Abstract
The early diagnosis of human immunodeficiency virus (HIV) infection in infants is critical to ensure the initiation of treatment before significant immunological compromise. Each year an estimated 300,000 HIV-exposed infants in South Africa require access to tests for the diagnosis of HIV infection. Currently, testing is performed at several facilities by using PCR amplification of HIV DNA at 6 weeks of age by the use of dried blood spots (DBSs) and whole blood (WB). The Gen-Probe Aptima HIV type 1 (HIV-1) screening assay (the Aptima assay) is a qualitative nucleic acid test based on transcription-mediated amplification (TMA), a technology routinely used in blood banks in South Africa. The performance characteristics of Gen-Probe's TMA technology compared well to those of the Roche Amplicor HIV-1 DNA (version 1.5) assay. The sensitivity of the assay with WB and DBS samples was 100%, and the specificities were 99.4% and 99.5% for DBSs and WB, respectively. The detection of HIV by the Aptima assay at greater levels of dilution in samples negative by the comparator assay indicates an improvement in sensitivity by the use of the TMA technology. The ability to process 1,900 samples in a 24-h period on the Tigris instrument makes the Aptima assay an attractive option for high-volume, centralized laboratories.
Most human immunodeficiency virus (HIV)-infected infants reside in sub-Saharan Africa (12, 21). Disease progression is generally rapid, and that rate of mortality among infected infants is high (1), with more than half of infants dying within the first 2 years of life (12). Recent studies have revealed the importance of early intervention with antiretroviral treatment before significant immunological deterioration, reinforcing the need for the early diagnosis of HIV infection (22). The persistence of maternal antibodies against HIV in exposed infants up to 18 months of age prevents the use of the enzyme-linked immunosorbent assay for the early diagnosis of HIV infection. Thus, approaches to the early diagnosis of infection in infants have included ultrasensitive heat-denatured p24 antigen quantitation (4, 16, 23) and nucleic acid amplification techniques that target HIV DNA (15), RNA (14), or total nucleic acid (3, 10, 18), most commonly with samples obtained at 6 weeks of age. It has also been established that the collection of blood as dried blood spots (DBSs) improves access to tests for the early diagnosis of HIV infection among infants in resource-limited settings (2, 17).
South Africa bears an enormous burden of individuals infected with HIV, with an estimated prevalence of 28% in an antenatal clinic in 2007(11). Over 1 million births are recorded annually, which translates into an estimated 300,000 HIV-exposed infants requiring access to early testing. Testing is performed in a limited number of laboratories nationally by using PCR amplification of HIV DNA from DBSs and whole blood (WB) collected at 6 weeks of age. The platform most widely used in the public health sector is the Roche Amplicor HIV type 1 (HIV-1) DNA (version 1.5) assay (the Roche Amplicor assay) with manual or automated extraction with the Roche MagNaPure analyzer (19). Recently, the new Cobas Ampliprep/Cobas TaqMan assay option has been evaluated as a means of improving throughput and reducing the need for user intervention (18).
To further address automation issues and to investigate the use of total nucleic acid detection for the diagnosis of HIV infection in infants, the Gen-Probe Aptima HIV-1 screening assay (the Aptima assay; a derivative of the Procleix Ultrio assay blood donor screening test used in South African blood banks; Gen-Probe, Inc., San Diego, CA) was investigated. Nucleic acid amplification assays were widely implemented by U.S. blood banks as early as 1999 (6, 20) and were implemented in South Africa, by use of the Procleix Tigris system, in 2005 (7). Two versions of the assay exist: the Procleix Ultrio assay (Chiron, Emeryville, CA; Gen-Probe) for screening for HIV-1 RNA, hepatitis B virus DNA, and hepatitis C virus RNA for testing of human donor blood and the discriminatory assays specific for each infection (5, 9). Screening is generally conducted with the Ultrio assay, and confirmation of any positive result is done with the respective discriminatory assay (7). Both assays are qualitative tests based on nucleic acid technology and transcription-mediated amplification (TMA) and, in the case of HIV detection, target two regions of the HIV-1 genome. The Procleix Ultrio assay has FDA approval for the detection of HIV-1, hepatitis B virus, and hepatitis C virus in plasma and serum specimens from human donors. In addition, the same technology (Aptima HIV-1 RNA qualitative assay; Gen-Probe) has FDA approval for use for the diagnosis of acute HIV-1 infection.
