Abstract
Infection is the main treatment-related cause of mortality in cancer patients. Rapid and accurate diagnosis to facilitate specific therapy of febrile neutropenia is therefore urgently warranted. Here, we evaluated a commercial PCR-based kit to detect the DNA of 20 different pathogens (SeptiFast) in the setting of febrile neutropenia after chemotherapy. Seven hundred eighty-four serum samples of 119 febrile neutropenic episodes (FNEs) in 70 patients with hematological malignancies were analyzed and compared with clinical, microbiological, and biochemical findings. In the antibiotic-naïve setting, bacteremia was diagnosed in 34 FNEs and 11 of them yielded the same result in the PCR. Seventy-three FNEs were negative in both systems, leading to an overall agreement in 84 of 119 FNEs (71%). During antibiotic therapy, positivity in blood culture occurred only in 3% of cases, but the PCR yielded a positive result in 15% of cases. In six cases the PCR during antibiotic treatment detected a new pathogen repetitively; this was accompanied by a significant rise in procalcitonin levels, suggestive of a true detection of infection. All patients with probable invasive fungal infection (IFI; n = 3) according to the standards of the European Organization for Research and Treatment of Cancer had a positive PCR result for Aspergillus fumigatus; in contrast there was only one positive result for Aspergillus fumigatus in an episode without signs and symptoms of IFI. Our results demonstrate that the SeptiFast kit cannot replace blood cultures in the diagnostic workup of FNEs. However, it might be helpful in situations where blood cultures remain negative (e.g., during antimicrobial therapy or in IFI).
While systemic infection is the most common cause of a febrile neutropenia episode (FNE) with significant effects on morbidity and mortality, only 30% of blood cultures taken at the onset of fever are positive (11, 15). Nonetheless, patients with FNEs are treated with broad-spectrum antimicrobial agents regardless of the result of their blood culture (7) because potentially life-threatening infections need early treatment to ensure better clinical outcome. Noninfective causes of a systemic reaction culminating in a rise in temperature such as tumor fever, drug fever, or transfusion reactions complicate the diagnostic challenge in cancer patients. In addition, the etiology of a deterioration of an FNE during antimicrobial therapy is often difficult to elucidate, since blood cultures are infrequently positive once effective antimicrobial therapy has started (4). Pathogens such as molds which are rarely found in blood cultures are not uncommon in patients with FNEs, particularly if they suffer from hematological malignancies. For these reasons, FNE is one of the conditions where new diagnostic tools to distinguish an infection from a nonmicrobial cause for fever or to identify rare pathogens are most urgently needed. In the past, raised levels of indirect markers such as procalcitonin (PCT) and interleukin 6 (3, 16) have been shown to be associated with bacteremia. Ideally, though, the cause should be identified directly and improvements in the detection of pathogens in the bloodstream should be made. In addition to refinements of the classical blood culture systems, attempts have been made to detect pathogen DNA by means of PCR. Initially, this involved conventional PCR techniques (9) detecting the gene for the 16S subunit of bacterial rRNA for the presence of bacterial DNA. Specification was then carried out by sequencing the PCR product. Later, more rapid methods were developed when real-time PCR became available (1, 14, 17). PCR results are more readily available, and the method also detects remnants of bacteria, which might make it more robust to the influence of antibiotic treatment, while potentially detecting pathogens which do not grow in the blood cultures. However, the main disadvantage, apart from higher costs, a potential for false-positive results during transient bacteremia/fungemia (e.g., during brushing of teeth), and laboratory workload, is the restriction of the spectrum of species detected. In addition, because most PCR methods use smaller sample volumes (commonly 1 to 4 ml [9, 17]), this method depends on a higher concentration of bacteria than that for the blood culture, which theoretically can reveal positive results after one living and propagating bacterial cell has been injected into the culture bottle.
