Abstract
Cefepime (FEP) and ceftazidime (CAZ) are broad-spectrum cephalosporins that display similar MICs for wild-type Pseudomonas aeruginosa strains. Recently, P. aeruginosa isolates showing a discordance in susceptibility to CAZ and FEP have been noted at the Hospital de Bellvitge in Barcelona, Spain, and a clustering was suspected. During the study period (March to December 2007), 51 patients, particularly those in an intensive care units (ICUs) (n = 29 [57%]), infected or colonized with at least one P. aeruginosa non-FEP-susceptible and CAZ-susceptible (Fepns Cazs) phenotype strain were detected. Twenty-three (45%) patients were infected, and the respiratory tract was the most frequent site of infection. Changes in the consumption of antimicrobials in the ICUs were observed over time: a progressive reduction in the levels of consumption of carbapenems (247 defined daily doses [DDD]/1,000 patient days to 66 DDD/1,000 patient days; P = 0.008), after restriction of its use in 2006, and an expected increase in the rate of piperacillin-tazobactam use (42 DDD/1,000 patient days in 2004 to 200 DDD/1,000 patient days in 2007; P < 0.001). Throughout the whole study period, only a single clone of a P. aeruginosa Fepns Cazs phenotype strain was identified by pulsed-field gel electrophoresis analysis to be associated with the hyperexpression of MexXY-OprM and the production of an integron-borne PSE-1 ß-lactamase. In conclusion, we identified an epidemic P. aeruginosa clone of an Fepns Cazs phenotype strain involving 51 patients, in particular, ICU patients. The combination of the overexpression of an efflux pump and PSE-1 ß-lactamase production is associated with the multidrug-resistant phenotype. The dominant use of a single class of antibiotics could have provided the selective pressure required for the emergence and spread of this P. aeruginosa strain.
Pseudomonas aeruginosa is an opportunistic pathogen that causes a variety of nosocomial infections. Cefepime (FEP) and ceftazidime (CAZ) are broad-spectrum cephalosporins that display similar MICs for wild-type P. aeruginosa strains. Although the rates of susceptibility of these two antipseudomonal cephalosporins appear to be identical for P. aeruginosa isolates in the United States (20), several European studies have recently reported an unusual discordance in the MICs of FEP and CAZ (4, 6, 7), with P. aeruginosa isolates being less susceptible to the former cephalosporin.
The most frequent mechanisms of resistance to extended-spectrum cephalosporins in P. aeruginosa are derepression of the chromosomal AmpC ß-lactamase, the impermeability of the outer membrane, and increased efflux (9). FEP, in contrast to CAZ, is efficiently extruded by various efflux pumps, including the MexCD-OprJ and MexXY-OprM efflux pumps (16, 22). In addition, previous studies have noted the selectively reduced susceptibility to FEP among P. aeruginosa strains producing OXA ß-lactamases (2), as opposed to those that produce class A extended-spectrum ß-lactamases (32), which usually confer resistance to CAZ and FEP.
In response to an increase in the incidence of carbapenem-resistant and multidrug-resistant P. aeruginosa isolates in the Hospital de Bellvitge, Barcelona, Spain, since 2004, a microbiology surveillance program was introduced. Recently, however, P. aeruginosa isolates showing variable susceptibilities to carbapenems and a discordance in susceptibility between CAZ and FEP have been noted. A clustering of the P. aeruginosa non-FEP-susceptible and CAZ-susceptible (Fepns Cazs) phenotype was suspected in the hospital's intensive care unit (ICUs). Here we elucidate the clinical and molecular epidemiology of the isolates with this discordant phenotype and the resistance mechanisms involved.
MATERIALS AND METHODS
Hospital setting.
The Hospital de Bellvitge is a university-affiliated public institution for adult patients located in Barcelona, Spain. It contains 900 beds for acute medical and surgical care but no wards for pediatrics, obstetrics, or burns. The hospital has two 12-bed medical-surgical ICUs managed by separate teams of medical and nursing staff. The paramedical care providers (physiotherapists, radiographic personnel, nutrition support team) have contact with patients in both ICU units. All rooms in the ICUs are single.
Data collection-outbreak detection.
The surveillance program was initiated in March 2007 after the detection of several P. aeruginosa Fepns Cazs phenotype isolates in the ICUs. A retrospective revision indicated that sporadic Fepns Cazs phenotype isolates occurred in 2005 and 2006.
