Abstract
A reverse transcription-PCR (RT-PCR) assay was designed, according to previously determined and newly derived genetic data, to target S genomic segments of 47 viruses, including 29 arthropod-borne human pathogens, of the family Bunyaviridae. The analytical sensitivity of the presented assay was evaluated through its application to RNAs extracted from quantitated dilutions of bunyaviruses of interest. Additionally, the assay's analytical specificity was determined through the evaluation of RNAs extracted from selected bunyaviruses and other representative arthropod-borne viruses isolated from a diverse group of host species and temporal and geographic origins. After RT-PCR amplification, DNAs amplified from bunyaviruses of interest were subjected to a novel multiplex sequencing method to confirm bunyavirus positivity and provide preliminary, species-level S segment identification. It is our goal that this assay will be used as a tool for identification and characterization of emergent arthropod-borne bunyavirus isolates of medical import as well as related viruses of the family Bunyaviridae that have not been associated with human illness.
The virus family Bunyaviridae includes more than 300 distinct members, with more than 60 viruses associated with human illness predominantly organized into four genera, Orthobunyavirus, Hantavirus, Nairovirus, and Phlebovirus, according to structural, genetic, and antigenic characteristics (2, 9, 25). Exemplifying the great diversity of species classified within the family, the fifth genus of the family Bunyaviridae, the genus Tospovirus, contains viruses that are known to cause diseases in plants and not animals (2, 9, 29). Viruses of the family Bunyaviridae are highly diverse, with members of each genus additionally classified into individual groups, subtypes, and complexes according to serological character (2, 6, 9, 29). Most bunyaviruses are arthropod-borne, with vectors of transmission including mosquitoes, ticks, culicoid flies, and thrips species (2, 25). However, members of the genus Hantavirus are not associated with an arthropod vector and are thought to be transmitted from infected rodents to humans through aerosolized excreta (2). Bunyaviruses are associated with significant human illnesses worldwide and cause a range of clinical manifestations, including severe pediatric encephalitis (associated with La Crosse virus, of the genus Orthobunyavirus), retinal and hepatic disease (associated with Rift Valley fever virus, of the genus Phlebovirus), and hemorrhagic fever (associated with Crimean-Congo hemorrhagic fever (CCHF) virus, of the genus Nairovirus) (2, 25). Bunyaviruses are enveloped, containing a tripartite genome of mostly negative polarity. The three genomic segments, L, M, and S, can occur in end-hydrogen-bonded circularized forms and minimally encode an RNA-dependent RNA polymerase, envelope glycoproteins (Gn and Gc), and a nucleocapsid (N) protein, respectively (2, 28). The segmented nature of the bunyavirus genome allows the possibility of evolution through antigenic shift. In fact, bunyavirus segment reassortment has been associated with outbreaks of human disease (4, 5, 10). However, the role of segment reassortment in bunyavirus evolution and pathogenicity is largely unknown due to the lack of comprehensive sequence data for members of the family Bunyaviridae (10).
It is believed that arthropod-borne viruses of the family Bunyaviridae will continue to be agents of public health import throughout the 21st century, as has been suggested by their frequent and devastating emergence in recent years (3, 10, 31). In our reference laboratory, we have a specific mission to detect arthropod-borne human pathogens of the family Bunyaviridae as they emerge. To support surveillance, outbreak investigation, and diagnosis, virus isolation is an invaluable tool for the detection of arthropod-borne bunyaviruses. The isolation of arthropod-borne bunyaviruses is performed through the inoculation of either suckling mice or susceptible cells (e.g., Vero cells) with sera or supernatants of homogenates derived from tissues of infected individuals or mosquito pools. Following isolation, identification and characterization of newly derived bunyavirus isolates have been historically performed through the use of predominantly antibody-based methods, including immunofluorescence and plaque reduction neutralization assays (23, 30). While these methods are irreplaceable for the identification of bunyaviruses in a group-specific manner, they are time-consuming and can be limited in their ability to generate unequivocal, species-identifying results due to antibody cross-reactivity and/or limited diversity of available antibodies. In addition, the antibodies required for these assays can be difficult and time-consuming to generate as supplies deplete over time.
The advent of molecular technologies, such as reverse transcription-PCR (RT-PCR), and the application of these technologies in diagnostic and reference laboratories provides a time-efficient alternative to traditional serological methods for the identification of virus isolates. An attractive quality of nucleic acid-based assays is the inherent flexibility of design which allows a researcher to develop an assay with various degrees of specificity, depending on his or her laboratory needs. Additionally, the application of RT-PCR-based molecular consensus assays, designed to detect a group of viruses of interest, followed by nucleotide sequencing for result confirmation and virus speciation has proven a powerful tool for emergent virus identification and discovery when applied to virus isolates in our laboratory (20, 21, 22). However, historically, molecular identification of bunyavirus isolates has been prevented due to limited nucleotide sequence data available for many of these agents and the high level of diversity among bunyaviruses of determined genetic character. Despite these significant challenges, it is proposed that the majority of arthropod-borne human pathogens within the family Bunyaviridae can be identified and characterized by a single RT-PCR-based assay.