A new commercially available kit (SeptiFast) to detect DNA from 20 clinically relevant pathogens has recently been evaluated in a small cohort of neutropenic patients (10) with promising results. The aim of our study was to evaluate the usefulness of the SeptiFast kit in a larger cohort of patients with febrile neutropenia after chemotherapy for hematological malignancies. Also, we sought to determine a correlation between SeptiFast results and clinical findings. Altogether 784 samples from 119 FNEs in 70 patients were analyzed and compared with clinical, microbiological, and biochemical findings in the antibiotic-naïve setting and during antimicrobial therapy.
MATERIALS AND METHODS
After approval from the local ethics committee and written informed consent, FNEs of the patients treated between September 2001 and February 2002 and between April 2003 and January 2004 on the hematology ward of our tertiary care hospital were evaluated. Fever was defined as temperature of >38°C in the axilla, and an FNE was defined as the period from the first day of fever until the patient was afebrile for five consecutive days. If two febrile episodes of one patient were included, this was usually during two different admissions for different cycles of chemotherapy. Neutropenia was defined as an absolute leukocyte count of <1 × 109/liter.
At onset of fever, one set of blood cultures (aerobic, anaerobic, and fungal) was taken by sterile venipuncture and processed using the Bactec 9420 system (Becton Dickinson, Heidelberg, Germany), followed 30 min later by a follow-up set of blood cultures. A 10-ml EDTA-blood sample was collected for determination of bacterial PCR and PCT. Thereafter, in all patients antimicrobial therapy was started. During the entire FNE, 10 ml EDTA-blood was collected daily for PCR and PCT measurements. The EDTA-blood samples were centrifuged, and the plasma was collected and frozen at −80°C until measurement. PCT level was measured using the immunochemoluminometric assay by Brahms Diagnostika (Henningsdorf, Germany) (normal range, 0 to 0.2 ng/ml). Aspergillus antigen (galactomannan) in the serum was measured twice weekly (Aspergillus Platelia; Bio-Rad, Munich, Germany); the threshold for a positive result was determined as an optical density index of >0.7 (6, 13). A chest radiogram was performed on day 1 or 2 of the febrile episode. In line with the guidelines of the DGHO Infectious Diseases Working Party (12), pneumonia was assumed in cases of typical clinical and radiological findings on the chest radiogram at onset of fever and a positive chest radiogram was confirmed by a high-resolution computed tomography (HRCT) scan. In cases of persistence of fever, an HRCT was performed after 72 h irrespective of the result of the initial chest radiogram to investigate the causality of the FNE and to exclude invasive fungal infection (IFI). IFI was assigned according to the criteria for a probable/proven IFI in keeping with the European Organization for Research and Treatment of Cancer Mycoses Study Group criteria (2). If pulmonary infiltrates were detected at this stage, pneumonia was assumed for the entire episode. Bacteremia was defined by microbial growth in one blood culture bottle, and only for coagulase-negative Staphylococcus species were two positive blood culture bottles from different venipunctures required; otherwise the result was regarded as contamination. Drug-induced fever was assigned as cause if there was a clear clinical association with a blood transfusion or high-dose cytarabine in the absence of any other signs of infection. Mucositis was assumed as the cause of fever if mucositis of >II° was present with no other signs of infection, and urinary tract infection was diagnosed if >105 CFU/ml was present in the urine in the absence of other signs of infection. Fever of unknown origin (FUO) was assumed if no other cause could be identified.
Antibiotic therapy consisted of first-line piperacillin-tazobactam monotherapy in all patients. After 72 to 96 h, antibiotics were changed to second-line treatment (usually meropenem) if the patient was still febrile. Depending on the clinical picture, some patients' second-line therapy included vancomycin. Antifungal agents (mostly amphotericin B) were added if any signs of pneumonia were present or after 3 to 5 days if the patient had persistent fever.
Preparation of DNA and PCR testing were performed as recommended by the manufacturer and as published previously (8), using the SeptiFast Lys kit, SeptiFast Prep kit, and LightCycler SeptiFast kit in a 1-ml whole-blood sample. A complete SeptiFast workflow included seven patient samples and was analyzed in less than 6 hours. The analytical sensitivity of the assay as determined by the manufacturer was between 3 and 100 CFU/ml depending on the microorganism (8). All reagents, instruments, and disposables were obtained from Roche Molecular Diagnostics (Roche Diagnostics GmbH, Penzberg, Germany). The SeptiFast kit detects 20 different pathogens (eight species of gram-negative bacteria, six species of gram-positive bacteria, and six species of fungi; for details, see reference 8). Because the PCR testing was performed retrospectively, there was no influence of PCR results on antimicrobial treatment of febrile neutropenia.
