Abstract
In this study, we developed a simple, reliable, serotype-specific PCR method to differentiate Streptococcus pneumoniae serotypes 6A, 6B, and 6C. It was more efficient and practical than the assays currently being used to identify serotypes 6A, 6B, and 6C. Of 120 selected serogroup 6 isolates from subjects with invasive (n = 101) and noninvasive (n = 19) pneumococcal disease, most of which were collected after 2003 in New South Wales, 45 had been identified as 6A and 75 had been identified as 6B by the Quellung reaction. PCR analysis confirmed the results for serotype 6B isolates and identified two different subtypes. Fourteen of 45 isolates that had been identified as serotype 6A actually belonged to serotype 6C.
Streptococcus pneumoniae is a well-characterized human pathogen and a major etiologic agent of pneumonia, meningitis, otitis media, and sepsis, primarily among very young children and older adults. With the identification of serotype 6C in 2007, there are now 91 recognized S. pneumoniae serotypes (20), some of which are more clinically significant and more extensively studied than others. For example, isolates belonging to serogroup 6 (especially 6B) consistently rank among the top three pathogens causing invasive pneumococcal disease (IPD) worldwide (5, 22, 24). Classically, S. pneumoniae is serotyped by using the Quellung reaction (6, 12), but the high cost of antisera and the technical expertise required have encouraged the development of PCR-based serotyping systems (1, 2, 10, 11, 15, 16, 21, 23). However, most of these systems cannot distinguish some closely related cross-reacting serotypes, including 6A and 6B.
The new serotype, 6C, is closely related to serotype 6A; they are serologically cross-reactive with polyclonal antisera (20). Serotype 6C was discovered because of differences in reactivity with monoclonal antibodies between what were initially thought to be subtypes of serotype 6A (20). Since the widespread use of the 7-valent conjugate pneumococcal vaccine, there has been an increase in the proportion of cases of IPD due to serotype 6C in the United States, where it is now the predominant serogroup 6 serotype. Serotype 6C has been reported to exhibit reduced susceptibility to penicillin (3, 8).
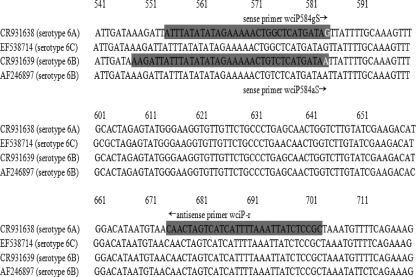
Previously, it has been shown that the capsular loci of serotypes 6A and 6B are almost identical, except for a single nucleotide polymorphism (SNP)—G584A (S195N)—in wciP (13), which is the basis of a pyrosequencing assay (17) developed to differentiate serotypes 6A and 6B. Serotype 6C appears to have originated by a single recombination event in which the 6A wciN gene was replaced by a different wciN gene of unknown origin (19). In serotype 6C, this gene, wciNbeta, is ∼200 bp shorter than the corresponding gene in 6A, wciN, and the genes show only ∼50% DNA sequence homology. However, serotypes 6A and 6C are identical at the site targeted by the pyrosequencing method (Fig. 1) (17).
FIG. 1.
Design of primers to differentiate serotypes 6A/6C and 6B. The figure shows part of the alignment of the CR931638, CR931639 and AF246897, and EF538714 sequences by the ClustalW tool provided in BioManager (Sydney Bioinformatics [http://www.angis.org.au/]). The gray highlights show primer regions. The white letters at the 3′ ends of the primers represent the key SNP, G584A, in serotypes 6A/6C and 6B. Two sense primers are positioned with their 3′ ends at the heterogeneity site. Primer pairs wciP584gS/wciP-r and wciP584aS/wciP-r are expected to amplify sequences of serotypes 6A/6C and 6B, respectively.
A multiplex PCR (mPCR) method for the identification of serotypes 6A and 6C has been described recently (7). The aim of our study was to use or, if necessary, modify this method to complement our existing molecular serotype identification system (9, 10) and to determine the prevalence of serotype 6C among IPD isolates in Australia.