A daily laboratory surveillance program was introduced by the infection control team to identify new cases. A clinical assessment was determined by following the definitions of the Centers for Disease Control and Prevention (13). Patients with samples from any body site positive for P. aeruginosa isolates of the Fepns Cazs phenotype but without related signs or symptoms of infection were considered colonized. Acquisition was considered to have occurred in the ICU when this phenotype was detected during a stay in an ICU or when it was detected during the first week after discharge from such a unit. Demographic characteristics and underlying diseases, according to the index of Charlson et al. (8) (the Charlson index), were recorded. We also assessed the patients for the devices that they had in place (intravascular catheters, urinary catheters, endotracheal tubes) and whether they had undergone surgery. Prior antibiotic therapy was defined as the administration of distinct groups of antibiotics for >48 h during the previous 3 months and/or from the time of admission until the time that P. aeruginosa Fepns Cazs phenotype colonization was noted. The groups of antibiotics analyzed were penicillins (piperacillin-tazobactam, amoxicillin [amoxicilline]-clavulanic acid), antipseudomonal cephalosporins (CAZ and FEP), other cephalosporins, aminoglycosides, fluoroquinolones, and carbapenems.
A case-control study was performed. The eligible case patients were all patients infected with P. aeruginosa strains with the Fepns Cazs phenotype identified from March to December 2007. The case patients were compared with patients from two control groups (15), according to the same type of infection and the closest date of admission. The members of control group 1 were selected from among patients with infections caused by P. aeruginosa strains susceptible to all antipseudomonal antimicrobials. Control group 2 consisted of patients who had had hospital stays equal to or longer than those of the case patients.
Antibiotic pressure.
Drug consumption was evaluated by using data from the hospital pharmacy records. Antibiotic pressure was determined from the consumption of several groups of antibiotics in ICU wards. The antimicrobials analyzed were carbapenems, antipseudomonal cephalosporins, fluoroquinolones, and piperacillin-tazobactam. Data were reported as the number of defined daily doses (DDDs) (24) per 1,000 patient days per month from January 2004 to December 2007 (the period from 2004 to 2006 was before the introduction of the surveillance program, and 2007 was during the study period). Time-series analysis was used to assess the trend in drug consumption. Segmented linear regression techniques were applied to show the patterns of consumption in the time periods before the study period (2004 to 2006) and during the study period (2007) by taking account of the trends over time.
In 2006, carbapenem consumption was restricted in ICUs to control the increase in the rate of carbapenem resistance among patients colonized or infected by P. aeruginosa. However, scheduled antimicrobial cycling was not implemented in these wards during the study period.
Bacterial strains and antimicrobial susceptibility studies.
P. aeruginosa strains were identified and tested for their antimicrobial susceptibilities by means of a MicrosScan automated microdilution system with CN1S and CO1S panels (Dade International, West Sacramento, CA). The antibiotics studied were piperacillin, ticarcillin, piperacillin-tazobactam, CAZ, FEP, aztreonam, imipenem, meropenem, ciprofloxacin, gentamicin, tobramycin, amikacin, and colistin. The criteria of the Clinical and Laboratory Standards Institute (CLSI) (10) were used to define susceptibility or resistance to these antimicrobial agents. Intermediate and resistant isolates were considered nonsusceptible.
The susceptibility profiles of strains with CAZ MICs of ≤8 μg/ml, FEP MICs of >8 μg/ml, piperacillin MICs of >64 μg/ml, piperacillin-tazobactam MICs of >64 μg/ml, and ticarcillin MICs of >64 μg/ml were confirmed by the disk diffusion method. These strains were examined for extended-spectrum ß-lactamse production by the double-disk synergy test (DDST). DDSTs were performed by placing disks of FEP, CAZ, and aztreonam (30 μg each) at a distance of 30 or 20 mm (center to center) from a disk containing amoxicillin-clavulanic acid (amoxicillin at 20 μg and clavulanic acid at 10 μg). For all strains, the FEP MICs (Bristol-Myers Squibb) were determined by the agar dilution method, as described in the CLSI guidelines (10). We used P. aeruginosa ATCC 27853 as a control strain.
Identification of the epidemic clone by PFGE.
Chromosomal DNA was prepared from 33 representative P. aeruginosa Fepns Cazs phenotype isolates for analysis by pulsed-field gel electrophoresis (PFGE) by a previously described method (12) and digested with XbaI. DNA fragments were separated with a CHEF DR III apparatus (Bio-Rad Laboratories, Hercules, CA). Another three epidemic multiresistant P. aeruginosa strains from our hospital were used as controls for PFGE (27). Electrophoresis was run at 6 V/cm and 14°C for 23 h, with the pulse times ranging from 5 to 25 s. The DNA restriction patterns generated by PFGE were interpreted according to the guidelines described previously (29, 31). Isolates with PFGE patterns that differed by more than four fragments were ascribed to distinct genotypes; isolates with differences of four restriction fragments or less were considered to be subtypes of a single genotype (31).