To evaluate this assertion, we determined both full- and partial-length S segment primary nucleotide sequence data for 16 bunyaviruses of previously unknown genetic character as targets for assay design and detection prior to the development of the presented assay (15; unpublished data). Here, we describe the development of an RT-PCR-based molecular consensus assay incorporating a diverse set of oligonucleotide primers for the detection of 47 viruses of the Orthobunyavirus, Nairovirus, and Phlebovirus genera of the family Bunyaviridae, including 29 human pathogens of newly derived and previously determined genetic character. Because of the high degree of characterization across genera and the relative conservation within genera, the S segment N open reading frame (ORF) is the genomic target of the presented assay.
The format of the presented RT-PCR-based molecular consensus assay includes the application of two supermixes of oligonucleotides for the detection of RNAs extracted from target viruses of (i) the Orthobunyavirus genus and (ii) the Nairovirus and Phlebovirus genera of the family Bunyaviridae. For S segment species determination, RT-PCR-amplified bunyaviral cDNAs were subjected to a novel multiplex sequencing method followed by NCBI BLAST analyses of newly derived nucleotide sequence data. Based on S segment species determination, this assay is being presented as a tool for the preliminary identification of medically important, and serologically related, viruses of the Orthobunyavirus, Nairovirus, and Phlebovirus genera of the family Bunyaviridae. While there are existing molecular consensus assays for the detection of serogroups of viruses within the family Bunyaviridae (11, 13, 27), to the best of our knowledge, the presented assay detects a larger diversity of bunyaviruses than any previously published effort. It is our hope that this assay will be used as a complement or alternative to standard antibody-based methods for the identification and characterization of bunyaviruses isolated from a variety of geographic locations and sources.
MATERIALS AND METHODS
All procedures were performed according to manufacturer instructions unless stated otherwise.
Viruses.
Viruses used in this study were provided by the Arbovirus Diseases Branch of the Division of Vector-Borne Infectious Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention (CDC), and from the World Health Organization arthropod-borne virus reference collection. Viruses selected for evaluation included human pathogens and serologically related viruses of the family Bunyaviridae (Table 1). Viruses were identified at the species level through serological analyses prior to the beginning of this study. In a few cases, regulations in handling live viruses prevented our ability to work with these agents in our laboratory. For alternatives, we obtained Nairobi sheep disease virus RNA and CCHF virus plasmid-derived RT-PCR controls from George Ludwig (USAMRIID) and Rosemary Sang (Kenya Medical Research Institute), respectively. For quantitation, human pathogens (Table 1) were subjected to plaque titration of 10-fold dilutions in Vero cells prior to molecular evaluation. Additional viruses of the family Bunyaviridae, as well as representative arthropod-borne viruses of the Alphavirus and Flavivirus genera, of the virus families Togaviridae and Flaviviridae, were evaluated to determine assay specificity (Tables 1 and 2).
TABLE 1.
Bunyaviruses that are targeted and amplified by the molecular consensus assaya
| Genus/serogroup | Virusb | Strain | Source, location, and yr of isolation | RT-PCR result/serological identity confirmed by nucleotide sequencing and BLAST analysis | Detection limit (PFU/0.1 ml)c |
|---|---|---|---|---|---|
| Orthobunyavirus | |||||
| Bunyamwera | Bunyamwera virus | Original | Aedes spp., Uganda, 1943 | POS/yes | 70 |
| Cache Valley virus | 6V633 | Culiseta inornata, Utah, 1956 | POS/yes | 600 | |
| Fort Sherman virus* | 86MSP18 | Human, Panama, 1985 | POS/yes | 100 | |
| Ilesha virus | KO/2 | Human, Nigeria, 1957 | POS/no | 300 | |
| Shokwe virus* | Sa Ar 4042 | Aedes cumminsii, South Africa, 1962 | POS/yes | 75 | |
| Wyeomyia virus* | Original | Wyeomyia melanocephala, Colombia, 1940 | POS/yes | 660 | |
| Xingu virus* | BeH 388464 | Human, Brazil, unknown date | POS/yes | 4 | |
| Ngari (Garissa) virus | Not evaluated | Not evaluated | ND | ND | |
| Germiston virus | Not evaluated | Not evaluated | ND | ND | |
| Maguari virus | 75V3429 | Aedes scapularis, Ecuador, 1974 | POS/no | ND | |
| Potosi virus | 89-3380 | Aedes albopictus, Missouri, 1989 | POS/yes | ND | |
| Northway virus | 234 | Aedes spp., Alaska, 1971 | POS/yes | ND | |
| Biroa virus | DakArB 2198 | Anopheles pharoensis, CAR, 1969 | POS/yes | ND | |
| Batai virus | 184 | Anopheles maculipennis, Czechoslovakia, 1950 | POS/yes | ND | |
| Bwamba | Bwamba virus* | M 459 | Human, Uganda, 1937 | POS/yes | 200 |
| Pongola virus* | Sa Ar 1 | Aedes circumluteolus, South Africa, 1955 | POS/yes | 200 | |
| Simbu | Oropouche virus | TRVL 9760 | Human, Tobago, 1955 | POS/yes | 0.1 |
