Abstract
A prototype, real-time reverse-transcription PCR assay, based on MultiCode-RTx technology, quantifying hepatitis C virus (HCV) RNA by targeting the HCV 3′ untranslated region demonstrated linearity over 7 logs, with a good correlation between the quantitative results of this assay and the results of two commercially available comparator assays for 466 clinical specimens comprising all six HCV genotypes.
Current commercially available laboratory methods for the detection and quantification of hepatitis C virus (HCV) RNA in clinical specimens include transcription-mediated amplification, branched-DNA, and real-time PCR (1, 2, 4-6, 9, 10, 13, 18). All of these methods target the highly conserved HCV 5′ untranslated region (UTR) rather than the similarly conserved 3′ UTR, which was not recognized during initial characterization of the HCV genome (3, 7) but was subsequently identified (8). Recently, a prototype real-time PCR assay based on MultiCode-RTx PCR technology (i.e., the RTx assay; EraGen Biosciences, Inc., Madison, WI) (11, 15-17) and targeting the HCV 3′ UTR was developed for HCV detection and quantification. Herein, we report the results of the initial analytical evaluation of this novel RTx assay along with preliminary correlation studies comparing the RTx assay to the Versant HCV RNA 3.0 assay (the bDNA assay; Siemens Healthcare Diagnostics, Inc., Tarrytown, NY) and a laboratory-developed assay based on the TaqMan HCV analyte-specific reagent (the TaqMan assay; Roche Molecular Systems, Inc.) (1, 2, 4, 6, 9).
For the initial analytical studies, individual RTx assay reaction mixtures contained 0.5 U/μl Moloney murine leukemia virus reverse transcriptase and 1× Titanium Taq DNA polymerase (Clontech, Mountain View, CA) in 1× ISOlution 1482 buffer (EraGen Biosciences, Inc.) along with primer sets to specifically amplify two targets. The first primer set, labeled with 6-carboxyfluorescein, targeted a 48-bp region of the HCV 3′ UTR, while the second primer pair, labeled with hexachlorofluorescein, targeted a 72-bp segment of an RNA internal control sequence added at 250 copies/reaction. Thermal cycling was performed with an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA), with the following parameters: 15 min at 50°C, 2 min at 95°C, followed by 50 cycles of 95°C for 5 s, 58°C for 10 s, and 72°C for 20 s. Following PCR, a 5-min thermal ramp (60°C to 95°C) was performed to determine melting point (Tm) values of the amplified products.
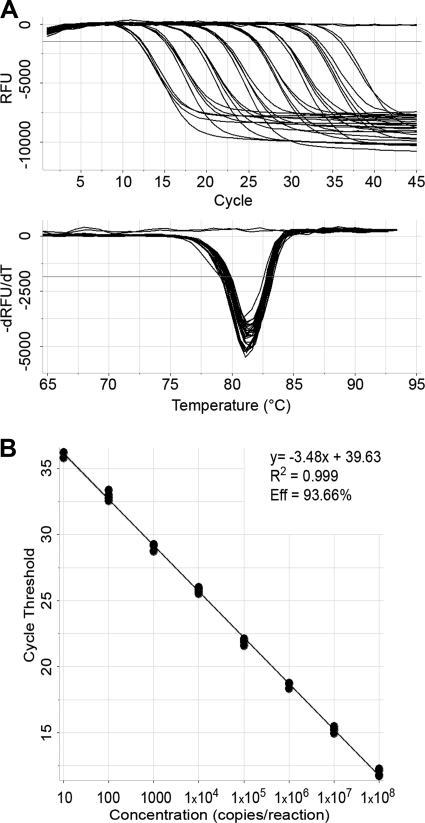
An in vitro RNA transcript containing both the HCV 5′ UTR and the 3′ UTR was used to prepare a series of RTx assay reaction mixtures containing 10-fold dilutions of this transcript ranging from 108 to 101 copies/reaction. Data obtained from RTx assay testing of these analytical samples in quadruplicate were exported to MultiCode-RTx analysis software (EraGen Biosciences, Inc.) for further analysis. The presence of HCV RNA was confirmed by a 6-carboxyfluorescein amplification curve with a Tm of 80.4 ± 1.0°C occurring within 50 cycles and crossing a cycle threshold (CT) set at 10 times the standard deviation (SD) of the baseline noise. Results showed that the RTx assay was linear over 7 logs (Fig. 1) and sensitive enough to detect two of four replicates at 10 copies/reaction.