The study described here investigated the novel use of the Aptima assay (a derivative of the Procleix HIV-1 discriminatory assay) and its application for high-throughput testing with the Tigris instrument for the early diagnosis of HIV infection in infants by the use of WB and DBSs.
MATERIALS AND METHODS
Sample population.
WB samples in potassium EDTA (n = 500) sent to the HIV PCR laboratory at the Charlotte Maxeke Academic Hospital (previously the Johannesburg Hospital), Johannesburg, South Africa, for assay for the routine diagnosis of HIV-1 infection in infants by the Roche Amplicor assay were randomly selected for this study. The venous blood samples selected had been collected, on average, between 2 and 7 days earlier.
For DBS analysis, 500 DBSs were selected. These DBSs had been collected and stored at room temperature for 8 to 10 months for a previous analysis that compared the performance of the Roche Amplicor assay and the Cobas Ampliprep/Cobas TaqMan total nucleic acid-based assay (18).
Sample preparation for Aptima assay.
Before they were tested, the WB samples (100 μl) were diluted in 900 μl reverse osmosis water in a sterile tube containing EDTA (EDTA tubes). The samples were then transported to the South African National Blood Service (SANBS) laboratories in Johannesburg, where the EDTA tubes were manually uncapped and loaded onto racks on the automated Tigris instrument. Positive and negative calibrators and controls were included in the assay kit and were prepared as indicated by the manufacturer before they were loaded onto the Tigris instrument racks.
The DBSs (diameter, 13 mm per spot), which had sample volumes that equated to between 50 and 70 μl, were cut in half, and both halves placed in a tube containing 1.0 ml of specimen transport medium (the supplier's lysis buffer). The samples were incubated by using an elution program (four cycles consisting of 5 min of incubation at 95°C and 10 s of vortexing) on Gen-Probe's SB-100 instrument. Following incubation, the eluate from each sample was aliquoted into a sterile EDTA tube, transported to SANBS, and loaded onto the Tigris instrument, together with the assay calibrators and controls.
Procleix discriminatory assay.
The Procleix discriminatory assay for HIV-1 was used to analyze WB and DBS samples according to the manufacturer's instructions. The Procleix discriminatory assays utilize the same three steps as the Procleix Ultrio assay (5), namely, target capture, TMA, and amplicon detection via the hybridization protection assay. Target capture involves a sample preparation step that releases viral RNA and DNA. The target is amplified by TMA with two enzymes, Moloney murine leukemia virus reverse transcriptase and T7 RNA polymerase. This is followed by hybridization to oligonucleotide probes with chemiluminescent labels; the probes are homologous to two separate highly conserved regions of the HIV-1 genome. The entire process is controlled by using an internal control, added at the target capture step, which can be distinguished from the target probe by using a different label. During detection, the chemiluminescent signal is measured in a luminometer and is reported as relative light units. An analyte signal/cutoff (S/CO) ratio is calculated for each sample by using a run-specific analyte cutoff value. An S/CO ratio of >1 is reported as a qualitative reactive result, and an S/CO ratio of <1 is reported as a nonreactive result.
Dilution studies with WB and DBSs.