MATERIALS AND METHODS
S. pneumoniae strains.
A set of 125 S. pneumoniae isolates, which had been identified by conventional serotyping (CS) as belonging to serogroup 6, was used for this study. These isolates included serotype 6A and 6B reference strains provided by the Statens Seruminstitut (SSI), Copenhagen, Denmark, and 123 clinical isolates provided by the Pneumococcal Reference Laboratory at the Centre for Infectious Diseases and Microbiology, Westmead Hospital, Westmead, Australia. Of the 123 clinical isolates, 101 were from blood or cerebrospinal fluid and 22 were from respiratory specimens; 46 had been identified as serotype 6A, and 77 had been identified as serotype 6B. Patients included children aged 5 years or less (n = 26), children aged 6 to 17 years (n = 4), and adults aged 18 years or older (n = 93). Most of the isolates (n = 117) were obtained after the introduction of routine childhood immunization in 2003.
Analysis of reference sequences from GenBank.
Four S. pneumoniae sequences in GenBank, with cps loci annotated, were used as reference sequences and included representatives of serotype 6A (an SSI reference strain sequence [accession number CR931638]), 6B (an SSI reference strain sequence [accession number CR931639] which is very similar to two other 6B sequences [accession numbers AF316640 and AF298581] and a sequence [accession number AF246897] which differs from the other 6B sequences), and 6C (accession number EF538714). The sequences were aligned using the ClustalW tool in BioManager (Sydney Bioinformatics [http://www.angis.org.au/]). Based on the alignment, the wciP sequences of serotypes 6A and 6C were highly homologous. The serotype 6B cps locus in the AF246897 sequence was longer than that in the CR931639 sequence due to a 308-bp insert in cps6bQ (GenBank access no. AF246897) between wciN and wciO, as described previously (13). To distinguish these serotype 6B variants, we designated the CR931639 and AF246897 sequences types 6B-I and 6B-II, respectively.
Serotype-specific PCR primer design and prediction of results.
Our alignment confirmed the presence of the G584A SNP, which distinguishes serotypes 6A/6C and 6B. We designed two SNP-specific sense primers, wciP584gS and wciP584aS, with the SNPs at the 3′ ends of the primers. These two primers and the previously described reverse primer wciP-r (17) were the basis of serotype 6A/6C- and 6B-specific amplification (Table 1).
TABLE 1.
Serotype 6A, 6B, and 6C oligonucleotide primers used in this study
| Primera | Specificity
|
GenBank accession no. | Sequence (5′-3′)c | |
|---|---|---|---|---|
| Serotype(s) | Target | |||
| wciNbetaS1 | 6C | wciNbeta | EF538714 | 6960 ATC TCT AAA TCT GAA TAT GAA GCG GCT CAA TC 6991 |
| wciNbetaS2 | 6C | wciNbeta | EF538714 | 6982 CGG CTC AAT CTT TAA AAA TAC CCC TTA AGA AAT TGA C 7018 |
| wciNbetaA1 | 6C | wciNbeta | EF538714 | 7290 CCA CCC ACC CTG TTA TAA AAA ATG AGC TTC G 7260 |
| wciNbetaA2 | 6C | wciNbeta | EF538714 | 7319 GAA CTG AGC TAA ATA ATC CTC TGG ATT ATC CAC C 7286 |
| wciP584gS | 6A, 6C | wciP | CR931638 | 8855 ATT TAT ATA TAG AAA AAC TGG CTC ATG ATA G 8885 |