Characterization of β-lactamases.
To detect the production of non-class C ß-lactamases, two approaches were followed: (i) the cloxacillin inhibition test, by which ß-lactamase activity was determined after the incubation of crude sonic extracts in 50 μM cloxacillin (a class C ß-lactamase inhibitor) for 15 min, as described previously (17), and (ii) isoelectric focusing of crude sonic extracts with Phast gels (pH gradient 3 to 9) in a Phast system apparatus (Pharmacia AB, Uppsala, Sweden). On the basis of the results of the preliminary phenotypic tests, the isolates were examined for the potential presence of genes encoding acquired ß-lactamases by PCR. Previously described primers and conditions were used to amplify the genes encoding PER-, CTX-M-, SHV-, TEM-, OXA-, and PSE-type ß-lactamases (11, 14). Strains from our laboratory collection producing the β-lactamases PER (PER-1), PSE (PSE-1), SHV (SHV-2), TEM (TEM-1), CTX-M (CTX-M-1, CTX-M-2, and CTX-M-9, as representatives of the most frequent groups of CTX-M enzymes), and OXA (OXA-1, OXA-2, and OXA-10, as representatives of the most frequent groups of OXA enzymes) were used as controls in the PCR experiments.
Characterization of genetic elements harboring horizontally acquired ß-lactamases.
To ascertain the presence of an integron carrying the potential horizontally acquired ß-lactamase genes, previously described primers and conditions were used to amplify the genes within the integron, namely, intl1, qacEΔ1, and the genes in between those two genes (14). After PCR amplification, sequencing reactions were performed with a BigDye Terminator kit (Perkin-Elmer Applied Biosystems, Foster City, CA), and the sequences were analyzed on an ABI Prism 3100 DNA sequencer (Perkin-Elmer Applied Biosystems). The resulting sequences were then compared with those available in the GenBank database (www.ncbi.nih.gov/BLAST).
Quantification of gene expression by RT-PCR.
To quantify the expression of the chromosomal ß-lactamase AmpC and several efflux pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM), the relative level of mRNA of each gene (ampC, mexB, mexD, mexF, or mexY) was determined by real-time reverse transcription-PCR (RT-PCR) by previously described protocols (17, 25). Strains were considered AmpC, MexCD-OprJ, MexEF-OprN, or MexXY-OprM hyperproducers when the relative level of expression of the corresponding gene was at least 10-fold higher than that by the reference strain PAO1. The breakpoint used for MexAB-OprM hyperproduction was a threefold higher level of expression than that by PAO1. Previously obtained or constructed PAO1 mutants that hyperproduce AmpC or the several efflux pumps were used as controls in the RT-PCR experiments (18, 21, 23).
Statistical analysis.
Continuous variables were compared by the t test. The chi-square test or Fisher's exact test was used to compare categorical variables. Multivariate logistic regression analysis was performed to determine variables that were independently associated with the risk of acquisition of a P. aeruginosa strain with the Fepns Cazs phenotype.
RESULTS
Description of outbreak.
The outbreak was initially detected in March 2007, when an unusual discordance in the MICs of FEP and CAZ was detected for a strain of P. aeruginosa isolated from a blood sample from a neutropenic patient in the ICU who had acute lymphoblastic leukemia. Four additional cases with this P. aeruginosa phenotype were detected during the same month in the ICUs.