Dilution studies with WB and DBSs were performed in two parts. The first used 21 DBSs prepared by serially diluting WB into negative plasma before 50 μl was spotted onto cards (Protein Saver 903; Whatman GmbH, Dassel, Germany). Negative plasma was obtained from the known HIV-1-negative plasma samples used in Gen-Probe's proficiency panel for the Aptima assay. The dilutions ranged from undiluted to 1:10,000 (n = 10) and undiluted to 1:100,000 (n = 11). Both the Roche Amplicor and the Aptima assays were performed with this DBS dilution series. The second part used 10 WB samples prepared in dilutions that ranged from undiluted to 1:100,000 and DBSs prepared from those dilutions. This dilution series was used to compare the WB dilutions to the DBS dilutions and was tested only by the Aptima assay. All diluted WB samples used 100 μl, to which 900 μl reverse osmosis water was added. One spot for each DBS was punched and processed as described above from the most diluted to the least diluted samples to reduce the risk of crossover contamination.
Statistical analysis.
Both the test assay (the Aptima assay) and the reference/comparator assay (the Roche Amplicor assay) generate discrete reportable results (either HIV positive or HIV negative, based on defined cutoff values). The results of the Roche Amplicor assay were taken as the reference and therefore the correct result against which the result of the Aptima assay was compared for calculation of the sensitivity and the specificity (including confidence intervals). This was performed by using the VassarStats online calculator (http://faculty.vassar.edu/lowry/clin1.html). A scatter plot presents the S/CO ratio values (continuous variable) for DBS and WB results from the Aptima assay only.
RESULTS
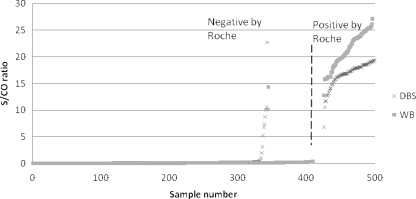
Of 500 WB samples referred for testing, 481 were available for analysis. Seventeen samples were not tested by the Roche assay due to poor sample integrity (they were clotted), one sample showed inhibition by the Roche Amplicor assay, and another sample had an internal control failure by the Aptima assay. The number of samples with reactive results by the Roche Amplicor assay was 72 (14.9%), and the number of samples with nonreactive results was 409 (85%). The sensitivity of the results of the Aptima assay were found to be 100% (Table 1). Two samples nonreactive by the Roche assay yielded reactive reportable values by the Aptima assay, making the specificity 99.5%. The S/CO ratio values for these discrepant samples were <15, with a clear separation between S/CO ratio values for reactive samples (>10) and nonreactive samples (<1), as shown in Fig. 1.
TABLE 1.
Sensitivity and specificity of the Aptima assay compared to the results of the Roche Amplicor assay performed with DBSs and WBa
| Sample | Sensitivity (%) | Specificity (%)b |
|---|---|---|
| DBS (n = 494) | 100 (96.9-100) | 97 (94.5-98.5) |
| Whole blood (n = 481) | 100 (93.6-100) | 99.5 (98-99.9) |
The Roche Amplicor assay was considered the reference assay. Values in parentheses are confidence intervals.
Ten reactive samples with discrepant results yielded reactive results in two samples upon repeat testing, increasing the specificity to 99.4% (confidence interval, 97.7% to 99.9%).
FIG. 1.
Scatter plot of the results for the WB and DBS samples for comparison of the Aptima assay and the Roche Amplicor assay results. The vertical axis is the S/CO ratio value from the Tigris platform, and the horizontal axis is the sample number sorted by samples with reactive and nonreactive results reported by the Roche Amplicor assay. The vertical line separates samples found to be negative and positive by the Roche Amplicor HIV-1 DNA assay. Two WB samples were reactive (S/CO ratio, >1) by the Aptima assay but nonreactive by the Roche Amplicor assay, with S/CO ratio values of <15. Ten DBSs were reactive by the Aptima HIV-1 assay and nonreactive by the Roche Amplicor assay. One sample had an S/CO ratio value of 22.75, and the remaining nine samples had S/CO ratio values of ≤10. The values for these samples obtained upon repeat testing are also shown in Table 2.