The data were analyzed using SPSS (version 14.0 for Windows; SPSS, Munich, Germany). The Kruskal-Wallis test was used to test for differences; a two-sided P value of less than 0.05 was considered significant.
RESULTS
Patient characteristics and results of blood cultures.
One hundred nineteen FNEs of 70 patients who entered the study were analyzed. Patient characteristics and clinical/microbiological causes of fever are listed in Table 1. All patients had blood cultures drawn on the first day of the febrile episode, giving a positive result in 34 FNEs.
TABLE 1.
Patient characteristics
| Patient characteristic | Value(s) |
|---|---|
| Median age, yrs (IQR) | 60 (49-66) |
| No. of males (%) | 38 (54) |
| No. (%) of patients with diagnosis | |
| Acute myeloid leukemia | 50 (71) |
| Acute lymphatic leukemia | 8 (11) |
| Non-Hodgkin lymphoma | 4 (6) |
| Multiple myeloma | 2 (3) |
| Myelodysplastic syndrome | 2 (3) |
| Aplastic anemia | 2 (3) |
| Metastatic carcinoma | 2 (3) |
| Median duration of fever, days (IQR) | 5 (2-9) |
| Median leukocyte count, 109/μl (IQR) | 0.42 (0.2-1.0) |
| No. (%) of patients with cause of fever; | |
| median PCT level, ng/ml (IQR) | |
| FUO | 46 (38); 0.27 (0.18-0.62) |
| Bacteremiaa | 34 (29); 0.77 (0.32-4.11) |
| Pneumonia | 25 (21); 0.48 (0.17-0.98) |
| Drug fever | 6 (5); 0.19 (0.13-0.25) |
| Mucositis | 5 (4); 0.11 (0.1-2.5) |
| Urinary tract infection | 3 (3); 0.15 (0.12-0.17) |
Eleven patients with bacteremia also had pneumonia, and three also had mucositis.
Correlation of blood cultures and PCR in the antibiotic-naïve setting with regard to bacteria.
Of the 119 FNEs, 34 were associated with a positive blood culture. Seven of them yielded two isolates and one yielded three isolates, resulting in 43 isolates overall detected in the blood cultures (Table 2). Of note, the kit detects Staphylococcus epidermidis and Staphylococcus haemolyticus as coagulase-negative staphylococci (CoNS) and Streptococcus mitis as Streptococcus species. Also, the SeptiFast kit does not detect Corynebacterium spp., Fusobacterium spp., and Morganella morganii, leaving five positive blood culture results in our cohort undetectable by PCR. Therefore, taking the blood cultures as a gold standard, we could expect 38 positive results. The PCR samples taken at the beginning of the FNEs yielded 29 positive results in 25 FNEs. In 11 FNEs the positive result in the PCR was concordant with the blood culture result; in two FNEs both PCR and blood cultures were positive but yielded different results. Thus, with blood cultures and PCR together, 61 pathogens were detected at the beginning of 46 FNEs whereas 73 FNEs were negative in both systems. Thirty-two pathogens were isolated by blood culture only (including those not detectable by SeptiFast), 18 by PCR only, and 11 by both systems. Table 2 depicts the overall agreement of results yielded at the beginning of the FNE. In 84 of 119 (71%) FNEs, there was an agreement between PCR and blood culture (either both negative or both yielding the same isolate). If the five FNEs with a blood culture isolate not included in the SeptiFast panel are excluded, the overall agreement changes to 84/114 FNEs (74%).
TABLE 2.