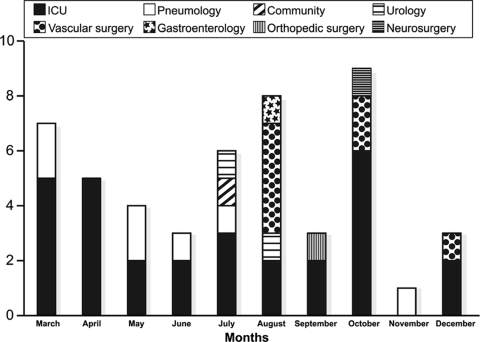
During the study period (March to December 2007), 51 patients with at least one clinical sample containing a P. aeruginosa isolate with the Fepns Cazs phenotype were detected. The genotype analysis confirmed the clonal origin of the strains selected. Among these 51 patients, 29 (57%) were ICU patients, 19 (37%) acquired this strain in a non-ICU setting, and the remaining 3 patients were considered to have acquired the strain in a health care setting: 1 frequently attended a urology outpatient clinic and 2 had previously been admitted to another hospital. The ICU wards were the epicenter of the outbreak, which finally reached six other wards. Figure 1 shows the distribution of patients colonized or infected by the strain according to the ward in which they were hospitalized. Nineteen patients were in non-ICU wards; eight of those patients in the vascular surgery ward and six were in the pneumology ward. The remaining five patients were sporadic cases without evidence of temporal clustering: urology ward (two cases), gastroenterology ward (one case), orthopedic surgery ward (one case), and neurology ward (one case). Epidemiological analysis of these 19 patients showed that 4 of them had previously been admitted to an ICU (2 patients from the pneumology ward, 1 patient from the orthopedic surgery ward, and 1 patient from a vascular surgery ward).
FIG. 1.
Ward locations of patients colonized by or infected with a P. aeruginosa strain hyperproducing MexXY-OprM and the PSE-1 ß-lactamase over the course of the study period.
Subsequent to the described study period, the incidence of cases colonized or infected by P. aeruginosa isolates with the Fepns Cazs phenotype remained at a similar level (47 cases) after reinforcement of the use of barrier precautions without an active antibiotic control program. The incidence of cases in the ICUs declined in 2008 (14/47 [30%]), while colonization or infection with such P. aeruginosa Fepns Cazs isolates in the general medical and surgical units persisted, particularly in the vascular surgery ward (12/47 [25.5%]).
Clinical data.
Among the 51 patients, 23 (45%) were infected and 28 (55%) were colonized. Of the 23 infected patients (the initial episode only), respiratory tract infection was the most frequent (nine episodes), followed by bacteremia of unknown origin (three episodes). The remaining infections were urinary tract infections (three cases), osteomyelitis (three cases), soft tissue infections (four cases), and vascular catheter-related bacteremia (one case).
The overall mortality rate as a result of infection with the P. aeruginosa strain with the Fepns Cazs phenotype was 35% (8 of 23 patients with infections). Of those eight patients, four of them had bacteremia (three of unknown origin and one vascular catheter-related bacteremia). Two patients with bacteremia of unknown origin died during the first 24 h after a positive blood culture. Of the other two patients, one received adequate antimicrobial therapy with imipenem and amikacin and the other was treated only by removing the vascular catheter; the patients died 23 days and 50 days, respectively, after P. aeruginosa was isolated in blood cultures. The remaining four patients had pneumonia, and all received adequate definitive antimicrobial therapy and died between 10 and 30 days after diagnosis of the infection.
Table 1 and Table 2 show the results of univariate and multivariate analyses of risk factors for P. aeruginosa Fepns Cazs phenotype acquisition, respectively.
TABLE 1.
Univariate analysis of risk factors for P. aeruginosa Fepns Cazs phenotype acquisition in 23 infected patients
| Factor | Value for case patients | Control group 1a (n = 23)
|
Control group 2b (n = 23)
|
||
|---|---|---|---|---|---|
| Value | P | Value | P | ||
| Age (yr) | 59.9 ± 16.2 | 65.5 ± 16.2 | 0.1 | 63.2 ± 16.0 | 0.3 |
| Sex (no. of M/no. of Fc) | 18/5 | 17/6 | 0.7 | 18/5 | 1.0 |
| No. of patients with underlying disease (Charlson index > 2) | 4 | 12 | 0.01 | 8 | 0.1 |
| Duration of prior hospitalization (days) | 33.5 ± 32.8 | 25.3 ± 15.1 | 0.3 | 34.0 ± 29.0 | 0.8 |
| No. of patients with: | |||||
| Catheter use | |||||
| Venous | 19 | 23 | 0.1 | 23 | 0.1 |
| Urinary | 17 | 18 | 1.0 | 17 | 0.7 |
| Mechanical ventilation | 13 | 14 | 0.9 | 13 | 0.8 |
| Surgery | 10 | 14 | 0.2 | 16 | 0.07 |
| Prior antimicrobial use | |||||
| Any antimicrobial | 23 | 23 | 1.0 | 22 | 1.0 |
| Amoxicillin-clavulanic acid | 10 | 10 | 1.0 | 13 | 0.3 |
| Piperacillin-tazobactam | 18 | 5 | <0.01 | 6 | <0.01 |
| Antipseudomonal cephalosporins | 6 | 4 | 0.4 | 1 | 0.09 |
| Other cephalosporins | 6 | 4 | 0.4 | 2 | 0.2 |
| Fluoroquinolones | 9 | 2 | 0.01 | 5 | 0.2 |
| Aminoglycosides | 10 | 3 | 0.02 | 1 | <0.01 |
| Carbapenems | 6 | 7 | 0.7 | 5 | 0.7 |
| Overall mortality | 8 | 10 | 0.5 | 3 | 0.08 |
Control group 1, patients infected with drug-susceptible P. aeruginosa isolates.
