Abstract
Bacteria belonging to the Enterobacter genus are frequently isolated from clinical samples but are unusual causative agents of orthopedic implant infections. Twelve genetic clusters (clusters I to XII) and one sequence crowd (sequence crowd xiii) can be distinguished within the Enterobacter cloacae nomenspecies on the basis of hsp60 sequence analysis, and until now, none of these clusters could be specifically associated with a disease. In order to investigate if specific genetic clusters would be involved in infections of orthopedic material, two series of bacterial clinical isolates identified as E. cloacae by routine phenotypic identification methods were collected either from infected orthopedic implants (n = 21) or from randomly selected samples of diverse anatomical origins (control; n = 52). Analysis of the hsp60 gene showed that genetic clusters III, VI, and VIII were the most frequent genetic clusters detected in the control group, whereas cluster III was poorly represented among the orthopedic implant isolates (P = 0.006). On the other hand, E. hormaechei (clusters VI and VIII), but not cluster III, is predominantly associated with infections of orthopedic implants and, more specifically, with infected material in the hip (P = 0.019). These results support the hypothesis that, among the isolates within the E. cloacae complex, E. hormaechei and hsp60 gene sequencing-based cluster III are involved in pathogenesis in different ways and highlight the need for more accurate routine Enterobacter identification methods.
Prosthetic joint infection (PJI) is, after aseptic loosening, the second most frequent complication of prosthetic joint replacement. Improvements in surgical techniques and the prevention of infection have lowered the risk of infection for primary hip or knee replacement to less than 1 and 2%, respectively, but the incidence of infection can increase 10-fold in the case of surgical revision (30). The population at risk for PJI continues to steadily increase, with an estimated 1 million arthroplasties being carried out worldwide each year (2), and the socioeconomic burden could become considerable (14-16). The average cost of combined medical and surgical treatment of an infected joint prosthesis is estimated to be $30,000, with important discomfort and substantial economic consequences for the patient (4). Besides joint prosthetic implants, other implantable devices such as screws and internal fixation devices are also at risk for infection in a wide range of clinical situations. Gram-positive cocci are the most frequent pathogens encountered, but members of the Enterobacteriaceae family can also be involved (29). Among these, Enterobacter species are major nosocomial pathogens often found in intensive care settings, and their role in PJI remains to be documented.
Bacteria of the Enterobacter genus are widely encountered in nature. These microorganisms are saprophytic in the environment and commensal in the enteric flora since they are found in soil and sewage, as well as in the human gastrointestinal tract (17, 22). The taxonomy of the Enterobacter genus has been iteratively updated (8-12, 19). Several species are described in this taxon (Enterobacter aerogenes, E. amnigenus, E. cancerogenus, E. cowanii, E. gergoviae, E. intermedius, E. pyrinus), as is a genetic complex, referred to as the “E. cloacae complex,” in which other species have been identified (E. asburiae, E. kobei, E. ludwigii, E. hormaechei, E. nimipressuralis, and E. cloacae). E. sakazakii strains, which belong to several genomospecies, were recently reassigned to the newly proposed genus Cronobacter (12).
Enterobacter species can act as pathogens, and the E. cloacae complex, commonly referred to as “E. cloacae,” represents the Enterobacter group most frequently encountered in human clinical samples. Although identification as “E. cloacae” is routinely performed by phenotypic methods in clinical laboratories, the accurate identification of isolates within this taxon is difficult. Analysis of the 16S rRNA gene is widely used for bacterial identification, but it is poorly discriminatory for closely related members of the Enterobacteriaceae family and, more specifically, for members of the Enterobacter genus (27). Other targets for use for the molecular identification of isolates within the Enterobacter genus have been described, such as the oriC locus (23), gyrB (6), rpoB (8, 21), and hsp60 (8). Sequence analysis of a fragment of the hsp60 gene showed that the E. cloacae nomenspecies could be divided into 12 genetic clusters (clusters I to XII) and one sequence crowd (sequence crowd xiii). Specific names could be attributed to some of the genetic clusters: E. asburiae (cluster I) (9), E. kobei (cluster II) (9), E. ludwigii (cluster V) (11), E. nimipressuralis (cluster X) (8), E. cloacae subsp. cloacae (cluster XI) (9), and E. cloacae subsp. dissolvens (cluster XII) (9). Although the name E. hormaechei was sometimes used as a generic name for strains belonging to different hsp60 gene sequencing-based clusters (20), clusters VI, VII, and VIII together formally constitute the E. hormaechei species, which has three subspecies: E. hormaechei subsp. oharae (cluster VI), E. hormaechei subsp. hormaechei (cluster VII), and E. hormaechei subsp. steigerwallti (cluster VIII) (10). Species names were not attributed to clusters III, IV, and IX and to sequence crowd xiii, although at least cluster III is of significant clinical importance (8, 26).