FIG. 1.
Linearity of the RTx assay. (A) The upper graph shows real-time PCR fluorescence data as baseline-adjusted relative fluorescence units (RFU) versus the number of PCR cycles. The lower graph shows Tm analysis of the negative value of the change in RFU over the change in temperature (−dRFU/dT) versus temperature (degrees Celsius). (B) Linear relationship between RTx assay CT values and 3′ UTR target copy numbers/reaction. Eff, efficiency.
For all subsequent studies, the RTx assay was modified to include two manual nucleic acid extraction procedures, both of which incorporate the use of an unrelated extractable sample processing reference (SPR; EraGen Biosciences, Inc.) that contains the internal control sequence used in the analytical studies. In a quantitative small-volume RTx protocol used for initial testing of specimens, 3.5 μl of SPR (1,000 copies/μl) was added to 136.5 μl of specimen and processed by use of the QIAamp viral RNA minikit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions, with a final elution volume of 70 μl. In a qualitative large-volume RTx protocol used for testing specimens yielding discordant viral load (VL) results, 3 μl of SPR was added to 497 μl of specimen and processed by use of the QIAamp MinElute virus vacuum kit (Qiagen, Inc.) according to the manufacturer's instructions, with a final elution volume of 60 μl. The small-volume RTx assay reaction mixture (25 μl) contained 5 μl of eluate, while the large-volume RTx assay reaction mixture (100 μl) contained 50 μl of eluate.
To create a standard curve for RTx assay HCV VL determination, serial fivefold dilutions (neat to 1:3,125) of a high-titer HCV RNA-positive serum specimen were prepared in HCV RNA-negative serum and tested in replicate (20 reactions/dilution) by the small-volume RTx protocol. The standard curve was then calibrated to WHO Second International HCV standard 96/798 by correlating the known HCV VL and experimentally determined mean CT value from 36 replicate amplifications of a single concentration of the standard. The resulting equation was used to determine the HCV VLs (in IU/ml) in clinical specimens based on the mean CT of duplicate RTx assay reactions.
Amplification equivalency among HCV genotypes was initially studied by using a 13-member worldwide HCV performance panel (WWHV302) (Boston Biomedica, Inc., West Bridgewater, MA), which was tested in triplicate by the quantitative small-volume RTx protocol. The small-volume RTx assay VL results were compared to VL data (provided in the WWHV302 product insert) from the bDNA assay and the Cobas Amplicor HCV Monitor test, version 2.0 (CAM test; Roche Molecular Systems, Inc., Branchburg, NJ).
Frozen aliquots of 466 leftover clinical specimens submitted for routine HCV VL testing at three different clinical laboratories between January 2006 and January 2008 were deidentified, handled in a blind manner, and sent to EraGen Biosciences, Inc., for testing by the RTx assay. These specimens included 200 serum and plasma specimens from Dynacare Laboratories, Milwaukee, WI; 110 serum specimens from the Mayo Clinic, Rochester, MN; and 156 plasma specimens from the Washington University School of Medicine, St. Louis, MO. The 200 clinical specimens from Dynacare Laboratories consisted of 179 specimens with measurable HCV RNA by the bDNA assay and 21 specimens without detectable HCV RNA by the bDNA assay, with HCV genotypes 1a (n = 65), 1b (n = 21), 2 (n = 4), and 3 (n = 7) determined for 97 of the HCV RNA-positive specimens by the Trugene HCV 5′NC genotyping kit (Trugene; Siemens Healthcare Diagnostics Inc.). The 110 sera collected at the Mayo Clinic included 100 specimens with measurable HCV RNA by the bDNA assay and 10 HCV RNA-negative specimens from healthy blood donors. These 100 HCV RNA-positive sera consisted of samples with HCV genotypes 1 (n = 25), 2 (n = 25), 3 (n = 20), 4 (n = 20), 5 (n = 5), and 6 (n = 5), as determined by Trugene. All bDNA and Trugene testing was performed according to the manufacturer's instructions. The 156 specimens from Washington University School of Medicine consisted of 141 and 15 specimens with and without detectable HCV RNA according to the TaqMan assay, respectively. Among the 141 specimens with measurable HCV RNA, 86 had HCV genotype determination performed by the Versant HCV Genotype 2.0 assay (Siemens Healthcare Diagnostics Inc.), following the manufacturer's instructions, which yielded HCV genotypes 1a (n = 46), 1b (n = 21), 1 (n = 1), 2a/c (n = 2), 2b (n = 3), and 3a (n = 13). The TaqMan assay was performed with the Cobas TaqMan 48 analyzer (Roche Molecular Systems, Inc.); sample preparation was done by use of the QIAamp viral RNA minikit, and calibration to OptiQuant HCV RNA quantification panels (AcroMetrix Corp., Benicia, CA) was performed as previously described (2, 14).