Control group 2, patients with non-P. aeruginosa infections.
M, males; F, females.
TABLE 2.
Multivariate analysis of risk factors for acquisition of P. aeruginosa with the Fepns Cazs phenotype in 23 infected patients
| Risk factor | Control group 1
|
Control group 2
|
||
|---|---|---|---|---|
| OR (95% CI)a | P | OR (95% CI) | P | |
| Underlying disease (Charlson index > 2) | 0.2 (0.03-1.2) | 0.08 | 0.4 (0.07-2.2) | 0.2 |
| Receipt of piperacillin-tazobactam | 22.4 (3.5-141.1) | <0.01 | 7.3 (1.5-34.0) | 0.01 |
| Receipt of fluoroquinolones | 12.3 (1.3-113.4) | 0.02 | ||
| Receipt of aminoglycosides | 1.5 (0.1-12.4) | 0.6 | 6.8 (0.6-70.1) | 0.1 |
OR, odds ratio; CI, confidence interval.
Changes in the levels of consumption of antimicrobials in the ICUs were observed over time. A progressive reduction in the level of carbapenem use was found after the restriction of its use was introduced in 2006 (from 247 DDD/1,000 patient days in 2004 to 66 DDD/1,000 patient days in 2007; P = 0.008); an expected increase in the level of piperacillin-tazobactam use was observed (from 42 DDD/1,000 patient days in 2004 to 200 DDD/1,000 patient days in 2007; P < 0.001). Concomitantly, the levels of use of antipseudomonal cephalosporins (CAZ and FEP) decreased (P = 0.05), and fluoroquinolones were used regularly during the period analyzed (P = 0.6).
Antibiotic susceptibility testing.
The FEP MIC range for the non-FEP-susceptible isolates was from 16 to 32 μg/ml (for 42 [82%] strains the MICs were 16 μg/ml, and for 9 [18% strains the MICs were >16 μg/ml), and the CAZ MIC range was ≤1 to 8 μg/ml. The antibiotic resistance pattern for the epidemic strain was stable during the spread of the isolate, and all isolates were susceptible to amikacin and colistin. In addition, all isolates were resistant to aminoglycosides (gentamicin and tobramycin MICs, >16 μg/ml), fluoroquinolones (ciprofloxacin MICs, >32 μg/ml), piperacillin (MICs, >64 μg/ml), and piperacillin-tazobactam (MICs, >64 μg/ml). The aztreonam MICs were ≤8 mg/liter, although two of the isolates with these MICs showed intermediate resistance to aztreonam (MICs, 16 μg/ml). The only variation in the antibiotic resistance pattern observed concerned susceptibility to carbapenems; in 20 (39%) patients, the isolates in the first clinical sample from which P. aeruginosa isolates with the Fepns Cazs phenotype were isolated were imipenem resistant (MIC, ≥8 μg/ml), although four of the isolates from those samples were susceptible to meropenem (MICs, <4 μg/ml).
The DDST method indicated synergy between the FEP disk and the amoxicillin-clavulanic acid disk, but the Etest method did not show differences between the MICs of FEP alone and those of FEP plus clavulanic acid.
Characterization of resistance mechanisms.
The resistance mechanisms involved were studied in four random isolates (from different patients) of the epidemic clone. Cloxacillin inhibition tests revealed the presence of non-class C ß-lactamases, and the results of isoelectric focusing showed the presence of a unique band with a pI of approximately 5.7. All the PCRs performed for the detection of acquired ß-lactamases were negative, with the exception of that for blaPSE-1 and related genes. PCRs performed to characterize the integron potentially involved yielded positive results, and DNA sequencing revealed the presence of a class 1 integron, which contained (in addition to the integrase-encoding gene intl1) three gene cassettes: aacA4 (which encodes the aminoglycoside-modifying enzyme AAC6′-lb, which modifies susceptibility to tobramycin, netilmicin, and gentamicin but not amikacin), blaPSE-1 (which encodes the class A carbenicillinase PSE-1 or CARB-2), and aadA2 [which encodes the aminoglycoside-modifying enzyme ANT(3′)-l, which hydrolyzes streptomycin and spectinomycin].