| wciP584aS | 6B | wciP | CR931639 | 8747 AAG ATT ATT TAT ATA TAG AAA AAC TGT CTC ATG ATA A 8783 |
| wciP-rb | 6A, 6C, 6B | wciP | AF246897 | 8248 GCG GAG ATA ATT TAA AAT GAT GAC TAG TTG 8219 |
| 5106b | 6A, 6B, 6C | wchA | AF246897 | 4798 TAC CAT GCA GGG TGG AAT GT 4817 |
| CR931639 | 5795 TAC CAT GCA GGG TGG AAT GT 5814 | |||
| CR931638 | 5897 TAC CAT GCA GGG TGG AAT GT 5916 | |||
| EF538714 | 6143 TAC CAT GCA GGG TGG AAT GT 6162 | |||
| 5101b | 6A, 6B | wciN | AF246897 | 5883 ATT TGG TGT ACT TCC TCC 5900 |
| CR931639 | 6847 ATT TGG TGT ACT TCC TCC 6864 | |||
| CR931638 | 6949 ATT TGG TGT ACT TCC TCC 6966 | |||
| 3101b | 6A, 6B, 6C | wciO | AF246897 | 7149 CCA TCC TTC GAG TAT TGC 7132 |
| CR931639 | 7803 CCA TCC TTC GAG TAT TGC 7786 | |||
| CR931638 | 7905 CCA TCC TTC GAG TAT TGC 7888 | |||
| EF538714 | 7958 CCA TCC TTC GAG TAT TGC 7941 | |||
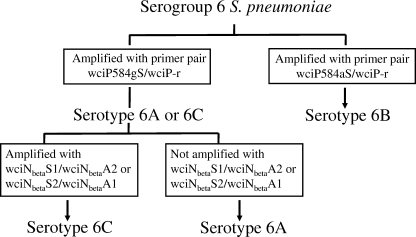
We designed two sets of primers, wciNbetaS1/wciNbetaA2 (outer primer set) and wciNbetaS2/wciNbetaA1 (inner primer set), targeting wciNbeta for 6C-specific amplification, with predicted PCR product lengths of 359 and 308 bp, respectively (Table 1). We planned to use these primer sets sequentially to distinguish serotype 6A/6C from 6B and then serotype 6C from among serotype 6A/6C isolates (Fig. 2).
FIG. 2.
Algorithm for identification of serogroup 6 serotypes. Sequences of primers wciP584gS/wciP-r, wciP584aS/wciP-r, wciNbetaS1/wciNbetaA2, and wciNbetaS2/wciNbetaA1 are listed in Table 1.
All primer sequences were evaluated using the Sigma DNA calculator (http://www.sigma-genosys.com/calc/dnacalc.asp) and compared against all GenBank sequences by using Blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure their specificity.
DNA extraction.
S. pneumoniae isolates were retrieved from storage, subcultured on 5% horse blood agar (Columbia II agar base), and incubated at 37°C for 24 h. Approximately five separate colonies were suspended in 200 μl of molecular-biology-grade water and boiled (100°C) for 15 min. The suspension was centrifuged for 5 min at 13,200 rpm to pellet the cell debris and stored at −20°C until required.
PCR method.
A 25-μl PCR volume was prepared as follows: 2 μl of template DNA (2.9 μg/ml), 0.125 μl of each forward and reverse primer (50 pmol μl−1), 1 μl of deoxynucleoside triphosphates (2.5 mM each), 2.5 μl of 10× PCR buffer (Qiagen), and 0.1 μl of Qiagen HotStarTaq polymerase (5 U μl−1) were mixed, and molecular-biology-grade H2O (Eppendorf) was added to obtain the final volume of 25 μl. The PCR program was performed according to the instructions provided with the Qiagen HotStarTaq polymerase kit, as follows: 95°C for 15 min; 35 cycles of 94°C for 30 s, 62°C for 60 s, and 72°C for 60 s; 72°C for 10 min; and holding at 22°C. PCR products were detected on a 2% agarose gel. mPCR was performed as described previously (7).
Data analysis.
Statistical analyses were performed in SPSS 10.0 by using the chi-square test, as appropriate, for categorical variables. A P value of less than 0.05 was considered to indicate a statistically significant difference between groups.
RESULTS AND DISCUSSION
Modification of the previously described identification methods for serogroup 6.