The degree of genomic diversity within the E. cloacae complex was recently reassessed by more global genotypic methods. Multilocus sequence analysis (MLSA) identifies seven clusters within the E. cloacae complex, and each of these corresponds to one or more hsp60 gene sequencing-based genetic clusters. Microarray-based comparative genomic hybridization analysis (CGH) showed two genetically distinct clades (21). Most strains associated with clinical disease belong to the youngest CGH-based clade, to which strains of hsp60 gene sequencing-based clusters III, VI, and VIII also belong. The second, and older, CGH-based clade comprises heterogeneous strains, some of which are associated with commensalism.
Single-locus-based molecular methods, as well as global approaches, such as MLSA or CGH, suggested that some genetic clusters are more prone to cause infection, although no specific epidemiological association could be demonstrated (8, 21). The repeated occurrence in our clinical laboratory of orthopedic implant infections due to E. cloacae isolates prompted us to further investigate these strains. We analyzed strains isolated from infected orthopedic devices and compared them to randomly selected strains isolated from clinical specimens taken from diverse anatomical sites. Analysis of the hsp60 gene showed that among the genetic clusters of the Enterobacter cloacae complex, clusters VI and VIII, but not cluster III, are predominantly associated with infections of orthopedic implants and, more specifically, with hip implants. These data support the hypothesis that genetic clusters of the E. cloacae complex are involved in pathogenesis in different ways and highlight the need for more accurate routine methods for the identification of Enterobacter species.
MATERIALS AND METHODS
Bacterial strains and epidemiological data.
For the study group, strains were collected from three large academic hospitals in the Paris, France, area (Ambroise Paré Hospital, Raymond Poincaré Hospital, and Cochin Hospital, AP-HP) involved in the management of osteoarticular infectious conditions. In each hospital, all bacterial strains isolated from orthopedic device-related surgical samples are systematically and prospectively collected and stored, irrespective of the bacterial species, the anatomical site of isolation, or the type of infection. For the study group, the collections were submitted to identical screening criteria (1999 to 2006 for Ambroise Paré Hospital, 2000 to 2006 for Raymond Poincaré Hospital, 2005 to 2007 for Cochin Hospital) for the selection of strains belonging to the E. cloacae complex that were responsible for the infection of the orthopedic implants. The following criteria were applied for inclusion of the strains in the study: (i) identification of the infecting organism as E. cloacae with routine phenotypic identification systems (the API 20E or the Vitek 2 system; BioMerieux, Marcy l'Etoile, France), (ii) clinical evidence of infection of the orthopedic implant in an adult patient, (iii) isolation of the E. cloacae strain from surgical samples collected at the point of contact between surrounding tissue and the implanted material (or from the implant itself) at the time of surgical treatment, and (iv) involvement of the E. cloacae strain as a probable causative agent for the infection and clinical management as such. Isolation of an E. cloacae strain from a wound, sinus tract, or drainage was not sufficient to ascertain its involvement in the implant infection; and such strains were not included in the study. One strain per patient was included, and in the case of the isolation of multiple isolates, the most clinically relevant isolate (i.e., from the implant itself rather than from the surrounding tissue) was used. Twenty-one strains were selected and are referred to as strains isolated from orthopedic implanted material (Table 1). Since different types of implants were involved, only the anatomical site of infection was considered for data analysis. All selected strains recovered from storage at −80°C were viable for subculture. Personal data (age, sex, site of infection, mono- or polymicrobial infection) as well as the hospital and the date of isolation were anonymously collected for the purposes of this study.
TABLE 1.
Characteristics of strains isolated from orthopedic implanted materiala
| Strain | Sex | Age (yr) | Hospital | Yr of isolation | hsp60-based genetic cluster | Site of infection | Form of isolation |
|---|---|---|---|---|---|---|---|
| P1 | F | 70 | CCH | 2006 | I | Elbow | With associated flora |
| P2 | F | 37 | CCH | 2006 | III | Tibia | With associated flora |
| P3 | F | 23 | APR | 2006 | III | Tibia | Pure |
| P4 | F | 79 | APR | 1999 | V | Ankle | Pure |
| P5 | F | 61 | APR | 2006 | V | Knee | Pure |
| P6 | M | 35 | CCH | 2007 | XI | Ankle | With associated flora |
| P7 | F | 57 | RPC | 2005 | VI | Knee | With associated flora |
| P8 | F | 60 | APR | 2005 | VI | Knee | Pure |
| P9 | M | 33 | RPC | 2006 | VI | Hip | Pure |
| P10 | M | 79 | RPC | 2004 | VI | Hip | With associated flora |
| P11 | F | 53 | APR | 1999 | VI | Hip | Pure |
| P12 | M | 36 | CCH | 2006 | VIII | Femur | Pure |
| P13 | F | 72 | RPC | 2006 | VIII | Knee | With associated flora |
| P14 | F | 26 | CCH | 2007 | VIII | Hip | With associated flora |
| P15 | M | 42 | CCH | 2006 | VIII | Hip | Pure |
| P16 | M | 63 | RPC | 2002 | VIII | Hip | Pure |
| P17 | F | 66 | RPC | 2000 | VIII | Hip | Pure |
| P18 | M | 56 | APR | 2001 | VIII | Hip | Pure |
| P19 | M | 77 | APR | 2005 | VIII | Hip | With associated flora |
| P20 | M | 25 | CCH | 2005 | VIII | Tibia | Pure |
| P21 | M | 23 | APR | 2005 | VIII | Tibia | With associated flora |
The ratio of males (M) to females (F) was 0.9. The mean age ± standard deviation was 51 ± 20 years. CCH, Cochin Hospital; APR, Ambroise Paré Hospital; RPC, Raymond Poincaré Hospital.