Blood culture and PCR results in the first 24 h of the 119 FNEs with 61 isolates total
| Pathogen | No. of FNEs with positive result by:
|
|||
|---|---|---|---|---|
| Either method | Both methods | PCR only | Blood culture only | |
| None | 73 | |||
| Gram-positive bacteria | ||||
| Staphylococcus species (CoNS) | 16 | 1 | 5 | 10 |
| Staphylococcus aureus | 8 | 4 | 4 | 0 |
| Streptococcus species | 7 | 0 | 0 | 7 |
| Streptococcus pneumoniae | 1 | 0 | 1 | 0 |
| Enterococcus faecalis | 1 | 0 | 1 | 0 |
| Enterococcus faecium | 3 | 1 | 0 | 2 |
| Gram-negative bacteria | ||||
| Escherichia coli | 7 | 2 | 2 | 3 |
| Stenotrophomonas maltophilia | 2 | 1 | 1 | 0 |
| Klebsiella pneumoniae/oxytoca | 2 | 0 | 0 | 2 |
| Pseudomonas aeruginosa | 5 | 2 | 1 | 2 |
| Fungi | ||||
| Candida albicans | 1 | 0 | 1 | 0 |
| Aspergillus fumigatus | 2 | 0 | 2 | 0 |
| Candida glabrata | 1 | 0 | 0 | 1 |
| Not included in PCR list | ||||
| Corynebacterium species | 2 | 0 | 0 | 2 |
| Fusobacterium species | 1 | 0 | 0 | 1 |
| Morganella morganii | 2 | 0 | 0 | 2 |
Evaluation of PCR in the light of clinical infection markers.
In the antibiotic-naïve setting, none of the febrile episodes classified as drug fever, mucositis, or urinary tract infection yielded any positive results in the PCR. In contrast, 5 out of 45 patients with FUOs and 1 of 25 patients with pneumonia showed a positive result in the PCR at the beginning of their febrile episodes. As expected, PCT levels were significantly higher when the blood culture and/or the PCR gave a positive result (for positive blood culture, median PCT level was 0.7 ng/ml and interquartile range [IQR] was 0.3 to 4.1 ng/ml; for negative blood culture, median PCT level was 0.3 ng/ml and IQR was 0.2 to 0.9 ng/ml [P = 0.004]; for positive PCR, median PCT level was 1.8 ng/ml and IQR was 0.3 to 5.7 ng/ml; and for negative PCR, median PCT level was 0.3 ng/ml and IQR was 0.16 to 0.72 ng/ml [P < 0.001, Kruskal-Wallis test]).
PCR during antimicrobial therapy.
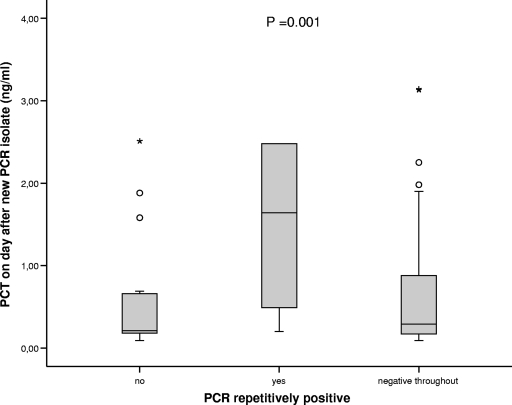
In addition to the blood cultures taken before initiation of antibiotic therapy, the first cohort of 53 episodes in 31 patients had blood cultures drawn on every day of fever (107 additional samples after start of antibiotic therapy). Here, two positive results on the second day of fever were obtained, one revealing CoNS and the other confirming a positive result on day 1 for Staphylococcus haemolyticus. Because of this low yield, the practice of daily blood cultures was abandoned in the following cohort and blood cultures during antimicrobial therapy were taken only if clinically indicated. In contrast to the blood cultures, which yielded 3% positive results during antibiotic therapy, the PCR was positive in 100 of 665 samples taken during antibiotic therapy (15% [Table 3]). Sixteen FNEs with initially positive results in the PCR remained positive for at least a second day (one FNE was double positive [Table 3]). Also, in nine FNEs with initially positive blood culture results and concordant PCR results, the PCR remained positive after the start of antibiotic therapy; in one case of Escherichia coli septicemia, the PCR was negative on day 1 but became positive on day 2 and day 3. Generally, if a pathogen was detected in the beginning of the febrile episode, the PCR remained positive for several days, in contrast to the blood culture results (data not shown). In 28 FNEs with a median duration of fever of 8 days (IQR, 6 to 14 days), an initially negative PCR result became positive or showed a new pathogen after a