Of the 500 DBSs prepared, 494 samples could be analyzed on both assay platforms. Six samples were excluded from the analysis because three were inhibited by the Roche Amplicor assay and three did not generate results by the Aptima assay and were flagged by the Tigris instrument as having insufficient volumes. According to the reference assay (the Roche Amplicor assay), the reportable results (considered the correct result) indicated that 151 (30.5%) samples were reactive and 343 (69.4%) samples were nonreactive. Four samples generated equivocal results (defined in the laboratory as absorbance values between 0.2 and 1.5) by the Roche Amplicor assay, but the assay was repeated and the result was negative (S/CO ratio, <0.8). The results for these samples were then included in the statistical analysis as negative results. The sensitivity of the Aptima assay compared to the results of the Roche Amplicor assay for the study with DBSs (n = 494) was 100%, as shown in Table 1. The specificity of the Aptima assay with DBSs compared to the results of the Roche Amplicor assay was 97%, with the results for 10 samples reactive by TMA being discrepant with the results by their comparator Roche Amplicor assay, by which they were negative. These results are presented in Fig. 1. Retesting of these samples by the Aptima assay yielded nonreactive results for eight samples and reactive results for two samples. This improved the specificity of the Aptima assay compared to the results of the Roche Amplicor assay to 99.4%. Among the 10 samples with discrepant results, 1 sample had an S/CO ratio of 22.75 (which on duplicate repeat testing yielded nonreactive results and S/CO ratios of 0.2 and 0.18) and 9 samples had S/CO values that ranged from 1 to 10.48. The S/CO ratio values for the samples with reactive results by the Roche Amplicor assay had S/CO values that were >10 for all except one sample, which had an S/CO ratio of >6 (Fig. 1). This is lower than the value of >15 obtained with the WB samples. Table 2 lists the results and S/CO values obtained by retesting by the Aptima assay for the 10 discrepant samples. Two samples invariably produced reactive results on retesting but yielded S/CO ratio values that were <5. Evaluation of the sequence in which the DBS samples were prepared showed that only four samples with discrepant Aptima assay reactive results had samples next to them that were confirmed to be reactive. The remaining discrepant samples were randomly distributed in relation to the locations of the other reactive samples.
TABLE 2.
Aptima assay results with DBSs and results of two repeat assays for 10 samples that were nonreactive by the Roche Amplicor assay
| Roche Amplicor assay result | Initial Aptima assay
|
First repeat of Aptima assay
|
Second repeat of Aptima assay
|
|||
|---|---|---|---|---|---|---|
| Result | S/CO ratio | Result | S/CO ratio | Result | S/CO ratio | |
| Nonreactive | Reactive | 0.58 | Nonreactive | 0.03 | Nonreactive | 0.30 |
| Nonreactive | Reactive | 1 | Nonreactive | 0.05 | Reactive | 3.38 |
| Nonreactive | Reactive | 1.95 | Nonreactive | 0.03 | Nonreactive | 0.17 |
| Nonreactive | Reactive | 3.12 | Reactive | 1.26 | Nonreactive | 0.02 |
| Nonreactive | Reactive | 5.21 | Nonreactive | 0.29 | Nonreactive | 0.19 |
| Nonreactive | Reactive | 6.83 | Nonreactive | 0.20 | Nonreactive | 0.12 |
| Nonreactive | Reactive | 7.35 | Nonreactive | 0.04 | Nonreactive | 0.18 |
| Nonreactive | Reactive | 8.79 | Nonreactive | 0.02 | Nonreactive | 0.11 |
| Nonreactive | Reactive | 10.04 | Nonreactive | 0.04 | Nonreactive | 0.05 |
| Nonreactive | Reactive | 22.75 | Nonreactive | 0.22 | Nonreactive | 0.18 |
Comparison of the results by the Roche Amplicor assay and the Aptima assay for the diluted DBS samples showed that six (28.5%) samples generated reactive results by the Aptima assay that were 1 log unit more sensitive than the result obtained by the comparator assay and that four (19%) samples generated reactive results by the Aptima assay that were 3 log units more sensitive. The comparison of the results obtained with WB and DBSs for the same samples tested only by the Aptima assay showed that 6 of the 10 samples yielded equivalent values at all dilutions. The lowest S/CO ratio value generated from WB was 1.87, and the lowest S/CO ratio value generated from DBS was 4.13. Although the results were not consistent for all 10 samples, the majority of the values for DBSs were less than those for WB.