For the control group (Table 2), 52 randomly selected clinical strains routinely identified as E. cloacae in the clinical laboratory at the academic Cochin Hospital by use of the API 20E or the Vitek 2 identification system were prospectively collected from clinical samples taken for diagnostic purposes from adult patients during the year 2006; environmental isolates were excluded. Thus, the control group was representative of E. cloacae strains from an adult population routinely identified in a clinical laboratory. One isolate was included per patient, and the strains collected were registered on the basis of the anatomical site of isolation, as follows: skin and soft tissue (n = 14), upper and lower respiratory tract (n = 15), urine (n = 12), joint or bone (in the absence of infected material; n = 3), intravascular catheter (n = 2), blood (n = 2), and gastrointestinal suppuration with exclusion of feces (n = 4). Personal data (age, sex, site of isolation) were anonymously collected for the purpose of this study.
TABLE 2.
Characteristics of control strainsa
| Characteristic | Value |
|---|---|
| No. of patients | 52 |
| M/Fb ratio | 1.4 |
| Mean age at time of isolation ± SD (yr) | 65 ± 18 |
| Anatomical site of isolation (no. of isolates) | |
| Skin and soft tissue | 14 |
| Upper and lower respiratory tract | 15 |
| Urine | 12 |
| Joint or bone, in the absence of infected material | 3 |
| Intravenous catheter | 2 |
| Blood | 2 |
| Gastrointestinal tract | 4 |
One isolate from each patient was studied.
M, male; F, female.
Identification methods.
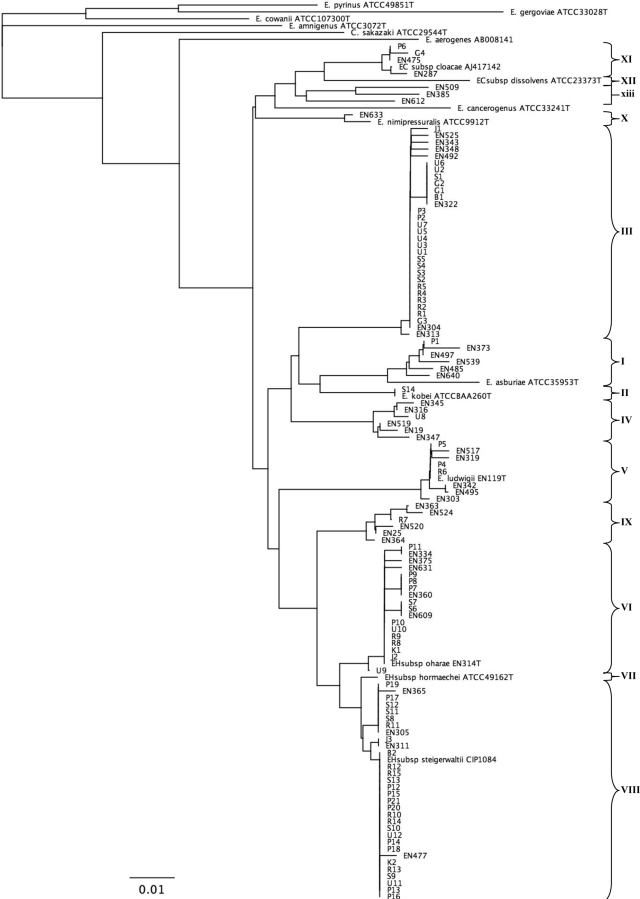
Bacterial DNA from bacterial colonies grown overnight at 37°C on 5% horse blood agar plates was prepared for PCR analysis by using the Instagene nucleic acid purification method (Bio-Rad, Marnes la Coquette, France). Partial sequencing of the hsp60 gene was performed by a previously described protocol (8). Briefly, oligonucleotide primers Hsp60-F (5′-GGTAGAAGAAGGCGTGGTTGC-3′) and Hsp60-R (5′-ATGCATTCGGTGGTGATCATCAG-3′) were used for genomic amplification of a 341-bp fragment of the hsp60 gene. A negative control containing all reagents except the target DNA (which was replaced by H2O) was included in each series. PCR was performed on a GeneAmp PCR system 9700 apparatus (Applied Biosystems) for 30 cycles by using the following conditions: 30 s at 94°C for denaturation, 30 s at 57°C for annealing, and 60 s at 72°C for elongation. Both strands of the purified amplified DNA fragment were sequenced by the BigDye Terminator cycle sequencing protocol with the same primers used for the PCR. Chromatograms of the complementary strands obtained with an ABI 313 apparatus (Applied Biosystems) were assembled by using the VectorNTi suite of programs (Invitrogen Corp.). A 272-bp fragment of the hsp60 gene was obtained for the 73 strains, and the sequence of the fragment was compared to reference sequences from strains previously described in taxonomic studies (8) by using the Clustal W algorithm (www.align.genome.jp). Sequence comparisons were exported as an unrooted neighbor-joining tree with proportional branch lengths.