The results of the real time RT-PCR experiments revealed that none of the isolates overexpressed AmpC, MexAB-OprM, MexCD-OprJ, or MexEF-OprN; in contrast, all four isolates were found to hyperproduce MexXY-OprM, with the relative mRNA levels ranging from being 24- to 32-fold higher than those in wild-type strain PAO1 (Table 3).
TABLE 3.
Expression of ampC, mexB, mexD, mexF, and mexY in selected isolates
| Strain | MIC (mg/liter)
|
Relative mRNA level with respect to that in reference strain PAO1a
|
|||||
|---|---|---|---|---|---|---|---|
| CAZ | FEP | ampC | mexB | mexD | mexF | mexY | |
| PAO1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 109351-07 | 4 | 16 | 0.9 ± 0.5 | 1.4 ± 0.6 | 4.1 ± 2.9 | 1.0 ± 0.4 | 27.2 ± 10.5 |
| 87999-07 | 2 | 16 | 3.1 ± 1.7 | 2.5 ± 0.1 | 5.0 ± 2.3 | 1.7 ± 0.1 | 25.4 ± 4.3 |
| 95818-07 | 8 | 16 | 1.7 ± 0.9 | 1.9 ± 0.6 | 4.0 ± 2.3 | 2.3 ± 0.7 | 23.8 ± 5.3 |
| 71702-07 | ≤1 | 16 | 1.2 ± 0.6 | 0.4 ± 0.1 | 2.5 ± 1.5 | 1.5 ± 0.8 | 32.3 ± 9.1 |
Strains were considered AmpC, MexCD-OprJ, MexEF-OprN, or MexXY-OprM hyperproducers when the relative level of expression of the corresponding gene was at least 10-fold higher than that in reference strain PAO1. The breakpoint used for MexAB-OprM hyperproduction was threefold higher expression than that for PAO1.
DISCUSSION
During routine infection control surveillance for carbapenem-resistant P. aeruginosa isolates in the Hospital de Bellvitge, a cluster of P. aeruginosa isolates with the Fepns Cazs phenotype was suspected. Our data from the study described here document the clonal nature of this phenotype on the basis of the results of PFGE analysis. ICU patients were mainly involved, although a few cases of acquisition were detected in a non-ICU ward, namely, the vascular surgery ward, where the epidemic caused by this P. aeruginosa clone spread and involved eight patients over a 4-month period.
The clone was initially recognized because of its particular profile of susceptibility to antipseudomonal cephalosporins. Regardless of the emergence of sporadic resistance to carbapenems, significant differences in the antibiotic susceptibility profiles were not observed. All isolates were susceptible to CAZ, amikacin, and colistin; in addition, a high percentage were susceptible to aztreonam. Eighty-two percent of the non-FEP-susceptible strains presented with intermediate resistance, as determined with an automated system. Concerns about the accuracies of automated systems for the detection of susceptibility to ß-lactam antipseudomonal agents and, consequently, false resistance to FEP have been reported (19). However, the FEP susceptibility data were confirmed by the CLSI reference method (10).
While a few previous studies have reported on the clonal spread of P. aeruginosa isolates overexpressing an efflux pump (3, 12), to the best of our knowledge, this is the first description of an outbreak caused by a P. aeruginosa strain that simultaneously hyperproduced MexXY-OprM and produced an integron-borne PSE-1 β-lactamase. These two resistance mechanisms have previously been reported among P. aeruginosa isolates that are more resistant to FEP than to CAZ (16). Our results suggest that the combination of these two resistance mechanisms is responsible for the multidrug resistance phenotype of our epidemic strain.
While MexXY-OprM confers moderate resistance to fluoroquinolones (6), other mechanisms that may explain the high levels of resistance to these antibiotics in the epidemic strain were not explored in this study. The role of the MexXY-OprM efflux pump in aminoglycoside resistance is also well documented (1, 22). Amikacin is a recognized substrate for the multidrug transporter MexXY, but amikacin alone is insufficient to promote the development of clinical (according to current breakpoints) amikacin resistance, since all the isolates were susceptible to this antibiotic. In contrast, the high levels of gentamicin and tobramycin resistance detected could be a result of the hyperexpression of MexXY-OprM, together with the production of AAC6′-lb.