| California | California encephalitis virus | BFS 283 | Aedes melanimon, California, 1944 | POS/yes | 6 |
| Inkoo virus | KN 3641 | Aedes communis punctor, Finland, unknown date | POS/yes | 1.3 | |
| Jamestown Canyon virus | 61V2235 | Culiseta inornata, Colorado, 1967 | POS/yes | 60 | |
| La Crosse virus | Original | Human, Wisconsin, 1964 | POS/yes | 200 | |
| Snowshoe hare virus | LEIV 13004AKH | Aedes communis; USSR, 1986 | POS/no | 34 | |
| Tahyna virus | Bardos 92 | Aedes caspius, Czechoslovakia, 1958 | POS/yes | 340 | |
| Serra do Navio virus | BeAr 103645 | Aedes spp., Brazil, 1966 | POS/yes | ND | |
| South River virus | NJ0-94F | Anopheles crucians, New Jersey, 1966 | POS/yes | ND | |
| Jerry Slough virus | BFS 4474 | Culiseta inorata, California, 1963 | POS/yes | ND | |
| Nairovirus | |||||
| CCHF virus | CCHF virus | S segment cDNA | Plasmid-derived PCR control | POS/yes | ND |
| Hazara virus | PakJC 280 | Ixodes redikorzevi, Pakistan, 1964 | POS/yes | ND | |
| NSD | Dugbe virus | IbAr 1792 | Amblyomma variegatum, Nigeria, 1964 | POS/yes | 2 |
| Nairobi sheep disease virus | RV082 | RNA control from Kenyan strain | POS/yes | ND | |
| Phlebovirus (SF) | Alenquer virus* | BeH 301101 | Human, Brazil, 1976 | POS/yes | 0.02 |
| Chandiru virus* | BeH 22511 | Human, Brazil, 1960 | POS/yes | 0.3 | |
| Chagres virus* | JW 10 | Human, Panama, 1960 | POS/yes | 0.0024 | |
| Punta Toro virus | D 4021A | Human, Panama, 1966 | POS/yes | 32 | |
| Rift Valley fever virus | MP 12 | Vaccine strain | POS/yes | 100 | |
| Sandfly fever Naples virus | Original | Human, Naples, Italy, 1944 | POS/yes | 0.23 | |
| Toscana virus | IssPhl 3 | Phlebotomus perniciosus, Tuscany, Italy, 1971 | POS/yes | 0.95 | |
| Sandfly fever Sicilian virus | Original | Human, Sicily, Italy, 1943 | POS/yes | 0.0045 | |
| Cacao virus* | VP 437R | Lutzomyia trapidoi, Panama, 1970 | POS/yes | ND | |
| Buenaventura virus | CoAr 3319 | Lutzomyia spp., Colombia, 1964 | POS/yes | ND | |
| Frijoles virus | VP 161A | Lutzomyia spp., Panama, 1969 | POS/yes | ND | |
| Itaituba virus | bean 213542 | Didelphis masupialis, Brazil, 1971 | POS/yes | ND | |
| Itaporanga virus* | Original | Sentinal mouse, Brazil, 1962 | POS/yes | ND | |
| Gabek Forest virus* | SudAn 754-61 | Acomys albigena, Sudan, 1961 | POS/yes | ND | |
| Turuna virus* | Bear352492 | Lutzomyia spp., Brazil, 1978 | POS/yes | ND | |
| Rio Grande virus* | TBM3-204 | Neotoma micropus, Texas, 1973 | POS/yes | ND | |
| Nique virus* | 9c | Lutzomyia panamensis, Panama, 1972 | POS/yes | ND |
Known human pathogens appear in bold. NSD, Nairobi sheep disease virus group; SF, Sandfly fever virus group; CAR, Central African Republic; POS, positive; PFU, PFU equivalent; ND, not determined.
Ngari and Germiston viruses were not evaluated due to a lack of availability (Ngari virus) and handling restrictions (Germiston virus). *, S segment sequence data were determined for this virus prior to this study.
PFU equivalent determined through plaque titration of virus dilutions in Vero cells.
TABLE 2.
Arthropod-borne human pathogens that are not amplified by the molecular consensus assaya
| Family (genus) | Serogroup | Virusb | Strain | Source, location, and yr of isolation |
|---|---|---|---|---|
| Bunyaviridae (Orthobunyavirus) | Nyando | Nyando virus | MP 401 | Eretmapodites chrysogaster, Cameroon, 1966 |
| California | Guaroa virus | CoH 352111 | Human, Colombia, 1956 | |
| Group C | Apeu virus | BeAn 848 | Cebus apella, Brazil, 1955 | |
| Caraparu virus | BeAn 3994 | Cebus apella, Brazil, 1956 | ||
| Itaqui virus | BeAn 1297 | Sentinal mouse, Brazil, 1959 | ||
| Madrid virus | BT 4075 | Human, Panama, 1961 | ||
| Marituba virus | BeAn 15 | Cebus apella, Brazil, 1954 | ||
| Nepuyo virus | BeAn 10709 | Sentinal mouse, Brazil, 1959 | ||
| Oriboca virus | BeAn 17 | Sentinel monkey, Brazil, 1954 | ||
| Restan virus | TRVL 51144 | Culex spp., Trinidad, 1963 | ||
| Guama | Catu virus | BeH 151 | Human, Brazil, 1955 | |
| Guama virus | BeAn 277 | Cebus apella, Brazil, 1955 | ||
| Flaviviridae | Dengue virus group | Dengue 2 virus | New Guinea C | Human, New Guinea, 1944 |
| (Flavivirus) | Yellow fever virus group | Yellow fever virus | 17D | Vaccine strain |
| Spondweni virus group | Zika virus | YAP-human, 2007 | RNA transcript derived from PCR-amplified DNA | |
| Japanese encephalitis virus group | St. Louis encephalitis virus | TBH-28 | Human, Florida, 1962 | |
| Japanese encephalitis virus group | West Nile virus | NY99 | Phoenicopterus ruber, New York, 1999 | |
| Japanese encephalitis virus group | Japanese encephalitis virus | SA-14-2-8 | Vaccine strain | |
| Togaviridae (Alphavirus) | Venezuelan equine encephalomyelitis virus complex | Venezuelan equine encephalomyelitis virus | TC83 | Vaccine strain |
| Western equine encephalitis virus complex | Western equine encephalitis virus | McMillan | Human, Ontario, Canada, 1941 | |
| Semliki Forest virus complex | ONNV | UgMP 30 | Human, Uganda, 1959 | |
| Semliki Forest virus complex | CHIKV | Original | Human, Tanganyika, 1953 |
All viruses were evaluated with both the Orthobunyavirus and Phlebovirus/Nairovirus primer mixes.