Statistical analysis.
Epidemiological associations were analyzed by use of the Fisher exact test and the corresponding two-tailed P value.
Nucleotide sequence accession numbers.
The sequences of the following type strains were retrieved from the GenBank database (the information in parentheses is the strain designation, GenBank accession number): E. asburiae (ATCC 35953, AJ417141), E. kobei (ATCC BAA260, AJ567899), E. cloacae subsp. dissolvens (ATCC 23373, AJ417143) (9), E. ludwigii (EN-119, AJ417114) (11), E. hormaechei subsp. oharae (EN-314, AJ543782), E. hormaechei subsp. hormaechei (ATCC 49162, AJ417108), E. hormaechei subsp. steigerwaltii (CIP108489, AJ543908) (10), E. nimipressuralis (ATCC 9912, AJ567900), E. cancerogenus (ATCC 33241, AJ567895), E. amnigenus (ATCC 3072, AJ567894), E. cowanii (ATCC 107300T, AJ567896), E. gergoviae (ATCC 33028, AJ567897), E. pyrinus (ATCC 49851, AJ567901), C. sakazaki (ATCC 29544, AJ567902) (12), and E. aerogenes (AB008141). For E. cloacae subsp. cloacae, no hsp60 sequence is available for strain ATCC 13047, but one can be found under GenBank accession number AJ417142 (strain ATCC 13049), which was used in another study (13). The GenBank accession numbers for previously described strains (8) are listed in the Fig. 1 legend. The GenBank accession numbers for the clinical strains described in this work are FJ595719 to FJ595791.
FIG. 1.
Neighbor-joining unrooted tree resulting from analysis of the hsp60 gene sequences of 73 clinical strains and previously reported sequences. Isolation site of clinical strains: P, infected orthopedic implant; B, blood; J, joint or bone, in the absence of implanted material; G, gastrointestinal tract; R, respiratory tract; S, skin and soft tissue; U, urine; K, intravascular catheter. For the previously described strains, type strains are indicated (8, 10-12). Strains labeled EN were reported previously (8) and correspond to sequences with GenBank accession numbers AJ417125, AJ417127, AJ543819, AJ567887, AJ543876, AJ543894, AJ567893, AJ543787, AJ543803, AJ543806, AJ543829, AJ543864, AJ543882, AJ543804, AJ543789, AJ543781, AJ543784, AJ567846, AJ543776, AJ543808, AJ543877, AJ543807, AJ543866, AJ543775, AJ543878, AJ543881, AJ543820, AJ567878, AJ567885, AJ543857, AJ543831, AJ543821, AJ543816, AJ543798, AJ567847, AJ543777, AJ543861, AJ543768, AJ543855, AJ543870, AJ567881, and AJ543837. EH, E. hormaechei; EC, E. cloacae. The bar indicates the number of substitutions per site. Genetic clusters are numbered according to the description provided previously (8).
RESULTS
Distribution of strains isolated from infected orthopedic implants within the genetic clusters of the Enterobacter cloacae complex.
Enterobacter is an uncommon causative agent of orthopedic implant infections. Investigation of the databases of three large academic orthopedic surgical centers in the Paris, France, area yielded 21 cases of orthopedic device infections due to E. cloacae on the basis of routine phenotypic identification methods (Table 1). The infections were found to be polymicrobial in 9 cases (43%), whereas isolates phenotypically identified as E. cloacae were found as the sole causative agent in 12 patients (57%). Partial sequencing of the hsp60 gene allowed the identification of all isolates as part of one of the clusters that form the E. cloacae complex (Fig. 1). Some genetic clusters appeared to account for most of the cases: 15 strains (71%) belonged to the E. hormaechei species (clusters VI and VIII), 2 (9%) belonged to cluster III, and 2 (9%) belonged to cluster V (E. ludwigii). E. cloacae subsp. cloacae and E. asburiae (clusters XI and I, respectively) were found only once; and clusters II (E. kobei), IV, VII (E. hormaechei subsp. hormaechei), IX, X (E. nimipressuralis), and XII (E. cloacae subsp. dissolvens) were absent from among the isolates in the study group.