The primers used in the pyrosequencing method for the identification of serotypes 6A and 6B (wciP-f/wciP-r) target a 25-bp sequence in wciP which includes the G584A (S195N) SNP (17). This SNP cannot be used to distinguish serotypes 6A and 6C, which are identical at this site; the cps loci of 6A and 6C are ∼98% homologous, except for wciN (in 6A) and wciNbeta (in 6C) (19) (Fig. 1). The previously described mPCR method developed to identify serotypes 6A and 6C targets the wciN region and produces amplicons of different sizes (7). Using this method, we obtained results somewhat different from those of Jacobs et al., who reported that primer pair 5101 and 3101 gave “products of 958 or 1267 bp with serotypes 6A and 6B, while no product is produced with serotype 6C” and that “primer pair 5106-3101 produces wciN PCR products of 2.0 and 1.8 kb with serotypes 6A and 6C, respectively, whereas no product is produced with serotype 6B” (7).
We performed Blastn comparisons of three primers used in the mPCR, 5106, 5101, and 3101, against GenBank sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and calculated the expected length of each PCR product (see the supplemental material). Our analysis showed that, theoretically, primer pair 5106-3101 in a single PCR would produce an amplicon of 1.8 kb from serotype 6C, 2.0 kb from 6A or 6B-I, or 2.3 kb from 6B-II. On the other hand, a single PCR using primer pair 5101-3101 would produce an amplicon of 956 bp from serotype 6A or 6B-I or 1.2 kb from 6B-II and no amplicon from serotype 6C. Apart from the minor differences in specificity (related to serotype 6B-II), we predicted that serotype 6A and 6B DNA would not be amplified in the mPCR by 5106-3101 (or would produce only very faint bands) because competition with primer pair 5101-3101 would lead to preferential amplification of the shorter PCR products.
mPCR results.
We performed mPCR as described by Jacobs et al. (7), using primers 5106, 5101, and 3101 for our 120 serogroup 6 clinical isolates (3 of our total of 123 clinical isolates were ultimately found to be non-serogroup 6 isolates). Of these 120 isolates, 14 produced amplicons of 1.8 kb and were therefore identified as serotype 6C. No amplicons of 2.0 kb or longer were produced, indicating that, as predicted, primers 5106 and 3101 did not amplify DNA from serotypes 6A and 6B. However, 29 isolates produced amplicons of 1.2 kb (and were identified as serotype 6B-II), and 77 isolates produced amplicons of 956 bp (and were identified as either serotype 6A or serotype 6B-I). These results indicate that the mPCR primers 5101 and 3101 described by Jacobs et al. can identify one subtype of serotype 6B (6B-II) but cannot distinguish the other (6B-I) from serotype 6A (Table 2). Since the two 6B subtypes occurred with similar frequencies among our isolates, this limitation is a significant shortcoming.
TABLE 2.
Comparison of results among serotype-specific PCR, CS, and mPCR
| Serotype | No. of isolates identified by:
|
Length of mPCR productd | ||
|---|---|---|---|---|
| Serotype-specific PCRa | CSb | mPCRc | ||
| 6A | 31 | 45 | 77 | 956 bp |
| 6B | 75 | 75 | 29 | 1.2 kb |
| 6C | 14 | 0 | 14 | 1.8 kb |
| Total | 120 | 120 | 120 | |
Serotype-specific PCR assay used in this study to differentiate S. pneumoniae serotypes 6A, 6B, and 6C.
The data presented are the final results obtained after the retesting of eight isolates which gave results discordant with those from PCR, including three which did not produce a PCR product and were shown to belong to non-serogroup 6 serotypes. Serotype 6C isolates were identified by CS as serotype 6A; these results were interpreted to be concordant with PCR results indicating 6A or 6C.
The mPCR assay was done as described previously (7). Fourteen isolates were consistently identified as serotype 6C by our PCR test and the mPCR method. Of the 77 isolates identified as serotype 6A by mPCR, 31 isolates were identified as serotype 6A and 46 were identified as serotype 6B-I by serotype-specific PCR.
The reference sequence for serotype 6B-II (GenBank accession no. AF246897) is longer than that for 6B-I (GenBank accession no. CR931639), due to a 308-bp insert in cps6Q between the wciN gene and the wciO gene.
Serotype-specific PCR results.