All evaluated viruses have a minimum titer of 10,000 PFU/ml in Vero cells. ONNV, O'nyong-nyong virus; CHIKV, Chikungunya virus.
Recently derived orthobunyavirus isolates of international origins.
A panel of recently derived orthobunyavirus isolates was assembled to evaluate the presented assays' ability to detect target RNAs from a diversity of origins (see Table 4). Isolates from Africa and China were derived from mosquito pools and described and characterized by others prior to their evaluation in our laboratory (8, 24).
TABLE 4.
Detection and identification of recently derived orthobunyavirus isolates by the molecular consensus assaya
| Genus/serogroup | Virus | Source, location/origin, and yr of isolation | RT-PCR result/serological identity confirmed by nucleotide sequencing |
|---|---|---|---|
| Orthobunyavirus | |||
| Bunyamwera | Bunyamwera virus | Aedes ochraceus, Kenya, 2006 | POS/yes |
| Aedes ochraceus, Kenya, 2006 | POS/yes | ||
| Aedes ochraceus, Kenya, 2006 | POS/yes | ||
| Aedes ochraceus, Kenya, 2006 | POS/yes | ||
| Aedes ochraceus, Kenya, 2006 | POS/yes | ||
| Pongola virus | Aedes mcintoshi, Kenya, 2006 | POS/yes | |
| California | Tahyna virus | Mosquito species, China, 2007 | POS/yes |
| Flavivirus | Zika virus | Human, YAP, 2007 | NEG |
| Alphavirus | Chikungunya virus | Human, traveler returning to United States, 2006 | NEG |
Viruses were isolated and serologically characterized at the CDC as part of international collaborations, with the exception of the Tahyna virus isolate from China. Tahyna virus RNA, extracted from Tahyna virus isolated and serologically characterized in China, was submitted to the CDC for confirmatory testing. Yap, Federated State of Micronesia; POS, positive; NEG, negative.
RNA extraction.
Viral RNA was extracted from virus seed by using the QIAamp viral RNA minikit (Qiagen). For specificity testing, extractions were performed on samples ranging in volume from 70 to 140 μl. RNA was eluted in a volume equal to the volume of the starting sample. For sensitivity analysis, extractions were performed on plaque-titrated virus dilutions of 220 μl in volume. RNA was eluted in 110 μl of elution buffer (Qiagen) for a 2× concentration of RNA. Eluted RNA was stored at −70°C until evaluation by RT-PCR amplification.
Primer design.
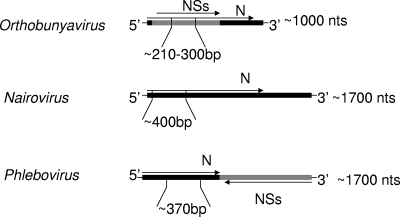
For amplification and sequencing, oligonucleotide primers (Table 3) were designed to target previously determined and newly derived S segment sequences of human pathogens of the Orthobunyavirus, Phlebovirus, and Nairovirus genera of the family Bunyaviridae (Tables 1 and 2). S segment sequences were aligned by serogroup and genus by using MegAlign software (DNASTAR). According to these alignments, primers were designed manually to target conserved regions shared within the overlapping N and nonstructural ORFs of analyzed orthobunyavirus sequences and the N ORFs of analyzed nairo- and phlebovirus sequences through visual inspection (Fig. 1; Table 3).
TABLE 3.
Oligonucleotide primers for the amplification and sequencing of the partial S segments of viruses of the family Bunyaviridae
| Targeted genus/genomic target | Forward primer
|
Reverse primer
|
Approx. amplicon size (bp) | ||
|---|---|---|---|---|---|
| Name | Sequence | Name | Sequence | ||
| Orthobunyavirus N ORF | Cal/Bwa group forward | GCAAATGGATTTGATCCTGATGCAG | Cal/Bwa group reverse | TTGTTCCTGTTTGCTGGAAAATGAT | 210 |
| Bun group forward | CTGCTAACACCAGCAGTACTTTTGAC | Bun group reverse | TGGAGGGTAAGACCATCGTCAGGAACTG | 250 | |
| Wyeomyia forward | ATGTCTGAAATTGTATTTGATGATATTGG | Wyeomyia reverse | TATTTCGATTCCCCGGAAAGT | 230 | |
| Oropouche forward | GGCCCATGGTTGACCTTACTTT | Oropouche reverse | ACCAAAGGGAAGAAAGTGAAT | 300 | |
| Nairovirus/phlebovirus | |||||
| Nairovirus S segment N ORF | Nairo forward | TCTCAAAGAAACACGTGCCGC | Nairo reverse | GTCCTTCCTCCACTTGWGRGCAGCCTGCTGGTA | 400 |
| Phlebovirus N ORF | Phlebo forward 1 | TTTGCTTATCAAGGATTTGATGC | Phlebo reverse | TCAATCAGTCCAGCAAAGCTGGGATGCATCAT | 370 |
| Phlebo forward 2 | TTTGCTTATCAAGGATTTGACC | ||||
FIG. 1.