median of 7 days (IQR, 5 to 8 days). Nine of these FNEs yielded two different isolates in different samples during the course of the episode. Again, the PCR results were evaluated in the light of clinical markers, and significantly higher PCT levels were detected on the day after a PCR detecting a gram-negative bacterium or fungus (for a PCR with a negative result, the median PCT level was 0.4 ng/ml and the IQR was 0.2 to 1 ng/ml; for a PCR detecting a gram-positive bacterium, the median PCT level was 0.4 ng/ml and the IQR was 0.3 to 1 ng/ml; for a PCR detecting a gram-negative bacterium, the median PCT level was 0.9 ng/ml and the IQR was 0.2 to 5.3 ng/ml; for a PCR detecting a fungus, the median PCT level was 1.4 ng/ml and the IQR was 0.8 to 2.3 ng/ml; P = 0.001 [Kruskal-Wallis test]). In 6 of the 28 FNEs with a new result during the course of the episode, the same new pathogen was detected in more than one sample. Interestingly, episodes with repetitively positive PCRs during antibiotic therapy showed a significant rise in PCT levels associated with the newly detected pathogen (Fig. 1).
TABLE 3.
Results of blood cultures and PCR during antimicrobial therapy
| Pathogen | No. (%) of positive results byc:
|
No. of FNEs (no. initially positive) | |
|---|---|---|---|
| Blood culture | PCR | ||
| None | 113 (97) | 565 (84) | |
| Staphylococcus aureus | 0 | 32 (4.8) | 19 (6) |
| Staphylococcus species (CoNS) | 2 (1.8)a | 13 (1.9) | 9 (2) |
| Enterococcus faecium | 0 | 14 (2.0) | 6 (1) |
| Enterococcus faecalis | 0 | 3 (0.4) | 1 (1) |
| Escherichia coli | 0 | 6 (0.9) | 4 (3) |
| Pseudomonas aeruginosa | 0 | 14 (2.0) | 3 (1) |
| Stenotrophomonas maltophilia | 1 (0.9)b | 4 (0.6) | 3 (2) |
| Candida krusei | 0 | 1 (0.1) | 1 (0) |
| Candida albicans | 0 | 8 (1.2) | 2 (0) |
| Aspergillus fumigatus | 0 | 9 (1.3) | 5 (1) |
One sample yielded a positive result with the same isolate in the accompanying PCR.
Follow-up result of previously positive blood culture; the accompanying PCR yielded the same isolate.
The numbers of samples taken on febrile days were 116 for blood culture and 665 (with 669 results) for PCR.
FIG. 1.
PCT levels in the group of patients with repetitively positive PCR results in follow-up (n = 6; median, 1.64 ng/ml; IQR, 0.49 to 2.48 ng/ml) compared to the group with only one positive PCR sample (n = 22; median, 0.24 ng/ml; IQR, 0.19 to 0.69 ng/ml) or with negative PCR/blood culture results throughout the FNE (n = 50; median, 0.29 ng/ml; IQR, 0.17 to 0.88 ng/ml; P = 0.001, Kruskal-Wallis test). Horizontal lines within bars represent medians; bars represent IQRs; whiskers represent confidence intervals; circles and asterisks represent outliers.
Detection of IFI by PCR.
The only patient with a proven IFI (Candida glabrata in one blood culture which also grew Klebsiella pneumoniae and Enterococcus faecium) yielded a negative result for fungus in the PCR, although the PCR did detect Enterococcus faecium. This might be due to the threshold for detection of Candida glabrata, as 100 CFU/ml are required to ensure 100% detection, a level which may well be above the yeast cell concentration in the patient's blood. Three patients with blood cultures negative for fungi had positive PCR results for Candida species during the course of their FNEs. In one case of a long (10-day) FNE with negative blood cultures, the PCR became positive for Candida albicans on day 5 and remained positive until the end of the episode. This course was associated with a convincing rise in PCT level despite negative blood cultures (Fig. 2A).