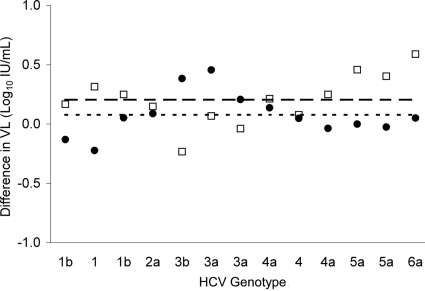
Initial testing of fivefold dilutions of the high-titer HCV-positive serum specimen by the small-volume RTx protocol yielded CT values below the established threshold in all 20 reactions for each dilution, except for the 3,125-fold dilution, which showed acceptable CT values in only 10 reactions. When calibrated to WHO Second International HCV standard 96/798, the standard curve derived by least-squares regression analysis of the CT and relative sample concentration (i.e., dilution factor) yielded a best-fit linear equation of CT = −3.8 × log10 IU/ml + 52.3 (r = 0.984, PCR efficiency = 83%). The mean CT values ± SDs for detectable replicates of the 625- and 3,125-fold dilutions translated to 3.91 ± 0.89 log10 IU/ml and 3.65 ± 1.13 log10 IU/ml, respectively. Among HCV genotypes contained in the worldwide HCV performance panel, overall mean VL differences ± SDs between the small-volume RTx assay and the bDNA assay and between the small-volume RTx assay and CAM test were 0.08 ± 0.19 log10 IU/ml (range, −0.22 to 0.46 log10 IU/ml) and 0.20 ± 0.22 log10 IU/ml (range, −0.23 to 0.59 log10 IU/ml), respectively (Fig. 2).
FIG. 2.
Differences in HCV VL results with a worldwide HCV performance panel of six HCV genotypes tested by three assay methods (•, small-volume RTx assay result − bDNA assay result; □, small-volume RTx assay result − CAM test result). The dotted line indicates the mean difference between small-volume RTx assay and bDNA assay results, and the dashed line indicates the mean difference between small-volume RTx assay and CAM test results.
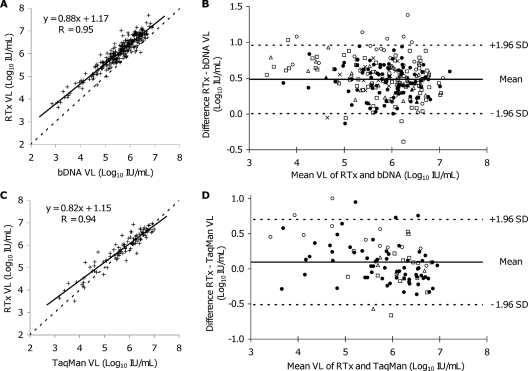
Four of 277 clinical specimens that were previously tested by the bDNA assay and had measurable HCV VLs by the bDNA assay were negative for HCV RNA by the small-volume RTx assay, while 2 specimens had VLs above the upper limit of quantification by the bDNA assay. Supplemental testing of the four specimens yielding qualitatively discordant results by the qualitative large-volume RTx protocol resolved two of four discordant results. However, the SPR failed to generate a hexachlorofluorescein signal in the third specimen despite multiple attempts with the large-volume RTx protocol (suggesting the presence of PCR inhibitors), while the fourth specimen was consistently negative by the large-volume RTx assay. Linear regression analysis of paired results from 273 specimens quantifiable by both the small-volume RTx assay and the bDNA assay indicated a good correlation (r = 0.95), with a slope of 0.88 (Fig. 3A). A Bland-Altman plot of VL differences between the small-volume RTx assay and the bDNA assay yielded a mean difference of 0.48 log10 IU/ml, with ±1.96 SD limits at ±0.51 log10 IU/ml and a range of −0.02 to 0.98 log10 IU/ml (Fig. 3B). Agreement between the small-volume RTx assay and the bDNA assay improved with increasing VLs, especially at concentrations of ≥5.0 log10 IU/ml. Specimens containing HCV genotype 3 (n = 27) yielded a mean difference of 0.75 log10 IU/ml between the small-volume RTx assay and the bDNA assay, compared to a mean difference of 0.48 log10 IU/ml for all 273 specimens. The difference in VLs between the two assays for genotype 3 was statistically significant (P <0.005), while differences for genotypes 1, 2, 4, and 5 were insignificant by one-way analysis of variance with Tukey-Kramer adjustment for multiple comparisons. Comparison between genotypes 3 and 6 was inconclusive due to the small number of genotype 6 specimens in this study.