Targeted regions of the presented assay shown within S segment viral cRNA N and nonstructural (NSs) ORF coding strategies of arthropod-borne genera of the family Bunyaviridae. N, nucleocapsid; nts, nucleotides.
RT-PCR molecular consensus assay amplification of bunyaviral cDNAs.
For the detection and amplification of medically important and serologically related viruses of the family Bunyaviridae, the following procedures were followed. RNAs extracted from bunyaviruses of interest and other arthropod-borne viruses (Tables 1 and 2) were subjected to RT-PCR amplification with a combination of virus- and serogroup-specific primers designed to detect viruses of the Orthobunyavirus, Nairovirus, and Phlebovirus genera (Table 1). RT-PCR assays were performed with 20 μl of extracted viral RNA and 50 pmol of each forward and reverse primer listed in mixes for detection of either (i) the Orthobunyavirus genus or (ii) the Nairovirus and Phlebovirus genera (Table 3) in a 50-μl total reaction volume by using Qiagen OneStep RT-PCR master mix (Qiagen). Reaction mixes were subjected to RT-PCR amplification using the following cycling conditions: 1 cycle of 50°C for 30 min and 95°C for 15 min, followed by 55 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 2 min. Reactions were terminated with a final extension step at 72°C for 10 min.
Primer selection and assay optimization.
Prior to the selection of presented primers (Table 3) and assay reaction conditions (see “RT-PCR molecular consensus assay amplification of bunyaviral cDNAs”), numerous alternatives were evaluated (data not shown). The presented oligonucleotide mixes and reaction conditions were selected based on the ability to detect the largest number of bunyaviruses with the highest relative sensitivity.
Molecular consensus assay result determination and DNA purification.
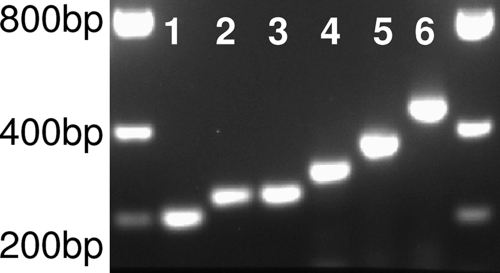
Products of RT-PCR amplification were evaluated by electrophoresis in a 2.0% agarose gel in 40 mM Tris-acetate-1 mM EDTA buffer. Five microliters of product was analyzed for each amplified sample. Visualization of DNA bands was achieved through ethidium bromide staining and UV transillumination. A sample was determined to be presumptively bunyavirus positive through size-determined migration and visualization of DNA bands, ranging from 210 to 400 bp (Fig. 2; Table 3), on the gel. For DNA purification, the remaining 45 μl of a positive sample was directly purified without the need for gel extraction using a PCR purification kit (Qiagen). As an exception, the presence of multiple bands necessitated gel extraction of the target-sized band from the amplification products of Gabek Forest virus by using a gel extraction kit (Qiagen).
FIG. 2.
Gel image of fragments amplified by the presented consensus assay. Fragments range in size from approximately 210 to 400 bp. Fragments from the amplification of La Crosse (lane 1), Wyeomyia (lane 2), Bunyamwera (lane 3), Oropouche (lane 4), Punta Toro (lane 5), and Dugbe (lane 6) viruses are represented. Five microliters of each DNA was loaded onto a 2% agarose gel containing an ethidium bromide stain.
Multiplex nucleotide sequencing of RT-PCR-amplified Orthobunyavirus, Nairovirus, and Phlebovirus DNAs for bunyavirus-positive result confirmation and preliminary species identification.
To determine the nucleotide sequence of purified cDNAs amplified from viral RNAs of the Orthobunyavirus, Nairovirus, and Phlebovirus genera, the following steps were taken. DNAs were sequenced with cocktails of either forward or reverse primers targeting (i) the Orthobunyavirus genus or (ii) the Nairovirus and Phlebovirus genera (Table 3). A total of 3.2 pmol of each listed primer in the appropriate forward or reverse primer mix (Table 3), along with approximately 50 ng of purified DNA, was added to each reaction by using the ABI BigDye Terminator V3.1 ready reaction cycle sequencing mix (Applied Biosystems). Sequencing reactions were purified using the DyEx 2.0 spin kit (Qiagen). Nucleotide sequences were determined by running purified sequencing reactions on the ABI 3130 genetic analyzer (Applied Biosystems). By these methods, partial S segment sequences were generated for all bunyaviruses amplified by the presented assay (Table 1).
S segment nucleotide sequence analysis of DNAs amplified by the consensus assay and preliminary species identification through the Nucleotide BLAST search mechanism of NCBI GenBank database.