DISCUSSION
This study represents the first evaluation of the Gen-Probe Aptima assay performed with the automated Tigris analyzer. The results obtained with Gen-Probe's TMA technology (the Aptima assay) compared well to the results of the Roche Amplicor assay with WB and DBSs for the early diagnosis of HIV-1 infection in infants in South Africa. The sensitivity of the Aptima assay obtained with WB and DBS samples was 100%, and the specificities were 99.4% and 99.5% with DBSs and WB, respectively. The detection by the Aptima assay of HIV-1 at greater levels of dilution in samples negative by the comparator assay indicates that an improvement in sensitivity is achieved by use of the TMA technology. These results compared favorably with those of the manual version of the assay with DBSs described by Kerr and colleagues, who reported a sensitivity of 99.2% and a specificity of 100% for the diagnosis of HIV-1 infection in infants by the use of DBSs (8).
Nucleic acid-based assays for the diagnosis of HIV-1 infection in infants will remain a critical component of the new plan for the accelerated prevention of mother-to-child transmission of HIV-1 proposed by the South African National Department of Health. PCR testing will be used for the diagnosis of HIV-1 infection in individual patients. Those results will facilitate the earlier initiation of antiretroviral therapy and will be a key tool for monitoring the success of the program (e.g., the proportion of infants accessing a PCR test at 6 weeks of age).The greatest advantage of the Tigris platform in the South African context would be the facilitation of a significantly higher throughput for centralized testing facilities. Following work flow optimization, 1,900 samples can be processed on one Tigris instrument within a 24-h period. When 150 samples are processed, comparisons of the work flows with the Aptima assay and the Roche assay showed a 6-h faster turnaround time with the Aptima assay.
Investigations are under way to ascertain whether the S/CO ratio obtained by the Aptima assay could predict a semiquantitative viral load. A clear separation of S/CO ratio readings between a reactive result (S/CO ratio, >1) and a nonreactive result (S/CO ratio, <1) with WB samples is evident, but the separation is less clear for DBS samples. This was also evident from the results of the tests with the dilution series, in which the S/CO ratio values for many DBSs were less than the values for their corresponding WB samples. The S/CO ratios values for samples with discrepant results were less than 15 for WB and less than 10 for DBSs and were higher for all samples with concordant results. Repeat testing may thus be indicated for samples with S/CO ratio values below 15 for WB (1.45% of cases in this study) and below 10 for DBSs (2.2% of cases in this study). The feasibility of using a sensitive qualitative RNA test as a marker of viral suppression has recently been explored and requires further validation (13). That study (13) found S/CO ratios similar to those found in the present study.
Further evaluation is required for the cases that showed discrepant results between the two assays. This is particularly relevant for those cases that were initially reactive by the Aptima assay and then negative by repeat testing of the same sample (Table 2). For cases that remained reactive on repeat testing, an enhanced sensitivity or subtype detection by the Aptima assay may need to be considered, and for those that were negative on repeat testing, the potential for contamination by the use of manual DBS sample preparation needs further investigation.
These preliminary data suggest that future studies are needed that include the evaluation of the performance of the Gen-Probe Aptima assay with samples from a pediatric cohort monitored longitudinally from birth until confirmation of the diagnosis of HIV infection at 18 months of age. This will establish a true “gold standard” for the detection of HIV infection to enable the comparison of assays, as well as allow investigation of whether testing could be implemented earlier than 6 weeks postdelivery. The use of this assay together with the automated processing of DBSs is also being investigated to further enhance throughput in a high-volume, centralized testing facility.