With the serotype 6A and 6B reference strains, our primer pairs wciP584gS/wciP-r and wciP584aS/wciP-r formed amplicons only from the corresponding serotype. These primer pairs were used to test the 123 clinical isolates (Table 2), of which 45 were identified as serotype 6A/6C and 75 were identified as serotype 6B; 3 isolates produced no PCR product. The comparison of these results with the original CS results showed that the data for 115 isolates were consistent (isolates identified as serotype 6A/6C by PCR had been identified as 6A by CS) and that the data for 8 isolates were discrepant. The eight isolates with discrepant results were retested by the Quellung reaction, and the PCR results were confirmed; three isolates previously identified as 6B were reassigned to 6A, two isolates previously identified as 6A were reassigned to 6B, and three isolates that gave negative PCR results did not to belong to serogroup 6.
The 45 isolates identified as serotype 6A (by CS) or 6A/6C (by PCR) were tested using primer pairs wciNbetaS1/wciNbetaA2 and wciNbetaS2/wciNbetaA1. These results were concordant with the mPCR results: 14 of 45 isolates (31.1%) were identified as serotype 6C. The concordance between the results obtained with our two serotype 6C primer sets indicates that either could be used for the identification of serotype 6C. A comparison of the results of PCR, mPCR, and CS analyses is shown in Table 2.
Others have reported that serotype 6C strains are more penicillin susceptible than serotype 6A strains (4, 14, 18). The rate of penicillin resistance among our serogroup 6 isolates was quite low, and there was no significant difference among the three serotypes (84, 81, and 93% of serotype 6A, 6B, and 6C isolates were fully susceptible to penicillin). However, 18 (25%) of 75 serotype 6B isolates were resistant to erythromycin, and this frequency was significantly higher than those of erythromycin-resistant serotype 6A isolates (3 of 31 [10%]; P = 0.009) and 6C isolates (1 of 14 [7%]; P = 0.001). The ages of patients from whom serotype 6C isolates were recovered were as follows: ≤5 years, 1; 6 to 17 years, 0; and ≥18 years, 13.
Conclusion.
In this study, we developed a serotype-specific PCR method to differentiate S. pneumoniae serotypes 6A, 6B, and 6C which is rapid (requiring ∼3.5 h for DNA extraction, PCR, and gel electrophoresis), simple, and cost-effective (∼US$0.35/specimen without labor costs). It is also more accurate than a previously described mPCR method because it distinguishes two subtypes of serotype 6B, one of which cannot be distinguished from serotype 6A by the previous method. The two serotype 6B subtypes were both present as considerable proportions of this small sample of serogroup 6 isolates (120), of which 75 (62%) were serotype 6B (of these, 46% were 6B-I and 29% were 6B-II), 31 (26%) were serotype 6A, and 14 (12%) were serotype 6C.
Supplementary Material
Footnotes
Published ahead of print on 17 June 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Batt, S. L., B. M. Charalambous, T. D. McHugh, S. Martin, and S. H. Gillespie. 2005. Novel PCR-restriction fragment length polymorphism method for determining serotypes or serogroups of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 432656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito, D. A., M. Ramirez, and H. de Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 412378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho Mda, G., F. C. Pimenta, R. E. Gertz, Jr., H. H. Joshi, A. A. Trujillo, L. E. Keys, J. Findley, I. S. Moura, I. H. Park, S. K. Hollingshead, T. Pilishvili, C. G. Whitney, M. H. Nahm, and B. W. Beall. 2009. PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J. Clin. Microbiol. 47554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.du Plessis, M., A. von Gottberg, S. A. Madhi, O. Hattingh, L. de Gouveia, and K. P. Klugman. 2008. Serotype 6C is associated with penicillin-susceptible meningeal infections in human immunodeficiency virus (HIV)-infected adults among invasive pneumococcal isolates previously identified as serotype 6A in South Africa. Int. J. Antimicrob. Agents 32(Suppl. 1)S66-S70. [DOI] [PubMed] [Google Scholar]
- 5.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30100-121. [DOI] [PubMed] [Google Scholar]