Multiple nucleotide sequences generated for each tested bunyavirus (Table 1) were aligned in both the 5′ and 3′ directions and then edited using SeqMan software (DNASTAR). The resultant consensus sequence was then queried using the NCBI BLAST function of the nucleotide collection database at http://blast.ncbi.nlm.nih.gov (1). For S segment species identification, queries were optimized for somewhat similar sequences (blastn) by using default algorithm parameters. With these methods, sequences that matched the queried sequence with the highest overall maximum value were considered to provide the closest identity to the S segment species of an evaluated DNA. From these data, preliminary species-level identifications of evaluated virus isolates were made.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the S segment sequences recently determined are EU564827 (Bwamba virus), EU564828 (Pongola virus), EU564829 (Fort Sherman virus), EU564830 (Xingu virus), and EU564831 (Shokwe virus), and newly determined accession numbers are FJ235921 (Wyeomyia virus), FJ235922 (Alenquer virus), FJ235923 (Chandiru virus), FJ235924 (Chagres virus), FJ235925 (Cacao virus), FJ235926 (Itaporanga virus), FJ235927 (Gabek Forest virus), FJ235928 (Turuna virus), FJ235929 (Rio Grande virus), FJ235930 (Nique virus), and GQ166188 (Itaituba virus).
RESULTS
The analytical sensitivity of the bunyavirus molecular consensus assay, as determined through its application to plaque-titrated human pathogens of the family Bunyaviridae, is reported in Table 1. For the Orthobunyavirus genus, the molecular consensus assay detects between 660 PFU/0.1 ml (for Wyeomyia virus) and 0.1 PFU/0.1 ml (for Oropouche virus). For the Phlebovirus and Nairovirus genera, detection limits range from 100 PFU/0.1 ml (for Rift Valley fever virus) to 0.0024 PFU/0.1 ml (for Chagres virus).
The analytical specificity of the bunyavirus molecular consensus assay, as determined through its application to viruses of the families Bunyaviridae, Flaviviridae, and Togaviridae, is presented in Tables 1 and 2. Forty-seven viruses, including 29 human pathogens, representing six major serogroups of the family Bunyaviridae, were targeted and amplified by the presented assay (Table 1). Demonstrating fidelity for bunyaviral RNA targets, no evaluated viruses of the Flaviviridae or Togaviridae families were detected by the consensus assay (Table 2). For all amplified bunyaviral DNAs, multiplex sequencing and NCBI BLAST analyses generated results for S segment nucleotide sequence data that confirmed the serologically derived identity, with the exceptions of Ilesha virus KO/2, Maguari virus 75V3429, and snowshoe hare virus LIEV 13004AKH strains (Table 1). The S segment species amplified and sequenced from Ilesha virus KO/2, Maguari virus 75V3429, and snowshoe hare virus LIEV 13004AKH strains shared the highest nucleotide sequence identities with various Batai, Cache Valley, and Chatanga virus strains, respectively, according to our analyses (data not shown).
The ability of the bunyavirus consensus assay to detect target viral RNAs extracted from temporally relevant samples was evaluated through its application to a panel of recent isolates of international origins (8, 24; Table 4). The presented molecular consensus assay amplified and identified Bunyamwera, Pongola, and Tahyna virus cDNAs of the Bunyamwera, Bwamba, and California serogroups of the genus Orthobunyavirus from viruses isolated from Kenya and China in 2006 and 2007, respectively (Table 4). Confirming fidelity for bunyaviral RNAs, other recently derived isolates of the Alphavirus and Flavivirus genera were not amplified by the presented consensus assay (Table 4).
DISCUSSION
To the best of our knowledge, the presented bunyavirus molecular consensus assay has been shown to detect a range of viruses more diverse than can be detected by any other molecular assay designed for the detection of arthropod-borne viruses. In addition, the assay has been shown to have sufficient sensitivity for the detection of virus strains with various titers isolated from a diverse range of host species and geographic and temporal origins (Tables 1 and 4). Also, when combined with multiplex sequencing and NCBI BLAST analyses, the presented assay was able to confirm serologically derived identities for all but 3 of 47 amplified viruses (Tables 1 and 4). Finally, the assay has been shown to have fidelity for bunyaviral RNAs when applied to arthropod-borne viruses of the families Flaviviridae and Togaviridae (Tables 2 and 4).
In addition to viruses of the Phlebovirus and Orthobunyavirus genera that were detected (Table 1), some evaluated orthobunyaviruses were not amplified by the presented assay (Table 2). While we were able to detect Nyando and Guaroa viruses by using alternative primer mixes and reaction conditions relative to those presented here, these alternatives compromised the sensitivity of detection for other medically important agents and, as a result, were abandoned in the final assay design (data not shown). For the group C and Guama serogroup viruses presented in Table 2, no efforts (presented or alternative) generated DNAs for sequencing. These findings are particularly confounding with regard to the group C agents, for which multiple amplification attempts, using oligonucleotide primers designed to target previously published group C, S, and M segments (26), failed to generate DNAs for sequencing. Having tried to eliminate other variables that might contribute to disparities in amplification efficiency, we believe that target nucleotide sequence heterogeneity is responsible for the inability to detect these agents by molecular methods.