Acknowledgments
We thank Tom Nugent of Gen-Probe Inc. for the development of the DBS extraction procedure. We also acknowledge SANBS for the use of their Tigris instruments throughout this study.
This publication was made possible by the generous support of the American people through the U.S. Agency for International Development.
The contents of this report are the responsibility of the authors and do not necessarily reflect the views of USAID or the U.S. government.
Footnotes
Published ahead of print on 27 May 2009.
REFERENCES
- 1.Brahmbhatt, H., G. Kigozi, F. Wabwire-Mangen, D. Serwadda, T. Lutalo, F. Nalugoda, N. Sewankambo, M. Kiduggavu, M. Wawer, and R. Gray. 2006. Mortality in HIV-infected and uninfected children of HIV-infected and uninfected mothers in rural Uganda. J. Acquir. Immune Defic. Syndr. 41504-508. [DOI] [PubMed] [Google Scholar]
- 2.Creek, T., A. Tanuri, M. Smith, K. Seipone, M. Smit, K. Legwaila, C. Motswere, M. Maruping, T. Nkoane, R. Ntumy, E. Bile, M. Mine, L. Lu, G. Tebele, L. Mazhani, M. K. Davis, T. H. Roels, P. H. Kilmarx, and N. Shaffer. 2008. Early diagnosis of human immunodeficiency virus in infants using polymerase chain reaction on dried blood spots in Botswana's national program for prevention of mother-to-child transmission. Pediatr. Infect. Dis. J. 2722-26. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham, C., T. Charbonneau, and K. Song. 1999. Comparison of human immunodeficiency virus-1 DNA polymerase chain reaction and qualitative and quantitative RNA polymerase chain reaction in human immunodeficiency virus 1-exposed infants. Pediatr. Infect. Dis. J. 1830-35. [DOI] [PubMed] [Google Scholar]
- 4.Fiscus, S. A., J. Wiener, E. J. Abrams, M. Bulterys, A. Cachafeiro, and R. A. Respess. 2007. Ultrasensitive p24 antigen assay for diagnosis of perinatal human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 452274-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gen-Probe. 2006. Procleix Ultrio assay package insert. IN0167EN-02 Rev. 1/ QCS 500453.2006-01. Gen-Probe, Inc., San Diego, CA.
- 6.Grant, P. R., and M. P. Busch. 2002. Nucleic acid amplification technology methods used in blood screening. Transfus. Med. 12229-242. [DOI] [PubMed] [Google Scholar]
- 7.Heyns, A. D. P., J. P. Swanevelder, P. N. Lelie, R. L. Crookes, and M. P. Busch. 2006. The impact of individual donation NAT screening on blood safety—the South African experience. ISBT Sci. Ser. 1203-208. [Google Scholar]
- 8.Kerr, R. J. S., G. Player, S. A. Fiscus, and J. A. E. Nelson. 2009. Qualitative human immunodeficiency virus RNA analysis of dried blood spots for diagnosis of infections in infants. J. Clin. Microbiol. 47220-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppelman, M. H. G. M., A. Assal, M. Chudy, P. Torres, R. Garcia de Villaescusa, H. W. Ressink, P. N. Lelie, and H. T. M. Cuypers. 2005. Multicenter performance evaluation of a transcription-mediated amplification assay for screening of human immunodeficiency virus-1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA in blood donations. Transfusion 451258-1266. [DOI] [PubMed] [Google Scholar]
- 10.Lambert, J. S., D. R. Harris, E. R. Stiehm, J. Moye, Jr., M. G. Fowler, W. A. Meyer III, J. Bethel, and L. M. Mofenson. 2003. Performance characteristics of HIV-1 culture and HIV-1 DNA and RNA amplification assays for early diagnosis of perinatal HIV-1 infection. J. Acquir. Immune Defic. Syndr. 34512-519. [DOI] [PubMed] [Google Scholar]
- 11.National Department of Health, South Africa. 2008. The national HIV and syphilis prevalence survey, South Africa, 2007. National Department of Health, Pretoria, South Africa. http://www.doh.gov.za/docs/index.html. Accessed November 2008.