FIG. 3.
Comparisons of small-volume RTx assay results to bDNA and TaqMan results for clinical serum and plasma specimens for quantification of HCV RNA. (A) Linear regression analysis of the small-volume RTx assay versus the bDNA assay for 273 specimens. (B) Bland-Altman plot of paired VL differences between the small-volume RTx assay and the bDNA assay. (C) Linear regression analysis of the small-volume RTx assay versus the TaqMan assay for 98 specimens. (D) Bland-Altman plot of paired VL differences between the small-volume RTx and TaqMan assay VLs. HCV genotypes in panels B and D are indicated by the following symbols: •, genotype 1; ▵, genotype 2; ○, genotype 3; ×, genotype 4; ▴, genotype 5; ▪, genotype 6; □, genotype unspecified.
Small-volume RTx assay yielded measurable HCV VLs for 98 of 100 specimens with VLs of ≥3 log10 IU/ml by the TaqMan assay, but HCV RNA was detectable in only 4 of 41 specimens with TaqMan assay VLs of <3 log10 IU/ml. Sufficient specimen volume was available for qualitative large-volume RTx assay testing of 13 of these 41 specimens, with HCV RNA detected in 10 of 13 (77%) specimens. Linear regression analysis of paired data from 98 specimens with HCV VLs of ≥3 log10 IU/ml by the TaqMan assay and measurable VLs by both methods showed a good correlation (r = 0.94), with a slope of 0.86 (Fig. 3C). A Bland-Altman plot of VL differences between the small-volume RTx assay and the TaqMan assay for these 98 specimens yielded a mean difference of 0.10 log10 IU/ml, with ±1.96 SD limits at ±0.61 log10 IU/ml and a range of −0.51 to 0.70 log10 IU/ml (Fig. 3D). Improved agreement with increasing VLs between the small-volume RTx assay and the TaqMan assay was also observed. Among specimens with known HCV genotypes, 82 yielded measurable VLs by the small-volume RTx assay, with a statistically significant (P <0.005) difference between the small-volume RTx assay and the TaqMan assay VLs among specimens containing genotype 3 (as opposed to genotypes 1 and 2) (one-way analysis of variance with Tukey-Kramer adjustment for multiple comparisons). There were no false-positive results with the small-volume RTx protocol among the combined total of 46 HCV RNA-negative specimens previously tested by the bDNA or TaqMan assay.
Analytical studies demonstrated that the RTx assay was capable of detecting HCV RNA present at a low copy number (i.e., 50% detection at 10 copies/reaction), and the RTx assay was linear over 7 orders of magnitude. However, the PCR efficiency of 94% determined with a synthetic HCV 3′ UTR target was higher than the 83% efficiency seen with the small-volume RTx protocol. While correlation between small-volume RTx and bDNA assay VLs was good, the sensitivity of the small-volume RTx protocol did not compare well to that of the TaqMan assay. The bias for VLs being higher with the small-volume RTx assay than with the bDNA assay at <5 log10 IU/ml may be due to differences in calibration, a predominance of specimens with VLs of ≥5 log10 IU/ml, or intrinsic performance characteristics of the RTx assay.
These preliminary studies suggest that an RTx-based assay targeting the HCV 3′ UTR may be used for quantification of HCV RNA in human serum or plasma. Despite the significantly higher VL for HCV genotype 3 when the small-volume RTx assay results were compared to the bDNA and TaqMan assay results, further optimization of assay conditions, increased sample input volume, and a more rigorous calibration of the RTx assay should be capable of achieving the increased sensitivity and accuracy necessary for detection and quantification of HCV RNA to meet clinical needs (12).
Acknowledgments
We acknowledge the technical assistance of Katherine Cavaletti and Magda Dwidar (Washington University Medical School, St. Louis, MO) and statistical analyses by Jayawant N. Mandrekar (Mayo Clinic, Rochester, MN).