FIG. 2.
Time course of examples with a positive result for fungal DNA. The left axis depicts the temperature in degrees Celsius; the right axis depicts PCT levels (ng/ml) and galactomannan optical density indices. (A) Episode with PCR positive for Candida albicans. (B) Episode with PCR positive for Aspergillus. This episode was classified as probable IFI with typical radiological features of fungal pneumonia and repetitively positive Aspergillus antigen.
In the entire cohort, 12 FNEs were associated with a galactomannan antigen level (optical density index) of >0.7 at least once during the FNE. With the use of SeptiFast, six FNEs were associated with a positive PCR result for Aspergillus fumigatus, five of which also had a galactomannan antigen level (optical density index) of >0.7. Two positive PCR results were detected in the beginning, and the other four were detected during the febrile episode. Five patients had pneumonia, and the remaining one was classified as drug fever clinically. Of note, the patient classified as drug fever did not reach a galactomannan antigen level (optical density index) of >0.7 and had only one positive PCR result. In contrast, of the patients with pneumonia, three were classified as probable IFIs according to HRCT findings and their galactomannan antigen levels (optical density index of >0.7 on two subsequent measurements; Fig. 2B shows an example). There were no more patients who fulfilled the criteria for a probable IFI; therefore, all of the patients with probable IFIs had positive results for Aspergillus in the PCR.
DISCUSSION
This study evaluated the usefulness of a multiplex PCR for the detection of microbial DNA in patients with FNEs prior to and during antibiotic therapy. PCR to detect microbial DNA is a method that has been receiving some attention throughout the past 10 years. It is an established method for certain pathogens such as cytomegalovirus, Epstein-Barr virus, hepatitis virus, and mycobacteria, and recent studies evaluated broad-range PCR for the detection of bacteria or fungi. In particular, one early study gave very promising results with regard to sensitivity, correctly identifying 9 of 11 blood culture-positive episodes and achieving positive results in another 20 blood culture-negative episodes (9). However, this was not a real-time PCR but instead a nested PCR with sequencing of the PCR product for identification and thus probably no less time-consuming than the blood cultures. Other groups tested different real-time PCR methods before the SeptiFast kit became commercially available. One study had a sensitivity of 10/13 (14), and one study had a sensitivity of only 3/10 (1). In contrast, both studies had high rates of positivity in blood culture-negative samples, 54/77 (14) and 10/35 (1), which were partially attributed to contamination during the processing procedure (14). There is little information as to how these positive results compare with the clinical setting with regard to any other markers of infection.
In our study, patients who had a positive PCR result in the antibiotic-naïve setting often remained positive for 1 or 2 more days. Unfortunately, the SeptiFast kit does not allow quantification of the DNA load, which would be interesting in the context of the clinical course. It is also likely that the PCR during antibiotic therapy detects remains of bacteria or nonviable microorganisms. In addition to information on the type of infection, detection of bacterial CpG motifs themselves might be clinically relevant because they have a proinflammatory effect (5). This is supported by our observation that patients with a positive PCR result have raised PCT levels.