In addition, several P. aeruginosa isolates with the Fepns Cazs phenotype were carbapenem resistant; this resistance could be explained by its decreased level of OprD porin expression during the course of carbapenem therapy (26, 30). The acquisition of carbapenem resistance in strains of the epidemic clone and its subsequent horizontal transmission determined that approximately 40% of the P. aeruginosa isolates with the Fepns Cazs phenotype were carbapenem resistant. The spread of carbapenem-resistant strains was mainly observed in a vascular surgery ward and involved seven of a total of eight patients; these seven patients were colonized directly by the epidemic strain that was resistant to carbapenem and had not previously received carbapenem.
Despite the complexity of factors that influence the incidence of antibiotic resistance, antibiotic surveillance revealed a progressive increase in the rate of piperacillin-tazobactam use from 2003 and a marked increase from 2006, after the restriction of carbapenem administration. The results of the analysis of the increased restriction of carbapenem use have not shown that a high prevalence of carbapenem-resistant P. aeruginosa isolates can be controlled; however, following increased piperacillin-tazobactam use for therapy in ICU patients, piperacillin-tazobactam-resistant P. aeruginosa strains emerged in our ICU wards. Despite the many factors that affect the incidence of antibiotic resistance among microorganisms, it is plausible that the application of the restriction of carbapenem use and the subsequent consumption of piperacillin-tazobactam potentially gave rise to the development of new and unexpected resistance patterns under conditions of high levels of selective antibiotic pressure (5). In addition, the information provided by the case-control study with two sets of controls may be complementary. Comparison of the results for the group infected or colonized with P. aeruginosa isolates with the Fepns Cazs phenotype with those for the group infected with susceptible P. aeruginosa isolates overestimated the risk associated with prior piperacillin-tazobactam use, while the analysis performed with the non-P. aeruginosa-infected control group showed that prior fluoroquinolone use was not a risk factor. In fact, prior fluoroquinolone consumption presented an independent association with the acquisition of isolates of the P. aeruginosa Fepns Cazs phenotype, but only when both the group infected with P. aeruginosa Fepns Cazs and the group infected with susceptible P. aeruginosa were compared. Thus, since the fluoroquinolones are universal substrates for the efflux systems of P. aeruginosa, the selection of strains expressing multidrug resistance may be expected following fluoroquinolone exposure (28). Finally, despite the sample size limitations of this study, the prior use of piperacillin-tazobactam was identified as an independent risk factor associated with the acquisition of P. aeruginosa isolates of the Fepns Cazs phenotype.
In conclusion, we identified an epidemic P. aeruginosa clone of the Fepns Cazs phenotype associated with the hyperexpression of MexXY-OprM and the production of an integron-borne PSE-1 ß-lactamase. This clone affected 51 patients, particularly those in ICUs, over a 9-month period. The dominant use of a single class of antibiotics may have provided the selective pressure required for the emergence of this Fepns Cazs phenotype. Efforts to encourage the variation and individualization of antibiotic use should be addressed by antibiotic use management policies.
Acknowledgments
This work was supported by National Health Service grant FIS 08/0276 from the Fondo de Investigación Sanitarias and was supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, Spanish Network for Research (grant REIPI C03/14), the Spanish Network for the Research in Infectious Diseases (grant REIPI RD06/0008), and the Ciber de Enfermedades Respiratorias (grant CB06/06/0037).
