA microarrays. Part A. Array platforms and wet-bench protocols. Methods Enzymol. 410386-400. [DOI] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Shearer, W. T., T. C. Quinn, P. LaRussa, J. F. Lew, L. Mofenson, S. Almy, K. Rich, E. Handelsman, C. Diaz, M. Pagano, V. Smeriglio, L. A. Kalish, et al. 1997. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N. Engl. J. Med. 3361337-1342. [DOI] [PubMed] [Google Scholar]
- 16.Shearer, W. T., H. M. Rosenblatt, R. S. Gelman, R. Oyomopito, S. Plaeger, E. R. Stiehm, D. W. Wara, S. D. Douglas, K. Luzuriaga, E. J. McFarland, R. Yogev, M. H. Rathore, W. Levy, B. L. Graham, and S. A. Spector. 2003. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol. 112973-980. [DOI] [PubMed] [Google Scholar]
- 17.Sherman, G. G., P. A. Cooper, A. H. Coovadia, A. J. Puren, S. A. Jones, M. Mokhachane, and K. D. Bolton. 2005. Polymerase chain reaction for diagnosis of human immunodeficiency virus infection in infancy in low resource settings. Pediatr. Infect. Dis. J. 24993-997. [DOI] [PubMed] [Google Scholar]
- 18.Sherman, G. G., G. Stevens, S. A. Jones, P. Horsfield, and W. S. Stevens. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J. Acquir. Immune Defic. Syndr. 38615-617. [DOI] [PubMed] [Google Scholar]
- 19.Tang, N., S. Huang, J. Salituro, W.-B. Mak, G. Cloherty, J. Johanson, Y. H. Li, G. Schneider, J. Robinson, J. Hackett, Jr., P. Swanson, and K. Abravaya. 2007. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J. Virol. Methods 146236-245. [DOI] [PubMed] [Google Scholar]
- 20.UNAIDS and WHO. 2007. AIDS epidemic update. WHO, Geneva, Switzerland.
- 21.Violari, A., M. F. Cotton, D. M. Gibb, A. G. Babiker, J. Steyn, S. A. Mahdi, P. Jean-Phillip, and J. A. McIntyre for the CHER Study Team. 2008. Early antiretroviral therapy and mortality among HIV-infected infants. N. Engl. J. Med. 3592233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. 2007. Early detection of HIV infection in infants and children. Guidance note on the selection of technology for the early diagnosis of HIV in infants and children. WHO, Geneva, Switzerland.
- 23.Zijenah, L. S., J. Humphrey, K. Nathoo, L. Malaba, P. Zvandasara, A. Mahomva, P. Iliff, and M. T. Mbizvo. 1999. Evaluation of the prototype Roche DNA amplification kit incorporating the new SSK145 and SKCC1B primers in detection of human immunodeficiency virus type 1 DNA in Zimbabwe. J. Clin. Microbiol. 373569-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]