The disappointingly low reproducibility of the blood culture results by PCR in our study could well be caused by the technique. A study testing the sensitivity of the SeptiFast kit on spiked blood samples showed a dose dependency of correct detection. Klebsiella pneumoniae, Candida glabrata, and Streptococcus spp. as well as CoNS have a detection rate ranging from 0% to 80% if they occur at the low concentration of 3 CFU/ml (8). Interestingly, SeptiFast failed to detect Klebsiella spp. (n = 2), Candida glabrata (n = 1), Streptococcus spp. (n = 7), and CoNS (n = 10) fairly frequently. It seems likely, therefore, that our low detection rate was caused by a low concentration of microbial DNA. One reason for a lower concentration of microbial DNA in neutropenic patients might also be the lack of neutrophils containing microbial DNA. Therefore, our PCR results strongly suggest that a blood culture prior to antibiotic therapy in febrile neutropenia cannot be replaced by a molecular method yet. The situation is different for persistent fever during antibiotic therapy and for diagnosis of IFI which may otherwise not be detected by blood cultures. In the setting of persistent fever during antibiotic therapy, the role for blood cultures is very small, since several studies show that a new pathogen is detected in <1% of cases (4, 15). Our study confirms this finding as we also had only one patient with a newly positive result after the start of antimicrobial therapy. On the other hand, the likelihood of an infection as the cause of persistent fever is very high in neutropenic patients, and it seems reasonable to assume that the low positivity rate is a false finding. Thus, the blood culture does not serve as a good standard in this setting. In contrast, the PCR technique results in a positivity rate of 15% during antimicrobial therapy and positive results were associated with increased PCT levels, rendering these findings clinically plausible. Nonetheless, detection of gram-positive pathogens like CoNS by PCR still needs to be interpreted with caution because of the risk of contamination (14), and further studies are required to evaluate the reliability of SeptiFast in this setting.
We were particularly intrigued by the detection of Aspergillus by PCR. All clinical courses but one make it likely that this is a clinically relevant finding as opposed to contamination. In addition, all patients qualifying for the diagnosis of a probable fungal infection according to the criteria of the European Organization for Research and Treatment of Cancer had at least one positive PCR. For this reason, the SeptiFast kit might contribute to a quicker and more accurate diagnosis. Obviously, larger prospective studies are needed to address this question in greater detail.
This is the first study to evaluate the SeptiFast kit in a larger cohort of patients with febrile neutropenia—probably the clinical situation where improved infection diagnostics are most urgently needed. It reflects practice in a typical clinical setting under realistic conditions in a large number of FNEs with a clear definition of the FNE and a thorough documentation of clinical parameters. We conclude from our study that the SeptiFast kit cannot as yet replace blood cultures in the initial diagnostic workup of febrile neutropenia but may provide helpful information in episodes with persistent fever during antimicrobial therapy and in the diagnosis of suspected IFI.
Acknowledgments
We thank Sabine Mering and Makbule Kobulay for excellent technical assistance and all the nurses and doctors of our leukemia ward for excellent clinical care.
This study was supported by the Leukämie-Initiative Bonn and by an unrestricted educational grant from Roche Diagnostics GmbH. Marie von Lilienfeld-Toal is supported by an EBMT-Amgen fellowship and a BONFOR research grant; Axel Glasmacher is an employee of Celgene.
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Ammann, R. A., F. Zucol, C. Aebi, F. K. Niggli, T. Kuhne, and D. Nadal. 2007. Real-time broad-range PCR versus blood culture. A prospective pilot study in pediatric cancer patients with fever and neutropenia. Support Care Cancer 15637-641. [DOI] [PubMed] [Google Scholar]
- 2.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 347-14. [DOI] [PubMed] [Google Scholar]