The identification of S segment species through multiplex sequencing and NCBI BLAST analyses is a powerful component of the presented molecular consensus assay. However, we regard the determination of virus species through GenBank-derived S segment identity as preliminary for the following reasons. (i) It is often impossible to verify the integrity of sequence data for a given virus species in GenBank. As a result, inaccurate data could potentially cause the misidentification of an S segment species by using the presented methodologies. However, our collective experience using GenBank queries for virus identification has indicated a high degree of integrity for these data in instances where we were able to verify virus identity by alternative methods (Table 2) (7, 21, 22, 23). (ii) The capacity for genomic segment reassortment among viruses of the family Bunyaviridae allows the possibility that a given isolate possesses genomic segments of heterologous ancestries. Although we did not evaluate them in this study, we consider our findings of Cache Valley, Batai, and Chatanga virus-like S segments, amplified from isolates serologically classified as Maguari, Ilesha, and snowshoe hare virus strains, respectively (Table 1), evidence of possible segment reassortment. While not yet undertaken, additional experiments will be performed by our group to elucidate the identities of the M and L segments of these virus strains to verify the ancestry of all three genomic segments. Finally, because of the above-described limitations on virus identification by GenBank querying, we recommend that the preliminary identification of a virus species through GenBank-derived S segment identity always be confirmed by additional molecular and/or serological analyses.
The molecular consensus assay is generally more sensitive for the detection of viruses of the Nairovirus and Phlebovirus genera than for the detection of viruses of the Orthobunyavirus genus (Table 1). We have considered several potential explanations for this difference in sensitivities. (i) From our collective laboratory experience developing and applying assays for the detection of arthropod-borne viruses (12, 14, 16, 17, 18, 19), it has been noted that, in general, increasing the number of different primers in a reaction causes a reduction in overall sensitivity of detection. To accommodate a greater diversity of nucleotide sequence targets, the Orthobunyavirus mix contains more primers than the Nairovirus/Phlebovirus mix does, possibly contributing to the noted differences in sensitivities of detection (Table 1). (ii) Oligonucleotides designed for the detection of orthobunyaviruses bind target nucleic acid less efficiently than those designed for the detection of nairo- and phleboviruses. In fact, to incorporate a broader diversity of targets, primers designed for the detection of viruses of the Orthobunyavirus genus are generally less specific for the target sequence than are those for the detection of viruses of the Nairovirus and Phlebovirus genera. Therefore, the Orthobunyavirus primers (Table 3) likely prime RT-PCR amplification less efficiently, resulting in reduced sensitivity. (iii) There could be a difference in the ratio of RNA copies to infectious viral particles in the Vero cell plaque assay for these viruses. If the evaluated phlebo- and nairoviruses have a higher ratio of RNA copies to PFU equivalents in the Vero cell culture system than the evaluated orthobunyaviruses, it would appear that the sensitivity of detection for the former agents is greater. The application of this assay to standards of known RNA copy number is supported by this conjecture. However, because PFU represent the most relevant unit for quantitating infectious virus, we consider experiments designed to determine assay sensitivity through its application to standards of known copy number beyond the necessary scope of this research.
The data presented here (Tables 1, 2, and 4) demonstrate the utility of the molecular consensus assay for the detection of virus isolates of the family Bunyaviridae. Most promising to us is the ability of the assay to correctly identify temporally relevant human pathogens of the genus Orthobunyavirus, isolated from mosquito pools as part of arbovirus surveillance programs in Kenya and China (Table 4) (8, 24). Unfortunately, we do not have a comparable panel of recent isolates for the characterization of the Nairovirus/Phlebovirus component of this assay. However, it is believed that the extremely high sensitivities reported for the detection of historically obtained viruses of these genera (Table 1) are conducive for the detection of newly emergent nairo- and phleboviruses as well. We recommend the incorporation of the presented molecular consensus assay in an algorithm that includes additional molecular and serological methodologies for the comprehensive identification of medically important, and serologically related, virus isolates of the family Bunyaviridae. Additionally, it is our intention that the presented assay might be combined with other previously published (11) or newly derived assays to target multiple segments of the bunyavirus genome. However, research to determine the feasibility of an assay with a designed capability for the detection of reassortant bunyaviruses was not performed in this study. Finally, because this assay has been designed to target conserved regions shared between genetically disparate agents, we believe that additional bunyavirus species besides those evaluated here will be amplified and detected by the presented assay. However, the paucity of genetic information available for many of these agents and extraordinary diversity among viruses of the family Bunyaviridae prevent absolute certainty that the assay will amplify uncharacterized viruses.
Acknowledgments
We thank Rosemary Sang of the Kenya Medical Research Institute (KEMRI) and George Ludwig (USAMRIID) for the provision of a CCHF virus plasmid and Nairobi sheep disease virus RNA, respectively, that were used in the evaluation of the presented assay. We also thank Jeff Chang for advice on consensus primer design, Mary Crabtree for ongoing collaborative discussion on bunyaviruses, Carol Blair for critical review of the manuscript, George F. Newell, Jr., for editorial advice, and Guo-Dong Liang for Chinese Tahyna virus DNA.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, A. D., and R. E. Shope. 2005. Bunyaviridae, p. 1025-1058. In B. Mahy and V. Ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, 10th ed. ASM Press, Washington, DC.