- 6.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 332759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs, M. R., S. Bajaksouzian, R. A. Bonomo, C. E. Good, A. R. Windau, A. M. Hujer, C. Massire, R. Melton, L. B. Blyn, D. J. Ecker, and R. Sampath. 2009. Occurrence, distribution and origins of serotype 6C Streptococcus pneumoniae, a recently recognized serotype. J. Clin. Microbiol. 4764-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs, M. R., C. E. Good, S. Bajaksouzian, and A. R. Windau. 2008. Emergence of Streptococcus pneumoniae serotypes 19A, 6C, and 22F and serogroup 15 in Cleveland, Ohio, in relation to introduction of the protein-conjugated pneumococcal vaccine. Clin. Infect. Dis. 471388-1395. [DOI] [PubMed] [Google Scholar]
- 9.Kong, F., M. Brown, A. Sabananthan, X. Zeng, and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay to identify 23 Streptococcus pneumoniae polysaccharide vaccine serotypes. J. Clin. Microbiol. 441887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong, F., W. Wang, J. Tao, L. Wang, Q. Wang, A. Sabananthan, and G. L. Gilbert. 2005. A molecular-capsular-type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA-cpsB sequencing and wzy- or wzx-specific PCR. J. Med. Microbiol. 54351-356. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence, E. R., D. B. Griffiths, S. A. Martin, R. C. George, and L. M. Hall. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J. Clin. Microbiol. 41601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund, E. 1960. Laboratory diagnosis of Pneumococcus infections. Bull. W. H. O. 235-13. [PMC free article] [PubMed] [Google Scholar]
- 13.Mavroidi, A., D. Godoy, D. M. Aanensen, D. A. Robinson, S. K. Hollingshead, and B. G. Spratt. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 1868181-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nahm, M. H., J. Lin, J. A. Finkelstein, and S. I. Pelton. 2009. Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J. Infect. Dis. 199320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Halloran, D. M., and M. T. Cafferkey. 2005. Multiplex PCR for identification of seven Streptococcus pneumoniae serotypes targeted by a 7-valent conjugate vaccine. J. Clin. Microbiol. 433487-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pai, R., R. E. Gertz, and B. Beall. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai, R., J. Limor, and B. Beall. 2005. Use of pyrosequencing to differentiate Streptococcus pneumoniae serotypes 6A and 6B. J. Clin. Microbiol. 434820-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, I. H., M. R. Moore, J. J. Treanor, S. I. Pelton, T. Pilishvili, B. Beall, M. A. Shelly, B. E. Mahon, and M. H. Nahm. 2008. Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J. Infect. Dis. 1981818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, I. H., S. Park, S. K. Hollingshead, and M. H. Nahm. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect. Immun. 754482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 451225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin, L. G., and A. Rizvi. 2004. PCR-based assays for detection of Streptococcus pneumoniae serotypes 3, 14, 19F and 23F in respiratory specimens. J. Med. Microbiol. 53595-602. [DOI] [PubMed] [Google Scholar]
- 22.Scott, J. A., A. J. Hall, R. Dagan, J. M. Dixon, S. J. Eykyn, A. Fenoll, M. Hortal, L. P. Jette, J. H. Jorgensen, F. Lamothe, C. Latorre, J. T. Macfarlane, D. M. Shlaes, L. E. Smart, and A. Taunay. 1996. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin. Infect. Dis. 22973-981. [DOI] [PubMed] [Google Scholar]
- 23.Slotved, H. C., M. Kaltoft, I. C. Skovsted, M. B. Kerrn, and F. Espersen. 2004. Simple, rapid latex agglutination test for serotyping of pneumococci (Pneumotest-Latex). J. Clin. Microbiol. 422518-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sniadack, D. H., B. Schwartz, H. Lipman, J. Bogaerts, J. C. Butler, R. Dagan, G. Echaniz-Aviles, N. Lloyd-Evans, A. Fenoll, N. I. Girgis et al. 1995. Potential interventions for the prevention of childhood pneumonia: geographic and temporal differences in serotype and serogroup distribution of sterile site pneumococcal isolates from children—implications for vaccine strategies. Pediatr. Infect. Dis. J. 14503-510. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.