Predominance of E. hormaechei in hip prosthetic infections.
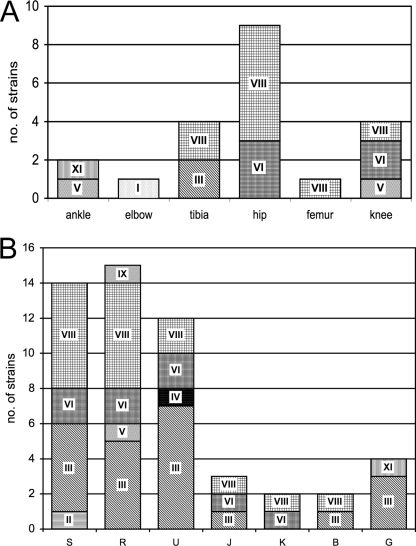
Various anatomical sites were involved in orthopedic implant infections. Surprisingly, E. hormaechei (clusters VI and VIII) was the species involved in all cases (n = 9) of prosthetic hip infections, whereas strains isolated from other anatomic locations also belonged to other clusters (Fig. 2). The epidemiological association of E. hormaechei with hip implants was statistically significant compared to its association with other anatomical implant sites (9/9 cases at hip implant sites versus 6/12 cases at other anatomical implant sites; P = 0.019), but E. hormaechei infection of hip implants could not be linked to other demographic or clinical factors, such as age, a history of wound infection, or an association with other causative agents of infection. E. hormaechei subsp. steigerwaltii (cluster VII) was the taxon that was more frequently involved (six of nine cases) than E. hormaechei subsp. oharae (cluster VI; three of nine cases). The subspecies E. hormaechei subsp. hormaechei was not found in this series.
FIG. 2.
Distribution of clinical strains within the genetic clusters of the E. cloacae complex. All strains could be assigned to one of the previously reported hsp60 gene sequencing-based genetic clusters of the E. cloacae complex. (A) Strains isolated from implanted orthopedic material at different anatomical sites (n = 21). Hip-associated strains exclusively belonged to the E. hormaechei species (clusters VI and VIII). (B) Randomly selected clinical strains of diverse anatomical origins (n = 52). S, skin and soft tissue; R, upper and lower respiratory tract; U, urine; J, joint or bone, in the absence of infected material; K, intravenous catheter; B, blood; G, gastrointestinal tract. Irrespective of the site if isolation, cluster III accounted for 42% of the control isolates.
Molecular identification of routinely identified clinical E. cloacae isolates.
In order to investigate the species distribution among the strains isolated from routine clinical samples, we performed sequence analysis of the hsp60 gene from the control group, which consisted of 52 randomly selected clinical strains (Table 2). Similar to strains isolated from infected osteoarticular implants, hsp60 sequencing showed that all strains phenotypically identified as E. cloacae belonged to one of the molecular clusters of the E. cloacae complex (Fig. 1).
Without distinction by the site of isolation (Fig. 2), three clusters (clusters III, VI, and VIII) accounted for 90% of all strains. Similar to the implant-associated strains, genetic clusters belonging to the E. hormaechei species (clusters VI and VIII) were predominant (48% of control strains). Interestingly, cluster VII (E. hormaechei subsp. hormaechei) was absent from our study, and E. hormaechei subsp. steigerwaltii (cluster VIII, n = 18) was predominant over E. hormaechei subsp. oharae (cluster VI, n = 8).
Cluster III accounted for 42% of control clinical isolates; thus, it occurred statistically more frequently within the control group of isolates than within the isolates from infected orthopedic implants (22/52 isolates versus 2/21 isolates; P = 0.006). These strains were recovered from various anatomical sites (respiratory, urinary and gastrointestinal tract, skin or soft tissue, and blood). The three bone and joint strains obtained in the absence of material (Fig. 2, series J [joint or bone]) were isolated from knee synovial fluid, knee soft tissue, and hallux valgus and belonged to clusters III, VI, and VIII, respectively; they thus represented each of the most frequently isolated clusters in the control group. The other clusters, cluster II (E. kobei, n = 1), cluster IV (n = 1), cluster V (E. ludwigii, n = 1), cluster IX (n = 1), and cluster XI (E. cloacae subsp. cloacae, n = 1), were poorly represented. Clusters I, VII, X, and XII and sequence crowd xiii were not found among the isolates in the control group. Taken together, these results show that cluster III, together with clusters VI and VIII, accounts for most of the strains routinely isolated from clinical specimens.