- 3.Bird, B. H., J. W. Githinji, J. M. Macharia, J. L. Kasiiti, R. M. Muriithi, S. G. Gacheru, J. O. Musaa, J. S. Towner, S. A. Reeder, J. B. Oliver, T. L. Stevens, B. R. Erickson, L. T. Morgan, M. L. Khristova, A. L. Hartman, J. A. Comer, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 2008. Multiple virus lineages sharing recent common ancestry were associated with a large Rift Valley fever outbreak among livestock in Kenya during 2006-2007. J. Virol. 8211152-11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen, M. D., S. G. Trappier, A. J. Sanchez, R. F. Meyer, C. S. Goldsmith, S. R. Zaki, L. M. Dunster, C. J. Peters, T. J. Ksiazek, and S. T. Nichol. 2001. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology 291185-190. [DOI] [PubMed] [Google Scholar]
- 5.Briese, T., B. Bird, V. Kapoor, S. T. Nichol, and W. I. Lipkin. 2006. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J. Virol. 805627-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calisher, C. H., and N. Karabatsos. 1988. Arbovirus serogroups: definition and geographic distribution, p. 19-57. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology. CRC Press, Boca Raton, FL.
- 7.Campbell, G. L., J. D. Mataczynski, E. S. Reisdorf, J. W. Powell, D. A. Martin, A. J. Lambert, T. E. Haupt, J. P. Davis, and R. S. Lanciotti. 2006. Second human case of Cache Valley virus disease. Emerg. Infect. Dis. 12854-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabtree, M., R. Sang, J. Lutomiah, J. Richardson, and B. Miller. Arbovirus surveillance of mosquitoes collected at sites of active Rift Valley fever virus transmission: Kenya, 2006-2007. J. Med. Entomol., in press. [DOI] [PubMed]
- 9.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Family Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel, C. M. Fauguet, and D. H. L. Bishop (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 10.Gerrard, S. R., L. Li, A. D. Barrett, and S. T. Nichol. 2004. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 788922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honig, J., J. Osborne, and S. Nichol. 2004. The high genetic variation of viruses of the genus Nairovirus reflects the diversity of their predominant tick hosts. Virology 31810-16. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, B. W., B. J. Russell, and R. S. Lanciotti. 2005. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 434977-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuno, G., C. J. Mitchell, G.-J. J. Chang, and G. C. Smith. 1996. Detecting bunyaviruses of the Bunyamwera and California serogroups by a PCR technique. J. Clin. Microbiol. 341184-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, A. J., O. Kosoy, J. O. Velez, B. J. Russell, and R. S. Lanciotti. 2007. Detection of Colorado tick fever viral RNA in acute human serum samples by a quantitative real-time RT-PCR assay. J. Virol. Methods 14043-48. [DOI] [PubMed] [Google Scholar]
- 15.Lambert, A. J., and R. S. Lanciotti. 2008. Molecular characterization of medically important viruses of the genus Orthobunyavirus. J. Gen. Virol. 892580-2585. [DOI] [PubMed] [Google Scholar]
- 16.Lambert, A. J., D. A. Martin, and R. S. Lanciotti. 2003. Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J. Clin. Microbiol. 41379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert, A. J., R. S. Nasci, B. C. Cropp, D. A. Martin, B. C. Rose, B. J. Russell, and R. S. Lanciotti. 2005. Nucleic acid amplification assays for detection of La Crosse virus RNA. J. Clin. Microbiol. 431885-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanciotti, R. S., and A. J. Kerst. 2001. Nucleic acid sequence based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J. Clin. Microbiol. 394506-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 384066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanciotti, R. S., O. L. Kosoy, J. J. Laven, A. J. Panella, J. O. Velez, A. J. Lambert, and G. L. Campbell. 2007. Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 13764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanciotti, R. S., O. L. Kosoy, J. J. Laven, J. O. Velez, A. J. Lambert, A. J. Johnson, S. M. Stanfield, and M. R. Duffy. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 141232-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 2862333-2337. [DOI] [PubMed] [Google Scholar]
- 23.Lanciotti, R. S., and T. F. Tsai. 2007. Arboviruses, p. 1486-1501. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. F. Pfaller (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.
- 24.Lu, Z., X.-J. Lu, S.-H. Fu, S. Zhang, Z.-X. Li, X. Yao, Y. Feng, A. Lambert, D. Ni, F. Wang, S. Tong, R. Nasci, Y. Feng, Q. Dong, Y. Zhai, X. Gao, H. Wang, Q. Tang, and G. Liang. 2009. Tahyna virus and human infection, China. Emerg. Infect. Dis. 15306-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 26.Nunes, M. R., A. P. Travassos da Rosa, S. C. Weaver, R. B. Tesh, and P. F. Vasconcelos. 2005. Molecular epidemiology of group C viruses (Bunyaviridae, Orthobunyavirus) isolated in the Americas. J. Virol. 7910561-10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Seco, M. P., J. M. Echevarría, L. Hernández, D. Estévez, J. M. Navarro-Marí, and A. Tenorio. 2003. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J. Med. Virol. 71140-149. [DOI] [PubMed] [Google Scholar]
- 28.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 29.Schmaljohn, C. S., and S. T. Nichol. 2007. Bunyaviridae, p. 1741-1778. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Shope, R. E., and O. R. Causey. 1962. Further studies on the serological relationships of group C arthropod-borne viruses and the application of these relationships to rapid identification of types. Am. J. Trop. Med. 26189-208. [DOI] [PubMed] [Google Scholar]
- 31.Vorou, R., I. N. Pierroutsakos, and H. C. Maltezou. 2007. Crimean-Congo hemorrhagic fever. Curr. Opin. Infect. Dis. 20495-500. [DOI] [PubMed] [Google Scholar]