- 12.Newell, M. L., H. Coovadia, M. Cortina-Borja, N. Rollins, P. Gaillard, and F. Dabis. 2004. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet 3641236-1243. [DOI] [PubMed] [Google Scholar]
- 13.Nugent, C. T., V. Nodelman, C. Giachetti, D. D. Richman, and D. J. Looney. 2009. Evaluation of a highly sensitive qualitative human immunodeficiency virus type 1 (HIV-1) RNA assay 1 for detection of HIV-1 suppression. J. Clin. Microbiol. 47833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouet, F., C. Montcho, C. Rouzioux, V. Leroy, P. Msellati, J. B. Kottan, B. You, I. Viho, F. Dabis, and Abidjan DITRAME Study Group (ANRS 049 clinical trial). 2001. Early diagnosis of paediatric HIV-1 infection among African breast-fed children using a quantitative plasma HIV RNA assay. AIDS 151849-1856. [DOI] [PubMed] [Google Scholar]
- 15.Sherman, G., P. Cooper, A. Coovadia, A. Puren, S. Jones, M. Mokhachane, and K. Bolton. 2005. HIV-1 DNA polymerase chain reaction for diagnosis of HIV infection in infancy in low resource setting. Paediatr. Infect. Dis. J. 24993-997. [DOI] [PubMed] [Google Scholar]
- 16.Sherman, G., G. Stevens, and W. Stevens. 2004. Affordable diagnosis of human immunodeficiency virus infection by p24 antigen detection. J. Pediatric Infect. Dis. 23173-176. [DOI] [PubMed] [Google Scholar]
- 17.Sherman, G. G., G. Stevens, S. A. Jones, P. Horsfield, and W. S. Stevens. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J. Acquir. Immune Defic. Syndr. 38615-617. [DOI] [PubMed] [Google Scholar]
- 18.Stevens, W., L. Erasmus, S. Sarang, and T. Taleng. 2008. The performance of a novel human immunodeficiency virus (HIV) type 1 total nucleic acid-based real-time PCR assay using whole blood and dried blood spots for diagnosis of HIV in infants. J. Clin. Microbiol. 463941-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens, W., G. Sherman, R. Downing, L. Parsons, C. Ou, S. Crowley, G. Gershy-Damet, K. Fransen, M. Bulterys, J. Homsy, L. Lu, T. Finkbeiner, and J. Nkengasong. 2008. Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: Consensus Statement on the Performance of Laboratory Assays for Early Infant Diagnosis. Open AIDS J. 217-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stramer, S. L., S. A. Glynn, and S. H. Kleinman. 2004. Detection of HIV and HCV infections among antibody-negative US blood donors by nucleic acid amplification testing. N. Engl. J. Med. 351760-768. [DOI] [PubMed] [Google Scholar]
- 21.UNAIDS/WHO. 2007. AIDS epidemic update. UNAIDS/WHO, Geneva, Switzerland. http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf.
- 22.Violari, A., M. F. Cotton, D. M. Gibb, A. G. Babiker, J. Steyn, S. A. Madhi, P. Jean-Philippe, J. A. McIntyre, and CHER Study Team. 2008. Early antiretroviral therapy and mortality among HIV-infected infants. N. Engl. J. Med. 3592233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zijenah, L., O. Tobaiwa, S. Rusakaniko, K. J. Nathoo, M. Nhembe, P. Matibe, and D. A. Katzenstein. 2005. Signal-boosted qualitative ultrasensitive p24 antigen assay for diagnosis of subtype C HIV-1 infection in infants under the age of 2 years. J. Acquir. Immune Defic. Syndr. 39391-394. [DOI] [PubMed] [Google Scholar]