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Barbeau, J. M., J. Goforth, A. M. Caliendo, and F. S. Nolte. 2004. Performance characteristics of a quantitative TaqMan hepatitis C virus RNA analyte-specific reagent. J. Clin. Microbiol. 423739-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caliendo, A. M., A. Valsamakis, Y. Zhou, B. Yen-Lieberman, J. Andersen, S. Young, A. Ferreira-Gonzalez, G. J. Tsongalis, R. Pyles, J. W. Bremer, and N. S. Lurain. 2006. Multilaboratory comparison of hepatitis C virus viral load assays. J. Clin. Microbiol. 441726-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 882451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colucci, G., J. Ferguson, C. Harkleroad, S. Lee, D. Romo, S. Soviero, J. Thompson, M. Velez, A. Wang, Y. Miyahara, S. Young, and C. Sarrazin. 2007. Improved COBAS TaqMan hepatitis C virus test (version 2.0) for use with the High Pure system: enhanced genotype inclusivity and performance characteristics in a multisite study. J. Clin. Microbiol. 453595-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira-Gonzalez, A., M. L. Shiffman, D. H. Lofland, M. R. Langley, D. W. Gainey, and C. T. Garrett. 1996. Assessing clinical utility of hepatitis C virus quantitative RT-PCR data: implications for identification of responders among alpha-interferon-treated patients. Mol. Diagn. 1109-120. [DOI] [PubMed] [Google Scholar]
- 6.Forman, M. S., and A. Valsamakis. 2004. Verification of an assay for quantification of hepatitis C virus RNA by use of an analyte-specific reagent and two different extraction methods. J. Clin. Microbiol. 423581-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houghton, M., Q. L. Choo, G. Kuo, A. J. Weiner, J. Han, M. S. Urdea, B. D. Irvine, and J. A. Kolberg. January 1998. Methods for detecting hepatitis C virus using polynucleotides specific for same. U.S. patent 5,712,088.
- 8.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 703363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konnick, E. Q., S. M. Williams, E. R. Ashwood, and D. R. Hillyard. 2005. Evaluation of the COBAS hepatitis C virus (HCV) TaqMan analyte-specific reagent assay and comparison to the COBAS Amplicor HCV Monitor V2.0 and Versant HCV bDNA 3.0 assays. J. Clin. Microbiol. 432133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majid, A. M., and D. R. Gretch. 2002. Current and future hepatitis C virus diagnostic testing: problems and advancements. Microbes Infect. 41227-1236. [DOI] [PubMed] [Google Scholar]
- 11.Moser, M. J., D. R. Christensen, D. Norwood, and J. R. Prudent. 2006. Multiplexed detection of anthrax-related toxin genes. J. Mol. Diagn. 889-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Institutes of Health. 2006. National Institutes of Health Consensus Development Conference statement. Management of hepatitis C: 2002-2004. National Institutes of Health, Bethesda, MD.
- 13.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 1221554-1568. [DOI] [PubMed] [Google Scholar]
- 14.Sábato, M. F., M. L. Shiffman, M. R. Langley, D. S. Wilkinson, and A. Ferreira-Gonzalez. 2007. Comparison of performance characteristics of three real-time reverse transcription-PCR test systems for detection and quantification of hepatitis C virus. J. Clin. Microbiol. 452529-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvaraju, S. B., M. Wurst, R. T. Horvat, and R. Selvarangan. 2009. Evaluation of three analyte-specific reagents for detection and typing of herpes simplex virus in cerebrospinal fluid. Diagn. Microbiol. Infect. Dis. 63286-291. [DOI] [PubMed] [Google Scholar]
- 16.Sherrill, C. B., D. J. Marshall, M. J. Moser, C. A. Larsen, L. Daude-Snow, S. Jurczyk, G. Shapiro, and J. R. Prudent. 2004. Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 1264550-4556. [DOI] [PubMed] [Google Scholar]
- 17.Svarovskaia, E. S., M. J. Moser, A. S. Bae, J. R. Prudent, M. D. Miller, and K. Borroto-Esoda. 2006. MultiCode-RTx real-time PCR system for detection of subpopulations of K65R human immunodeficiency virus type 1 reverse transcriptase mutant viruses in clinical samples. J. Clin. Microbiol. 444237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young, K. K., R. M. Resnick, and T. W. Myers. 1993. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol. 31882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]