- 3.Fleischhack, G., I. Kambeck, D. Cipic, C. Hasan, and U. Bode. 2000. Procalcitonin in paediatric cancer patients: its diagnostic relevance is superior to that of C-reactive protein, interleukin 6, interleukin 8, soluble interleukin 2 receptor and soluble tumour necrosis factor receptor II. Br. J. Haematol. 1111093-1102. [DOI] [PubMed] [Google Scholar]
- 4.Grace, C. J., J. Lieberman, K. Pierce, and B. Littenberg. 2001. Usefulness of blood culture for hospitalized patients who are receiving antibiotic therapy. Clin. Infect. Dis. 321651-1655. [DOI] [PubMed] [Google Scholar]
- 5.Hacker, H., R. M. Vabulas, O. Takeuchi, K. Hoshino, S. Akira, and H. Wagner. 2000. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J. Exp. Med. 192595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbrecht, R., V. Letscher-Bru, C. Oprea, B. Lioure, J. Waller, F. Campos, O. Villard, K. L. Liu, S. Natarajan-Ame, P. Lutz, P. Dufour, J. P. Bergerat, and E. Candolfi. 2002. Aspergillus galactomannan detection in the diagnosis of invasive aspergillosis in cancer patients. J. Clin. Oncol. 201898-1906. [DOI] [PubMed] [Google Scholar]
- 7.Hughes, W. T., D. Armstrong, G. P. Bodey, E. J. Bow, A. E. Brown, T. Calandra, R. Feld, P. A. Pizzo, K. V. Rolston, J. L. Shenep, and L. S. Young. 2002. 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34730-751. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann, L. E., K. P. Hunfeld, T. Emrich, G. Haberhausen, H. Wissing, A. Hoeft, and F. Stuber. 2008. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med. Microbiol. Immunol. 197313-324. [DOI] [PubMed] [Google Scholar]
- 9.Ley, B. E., C. J. Linton, D. M. Bennett, H. Jalal, A. B. Foot, and M. R. Millar. 1998. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 17247-253. [DOI] [PubMed] [Google Scholar]
- 10.Mancini, N., D. Clerici, R. Diotti, M. Perotti, N. Ghidoli, D. De Marco, B. Pizzorno, T. Emrich, R. Burioni, F. Ciceri, and M. Clementi. 2008. Molecular diagnosis of sepsis in neutropenic patients with hematological malignancies. J. Med. Microbiol. 57601-604. [DOI] [PubMed] [Google Scholar]
- 11.Marchetti, O., and T. Calandra. 2002. Infections in neutropenic cancer patients. Lancet 359723-725. [DOI] [PubMed] [Google Scholar]
- 12.Maschmeyer, G., T. Beinert, D. Buchheidt, H. Einsele, C. P. Heussel, M. Kiehl, and J. Lorenz. 2003. Diagnosis and antimicrobial therapy of pulmonary infiltrates in febrile neutropenic patients—guidelines of the Infectious Diseases Working Party (AGIHO) of the German Society of Hematology and Oncology (DGHO). Ann. Hematol. 82S118-S126. [DOI] [PubMed] [Google Scholar]
- 13.Orlopp, K., M. von Lilienfeld-Toal, G. Marklein, S. M. Reiffert, A. Welter, C. Hahn-Ast, I. Purr, M. Gorschluter, E. Molitor, and A. Glasmacher. 2008. False positivity of the Aspergillus galactomannan Platelia ELISA because of piperacillin/tazobactam treatment: does it represent a clinical problem? J. Antimicrob. Chemother. 621109-1112. [DOI] [PubMed] [Google Scholar]
- 14.Peters, R. P., T. Mohammadi, C. M. Vandenbroucke-Grauls, S. A. Danner, M. A. van Agtmael, and P. H. Savelkoul. 2004. Detection of bacterial DNA in blood samples from febrile patients: underestimated infection or emerging contamination? FEMS Immunol. Med. Microbiol. 42249-253. [DOI] [PubMed] [Google Scholar]
- 15.Serody, J. S., M. M. Berrey, K. Albritton, S. M. O'Brien, E. P. Capel, S. H. Bigelow, D. J. Weber, D. A. Gabriel, J. M. Wiley, M. J. Schell, P. H. Gilligan, and T. C. Shea. 2000. Utility of obtaining blood cultures in febrile neutropenic patients undergoing bone marrow transplantation. Bone Marrow Transplant. 26533-538. [DOI] [PubMed] [Google Scholar]
- 16.von Lilienfeld-Toal, M., P. Dietrich, A. Glasmacher, L. Lehmann, P. Breig, C. Hahn, I. G. Schmidt-Wolf, G. Marklein, S. Schroeder, and F. Stuber. 2004. Markers of bacteremia in febrile neutropenic patients with hematological malignancies: procalcitonin and IL-6 are more reliable than C-reactive protein. Eur. J. Clin. Microbiol. Infect. Dis. 23539-544. [DOI] [PubMed] [Google Scholar]
- 17.Warwick, S., M. Wilks, E. Hennessy, J. Powell-Tuck, M. Small, J. Sharp, and M. R. Millar. 2004. Use of quantitative 16S ribosomal DNA detection for diagnosis of central vascular catheter-associated bacterial infection. J. Clin. Microbiol. 421402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]