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 432624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, D., L. Poirel, J. Chevalier, S. Leotard, J. M. Pages, and P. Nordmann. 2001. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 451615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertrand, X., P. Bailly, G. Blasco, P. Balvay, A. Boillot, and D. Talon. 2000. Large outbreak in a surgical intensive care unit of colonization or infection with Pseudomonas aeruginosa that overexpressed an active efflux pump. Clin. Infect. Dis. 31e9-e14. [DOI] [PubMed] [Google Scholar]
- 4.Bontiglio, G., and F. Marchett. 2000. In vitro activity of ceftazidime, cefepime, and imipenem on 1,005 Pseudomonas aeruginosa clinical isolates either susceptible or resistant to beta-lactams. Chemotherapy 46229-234. [DOI] [PubMed] [Google Scholar]
- 5.Burke, J. P. 1998. Antibiotic resistance—squeezing the balloon? JAMA 2801270-1271. [DOI] [PubMed] [Google Scholar]
- 6.Cavallo, J. D., R. Fabre, F. Leblanc, M. H. Nicolas-Chanoine, and A. Thabaut. 2000. Antibiotic susceptibility and mechanisms of ß-lactam resistance in 1,310 strains of Pseudomonas aeruginosa: a French multicentre study (1996). J. Antimicrob. Chemother. 46133-136. [DOI] [PubMed] [Google Scholar]
- 7.Cavallo, J. D., D. Hocquet, P. Plesiat, R. Fabre, and M. Rousel-Delvallez on behalf of GERPA. 2007. Susceptibility of Pseudomonas aeruginosa to antimicrobials: a 2004 French multicentre hospital study. J. Antimicrob. Chemother. 591021-1024. [DOI] [PubMed] [Google Scholar]
- 8.Charlson, M. E., P. Pompei, K. L. Ales, and C. R. MacKenzie. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40373-383. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H. Y., M. Yuan, and D. M. Livermore. 1995. Mechanisms of resistance to ß-lactam antibiotics among Pseudomonas aeruginosa isolates collected in the UK in 1993. J. Med. Microbiol. 43300-309. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing, 18th informational supplement, document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Coque, T. M., A. Oliver, J. C. Pérez-Diaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum beta-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deplano, A., O. Denis, L. Poirel, D. Hocquet, C. Nonhoff, B. Byl, P. Nordmann, J. L. Vincent, and M. J. Struelens. 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 431198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1998. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16128-140. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez, O., C. Juan, E. Cercenado, F. Navarro, E. Bouza, P. Coll, J. L. Pérez, and A. Oliver. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 514329-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, A. D., T. B. Karchmer, Y. Carmeli, and S. H. Samore. 2001. Methodological principles of case-control studies that analyzed risk factors for antibiotic resistance: a systematic review. Clin. Infect. Dis. 321055-1061. [DOI] [PubMed] [Google Scholar]
- 16.Hocquet, D., P. Nordmann, F. El Garch, L. Cabanne, and P. Plesiat. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 501347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juan, C., M. D. Maciá, O. Gutierrez, C. Vidal, J. L. Pérez, and A. Oliver. 2005. Molecular mechanisms of ß-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 494733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juan, C., B. Moyá, J. L. Pérez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high level ß-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 501780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juretschko, S., V. J. LaBombardi, S. A. Lerner, P. C. Schreckenberger, and the Pseudomonas AST Study Group. 2007. Accuracies of ß-lactam susceptibility test results for Pseudomonas aeruginosa with four automated systems (BD Phoenix, MicroScan WalkAway, Vitek, and Vitek 2). J. Clin. Microbiol. 451339-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlowsky, J. A., M. E. Jones, C. Thornsberry, A. T. Evangelista, Y. C. Yee, and D. F. Sahm. 2005. Stable antimicrobial susceptibility rates for clinical isolates of Pseudomonas aeruginosa from the 2001-2003 tracking resistance in the United States today surveillance studies. Clin. Infect. Dis. 40(Suppl. 2)S89-S98. [DOI] [PubMed] [Google Scholar]
- 21.Maciá, M. D., N. Borrell, M. Segura, C. Gómez, J. L. Pérez, and A. Oliver. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsijimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 443322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulet, X., M. D. Maciá, A. Mena, C. Juan, J. L. Pérez, and A. Oliver. 2009. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob. Agents Chemother. 531552-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordic Council on Medicines. 1994. Nordic statistics on medicines. Nordic drug index with classification and defined daily doses. Nordic Council on Medicines, Uppsala, Sweden.
- 25.Oh, H., J. Stenhoff, S. Jalal, and B. Wretlind. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 8323-328. [DOI] [PubMed] [Google Scholar]
- 26.Pai, H., J. W. Kim, J. Kim, J. H. Lee, K.W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña, C., A. Guzmán, C. Suarez, M. A. Dominguez, F. Tubau, M. Pujol, F. Gudiol, and J. Ariza. 2007. Effects of carbapenem exposure on the risk for digestive tract carriage of intensive care unit-endemic carbapenem-resistant Pseudomonas aeruginosa strains in critically ill patients. Antimicrob. Agents Chemother. 511967-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 442233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 3452-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Belkum, A., P. T. Tassios, L. Dijkshoom, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens for the European Society of Clinical Microbiology and Infectious Diseases Study Group on Epidemiological Markers. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3)1-46. [DOI] [PubMed] [Google Scholar]
- 32.Weldhagen, G. F., L. Poirel, and P. Nordmann. 2003. Ambler class A extended-spectrum ß-lactamase in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob. Agents Chemother. 472385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]