DISCUSSION
In this work, we investigated the distribution of strains involved in infections of osteoarticular implanted material within the genetic clusters of the E. cloacae complex. On the basis of hsp60 analysis, we show that the cluster distribution for infected orthopedic implant-associated strains is different from that for randomly selected clinical strains of various anatomical origins. Our observation that all genetic clusters are not equally involved in pathogenesis highlights the need for more accurate routine bacterial identification tools and for a better understanding of the pathogenesis of the E. cloacae complex.
All strains evaluated in this study (n = 73) could be assigned to 1 of the 12 genetic clusters (clusters I to XII) of the E. cloacae complex. Only one strain was found to belong to cluster XI, the type strain of which is E. cloacae subsp. cloacae, which suggests that the widely used name E. cloacae is not representative of most clinical strains of the taxon. Preliminary identification as E. cloacae by conventional phenotypic identification methods (performed with the API 20E or the Vitek 2 system) was a prerequisite for the inclusion of strains in either the control or the study group. Although they might have led to possible underrepresentation of misidentified strains that authentically belong to the E. cloacae complex, the phenotypic identification methods performed with the API 20E and the Vitek 2 systems can be considered reliable tools for identification, as long as identification as “E. cloacae” is understood as “belonging to the E. cloacae complex.”
Although patient populations vary from one hospital to another, the results of our studies of the distribution of the control strains within genetic clusters are concordant with those published previously, since we observed that clusters III, VIII, and VI account for most of the clinical isolates (8). Strains belonging to hsp60 sequence analysis-based cluster III were shown to gather into the previously described MLSA-based cluster 1 as well as in CGH-based clade 2, with the latter being associated with strains that are the most frequently cultured in hospitals (21). Similarly, hsp60 sequence analysis-based cluster VI (E. hormaechei subsp. oharae) and cluster VIII (E. hormaechei subsp. steigerwaltii) were shown to gather in MLSA-based cluster 2 but also to belong to the clinically relevant CGH-based clade 2. Thus, our data showing a predominance of cluster III, VI, and VIII isolates among control clinical strains of different anatomical origins are consistent with data presented in previous reports on the genetic diversity of the strains within the E. cloacae complex and support the congruence of CGH-based clade 2 with clinically relevant samples.
Some genetic clusters were absent from our study. Cluster VII (E. hormaechei subsp. hormaechei) harbors the original species type strain and was also poorly represented in other studies (8). Cluster X (E. nimipressuralis) is found in potable water reservoirs but, to our knowledge, has never been associated with human disease (13). Cluster XII (E. cloacae subsp. dissolvens), formerly part of the genus Erwinia, was reassigned to the Enterobacter genus and forms a subspecies of the E. cloacae species. It is associated with plants (maize, coffee), but no human infections have been reported (7, 9).
Although it was the largest cluster within the group of control strains (42%), cluster III was poorly represented within the group of orthopedic implant-associated strains (9%), and this difference was statistically significant (P = 0.006). Low numbers of cluster III isolates emphasize the large proportion of cluster VI (E. hormaechei subsp. oharae) and cluster VIII (E. hormaechei subsp. steigerwaltii) isolates, both of which belong to the E. hormaechei species. Hip joint prosthesis infections appeared to be specifically associated with E. hormaechei, since, in our series, all cases of infections of implants at this site were due to isolates of either cluster VIII (n = 6) or cluster VI (n = 3). The third subspecies of the taxon, E. hormaechei subsp. hormaechei (cluster VII), was not found in our analysis.
The low prevalence of cluster III isolates within the group of isolates from infected orthopedic implants compared to their prevalence within the group of isolates from clinical samples of other origins suggests a specific pathogenicity and reinforces the need for a robust and discriminatory tool for the accurate identification of isolates within the E. cloacae complex. In this regard, hsp60 gene sequencing-based identification appeared to be both discriminatory and easily implementable, whereas other sequence-based molecular methods for the identification of Enterobacter were not as accurate. The absence of a consensus for the analysis of an rpoB DNA fragment (1 kb or 500 bp) has led to contradictory results, particularly for the genetic discrimination of isolates within hsp60 gene sequencing-based clusters III, VI, and VIII (8, 21), as well as to the confusing use of the species name E. hormaechei for strains that do not belong to hsp60 gene sequencing-based clusters VI to VIII (20). Similarly, sequence analysis of the gene encoding the DNA gyrase subunit B (gyrB) led to the hypothesis that most clinical isolates assigned to the E. cloacae complex by phenotypic semiautomated methods would belong to the E. hormaechei species (6). Analysis of the hsp60 gene sequence was not included in this study, but the absence of cluster III as a specific group within the group of clinical strains evaluated in this study might suggest that gyrB sequencing is not as discriminatory as hsp60 gene sequence analysis. Further investigation is needed to elucidate the latter point.
The epidemiological association between E. hormaechei and hip prosthetic implants allows new insights into previous observations to be made. First, the E. hormaechei species was reported in a case of prosthetic hip infection, although it was initially misidentified as Escherichia coli (25). This strain exhibited a phenotype of small-colony-variant formation, which was shown to be associated with regulation of the hemin uptake system (24). Although we did not systematically search for it, at least one of the strains involved in prosthetic infection displayed such a phenotype, with each step of subculture on solid medium leading to the emergence of fast- and slow-growing bacterial colonies. Second, although the study did not specifically refer to E. hormaechei, the ability of Enterobacter species to participate in biofilm formation on orthopedic implants was reported (1). Third, the ability of E. hormaechei to colonize implanted catheters as a biofilm and to be responsible for systemic infection was described (3, 5). The ability to grow as small-colony variants and the formation of biofilms are features frequently associated with the causative agents of orthopedic implant infections and contribute to the increased difficulty of diagnosis and treatment of such infections (18, 28). Taken together, the findings from the previous reports reinforce our observation showing the predominance of E. hormaechei as the cause of orthopedic implant infections. Further work is needed in order to identify the bacterial and host factors specifically involved in the bacterial colonization of the implanted material and in the pathophysiology of these infections.
Acknowledgments
We thank David Biau for precious advice on the statistical analysis.
This work was supported by Université Paris Descartes, Institut Cochin, and Assistance Publique—Hôpitaux de Paris.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Bartoszewicz, M., A. Rygiel, M. Krzeminski, and A. Przondo-Mordarska. 2007. Penetration of a selected antibiotic and antiseptic into a biofilm formed on orthopedic steel implants. Ortop. Traumatol. Rehabil. 9310-318. [PubMed] [Google Scholar]
- 2.Bernard, L., P. Hoffmeyer, M. Assal, P. Vaudaux, J. Schrenzel, and D. Lew. 2004. Trends in the treatment of orthopaedic prosthetic infections. J. Antimicrob. Chemother. 53127-129. [DOI] [PubMed] [Google Scholar]
- 3.Campos, L. C., L. F. Lobianco, L. M. Seki, R. M. Santos, and M. D. Asensi. 2007. Outbreak of Enterobacter hormaechei septicaemia in newborns caused by contaminated parenteral nutrition in Brazil. J. Hosp. Infect. 6695-97. [DOI] [PubMed] [Google Scholar]
- 4.Darouiche, R. O. 2004. Treatment of infections associated with surgical implants. N. Engl. J. Med. 3501422-1429. [DOI] [PubMed] [Google Scholar]
- 5.da Silva, C. L., L. E. Miranda, B. M. Moreira, D. Rebello, L. A. Carson, M. E. Kellum, M. C. de Almeida, J. L. Sampaio, and C. M. O'Hara. 2002. Enterobacter hormaechei bloodstream infection at three neonatal intensive care units in Brazil. Pediatr. Infect. Dis. J. 21175-177. [DOI] [PubMed] [Google Scholar]
- 6.Delmas, J., F. Breysse, G. Devulder, J. P. Flandrois, and M. Chomarat. 2006. Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene. Diagn. Microbiol. Infect. Dis. 55263-268. [DOI] [PubMed] [Google Scholar]
- 7.Frank, H. A., N. A. Lum, and A. S. Delacruz. 1965. Bacteria responsible for mucilage-layer decomposition in Kona coffee cherries. Appl. Microbiol. 13201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann, H., and A. Roggenkamp. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl. Environ. Microbiol. 695306-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann, H., S. Stindl, W. Ludwig, A. Stumpf, A. Mehlen, J. Heesemann, D. Monget, K. H. Schleifer, and A. Roggenkamp. 2005. Reassignment of Enterobacter dissolvens to Enterobacter cloacae as E. cloacae subspecies dissolvens comb. nov. and emended description of Enterobacter asburiae and Enterobacter kobei. Syst. Appl. Microbiol. 28196-205. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann, H., S. Stindl, W. Ludwig, A. Stumpf, A. Mehlen, D. Monget, D. Pierard, S. Ziesing, J. Heesemann, A. Roggenkamp, and K. H. Schleifer. 2005. Enterobacter hormaechei subsp. oharae subsp. nov., E. hormaechei subsp. hormaechei comb. nov., and E. hormaechei subsp. steigerwaltii subsp. nov., three new subspecies of clinical importance. J. Clin. Microbiol. 433297-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann, H., S. Stindl, A. Stumpf, A. Mehlen, D. Monget, J. Heesemann, K. H. Schleifer, and A. Roggenkamp. 2005. Description of Enterobacter ludwigii sp. nov., a novel Enterobacter species of clinical relevance. Syst. Appl. Microbiol. 28206-212. [DOI] [PubMed] [Google Scholar]
- 12.Iversen, C., N. Mullane, B. McCardell, B. D. Tall, A. Lehner, S. Fanning, R. Stephan, and H. Joosten. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 581442-1447. [DOI] [PubMed] [Google Scholar]
- 13.Kampfer, P., A. Nienhuser, G. Packroff, F. Wernicke, A. Mehling, K. Nixdorf, S. Fiedler, C. Kolauch, and M. Esser. 2008. Molecular identification of coliform bacteria isolated from drinking water reservoirs with traditional methods and the Colilert-18 system. Int. J. Hyg. Environ. Health 211374-384. [DOI] [PubMed] [Google Scholar]
- 14.Kurtz, S., K. Ong, E. Lau, F. Mowat, and M. Halpern. 2007. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J. Bone Joint Surg. Am. 89780-785. [DOI] [PubMed] [Google Scholar]
- 15.Kurtz, S. M., E. Lau, J. Schmier, K. L. Ong, K. Zhao, and J. Parvizi. 2008. Infection burden for hip and knee arthroplasty in the United States. J. Arthroplasty 23984-991. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz, S. M., K. L. Ong, J. Schmier, F. Mowat, K. Saleh, E. Dybvik, J. Karrholm, G. Garellick, L. I. Havelin, O. Furnes, H. Malchau, and E. Lau. 2007. Future clinical and economic impact of revision total hip and knee arthroplasty. J. Bone Joint Surg. Am. 89(Suppl. 3)144-151. [DOI] [PubMed] [Google Scholar]
- 17.Mazari-Hiriart, M., S. Ponce-de-Leon, Y. Lopez-Vidal, P. Islas-Macias, R. I. Amieva-Fernandez, and F. Quinones-Falconi. 2008. Microbiological implications of periurban agriculture and water reuse in Mexico City. PLoS One 3e2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neut, D., H. C. van der Mei, S. K. Bulstra, and H. J. Busscher. 2007. The role of small-colony variants in failure to diagnose and treat biofilm infections in orthopedics. Acta Orthop. 78299-308. [DOI] [PubMed] [Google Scholar]
- 19.O'Hara, C. M., A. G. Steigerwalt, B. C. Hill, J. J. Farmer III, G. R. Fanning, and D. J. Brenner. 1989. Enterobacter hormaechei, a new species of the family Enterobacteriaceae formerly known as enteric group 75. J. Clin. Microbiol. 272046-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paauw, A., M. P. Caspers, M. A. Leverstein-van Hall, F. H. Schuren, R. C. Montijn, J. Verhoef, and A. C. Fluit. 2009. Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 1551478-1488. [DOI] [PubMed] [Google Scholar]
- 21.Paauw, A., M. P. Caspers, F. H. Schuren, M. A. Leverstein-van Hall, A. Deletoile, R. C. Montijn, J. Verhoef, and A. C. Fluit. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS One 3e3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintanilha, A. G., B. Zilberstein, M. A. Santos, D. Pajecki, E. G. Moura, P. R. Alves, F. Maluf-Filho, and I. Cecconello. 2007. A novel sampling method for the investigation of gut mirobiota. World J. Gastroenterol. 133990-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roggenkamp, A. 2007. Phylogenetic analysis of enteric species of the family Enterobacteriaceae using the oriC-locus. Syst. Appl. Microbiol. 30180-188. [DOI] [PubMed] [Google Scholar]
- 24.Roggenkamp, A., H. Hoffmann, and M. W. Hornef. 2004. Growth control of small-colony variants by genetic regulation of the hemin uptake system. Infect. Immun. 722254-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roggenkamp, A., A. Sing, M. Hornef, U. Brunner, I. B. Autenrieth, and J. Heesemann. 1998. Chronic prosthetic hip infection caused by a small-colony variant of Escherichia coli. J. Clin. Microbiol. 362530-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stumpf, A. N., A. Roggenkamp, and H. Hoffmann. 2005. Specificity of enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction for the detection of clonality within the Enterobacter cloacae complex. Diagn. Microbiol. Infect. Dis. 539-16. [DOI] [PubMed] [Google Scholar]
- 27.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 363674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trampuz, A., and A. F. Widmer. 2006. Infections associated with orthopedic implants. Curr. Opin. Infect. Dis. 19349-356. [DOI] [PubMed] [Google Scholar]
- 29.Trampuz, A., and W. Zimmerli. 2005. Prosthetic joint infections: update in diagnosis and treatment. Swiss Med. Wkly. 135243-251. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerli, W., A. Trampuz, and P. E. Ochsner. 2004. Prosthetic-joint infections. N. Engl. J. Med. 3511645-1654. [DOI] [PubMed] [Google Scholar]