Abstract
Molecular methods such as PCR have become attractive tools for diagnosis of cutaneous leishmaniasis (CL), both for their high sensitivity and for their specificity. However, their practical use in routine diagnosis is limited due to the infrastructural requirements and the lack of any standardization. Recently, a simplified and standardized PCR format for molecular detection of Leishmania was developed. The Leishmania OligoC-TesT is based on simple and rapid detection using a dipstick with PCR-amplified Leishmania DNA. In this study, we estimated the diagnostic accuracy of the Leishmania OligoC-TesT for 61 specimens from 44 CL-suspected patients presenting at the leishmaniasis clinic of the Instituto de Medicina Tropical Alexander von Humboldt, Peru. On the basis of parasitological detection and the leishmanin skin test (LST), patients were classified as (i) confirmed CL cases, (ii) LST-positive cases, and (iii) LST-negative cases. The sensitivities of the Leishmania OligoC-TesT was 74% (95% confidence interval (CI), 60.5% to 84.1%) for lesion aspirates and 92% (95% CI, 81.2% to 96.9%) for scrapings. A significantly higher sensitivity was observed with a conventional PCR targeting the kinetoplast DNA on the aspirates (94%) (P = 0.001), while there was no significant difference in sensitivity for the lesion scrapings (88%) (P = 0.317). In addition, the Leishmania OligoC-TesT was evaluated for 13 CL-suspected patients in two different peripheral health centers in the central jungle of Peru. Our findings clearly indicate the high accuracy of the Leishmania OligoC-TesT for lesion scrapings for simple and rapid molecular diagnosis of CL in Peru.
Leishmaniasis is a vector-borne disease caused by obligatory intracellular parasites of the genus Leishmania. Several clinical manifestations are classified under the term leishmaniasis, but three are the most prominent: visceral leishmaniasis (VL), cutaneous leishmaniasis (CL), and mucocutaneous leishmaniasis (MCL), which result from replication of parasites in macrophages in the internal organs, dermis, and naso-oropharyngeal mucosa, respectively (15).
In Latin America, CL and MCL are important health problems, and Brazil and Peru are the two most affected countries (9). An increase of cases has been reported for Colombia, Ecuador, and Argentina (4, 25, 26). Human-made risk factors, such as migration, urbanization, and deforestation, likely contribute to the spread of the disease (10).
Over 10,000 CL cases per year are reported to occur in Peru, and more than a million people are at risk for infection (11, 22). Furthermore, this disease is endemic in 70% of Peruvian territory, causing high morbidity, lifelong scars, and major health problems for many communities (19).
Diagnosing CL is challenging because of its wide spectrum of clinical presentations. Lesions may vary in severity, clinical appearance, and duration (23). Moreover, differential diagnosis with other cutaneous diseases is often difficult (14, 15). In addition, CL can be caused by different Leishmania species. In Peru, the disease is mainly caused by Leishmania (Viannia) braziliensis, Leishmania (Viannia) peruviana, and Leishmania (Viannia) guyanensis, but Leishmania (Viannia) lainsoni and Leishmania (Leishmania) amazonensis infections have also been reported (1, 18).
Routine diagnosis of CL is still based on demonstration of amastigotes in skin lesion scrapings through microscopic analysis of direct smears or prior in vitro culture of the parasite (15, 23). Both methods require skilled personnel, and their sensitivities tend to be low and variable (12, 21, 27). Furthermore, in vitro culture is cumbersome and time-consuming. The leishmanin skin test (LST) detects cell-mediated immunity and is frequently used in Peru to support clinical diagnosis of CL. However, it cannot distinguish between past and present infections (12).
The PCR is a useful tool for detection of Leishmania parasites in clinical specimens, since high sensitivity and specificity have been reported. Attractive PCR targets are high-copy-number sequences, such as kinetoplast DNA (kDNA) (2, 8, 30) and the ribosomal small subunit (20, 28). Several PCR formats have been designed, but there is actually a demand for simplified and standardized approaches (24). Access to sophisticated equipment such as real-time PCR machines is often limited in Peru. Recently, a simple and rapid dipstick format for detection of amplified Leishmania DNA was developed (Leishmania OligoC-TesT) (7). The test is based on PCR amplification of a small sequence of the 18S rRNA gene followed by visualization of the PCR products on a dipstick by hybridization with a gold-conjugated probe. PCR product detection can be performed in 10 min, and no equipment other than a heating block and a pipette is needed. The test is a promising “low-tech” standardized PCR application for diagnosis of leishmaniasis and can be applied in a mid- to low-level-equipped laboratory (6).
In this report, we estimated the sensitivity of the Leishmania OligoC-TesT for 61 skin lesion scrapings from 44 Peruvian patients suspected of having CL. To assess the performance and to demonstrate the applicability of the test in low-level-equipped laboratories, two trials in rural hospitals in the Peruvian jungle were conducted.
MATERIALS AND METHODS
Patients.
Informed consent was obtained from the participants in the study. Ethical clearance for the study was obtained from the institutional review board of the Universidad Peruana Cayetano Heredia in Lima, Peru.
DNA extracts of aspirates and scrapings of 61 lesions from 44 patients were obtained from a previous study at the leishmaniasis clinic of the Instituto de Medicina Tropical Alexander von Humboldt in Lima, Peru, in the period between February and April 2007 (3). DNA extracts were stored at −70°C until testing in the current study, 6 months after initial specimen taking.
In addition, the test was evaluated for CL-suspected patients admitted at the peripheral health centers of San Martín de Pangoa and Kiteni in the Junín and Cusco regions, respectively. Both regions are among those with the highest CL endemicity in Peru, including MCL for the latter. In the Junín region, CL is mainly caused by L. (V.) guyanensis (1, 18), while L. braziliensis accounts for most CL and MCL cases in the Cusco region (18). Lesion scrapings were collected from 4 CL-suspected patients in San Martín de Pangoa in February 2008 and from 13 CL-suspected patients in Kiteni in July 2008.
Reference tests. (i) Smears.
Tissue was scraped from the lesion border by using a sterile lancet and spread on a glass slide. The slides were air dried, fixed in methanol, and stained with Giemsa, followed by microscopic analysis under light microscopy. Amastigotes in positive smears at the leishmaniasis clinic in Lima were quantified and classified into six grades according to the method of Chulay and Bryceson (5).
(ii) Culture.
In vitro culture of parasites was performed as described by Boggild et al. (3). Three different culture methods were used: (i) conventional culture in 10% RPMI medium, (ii) the microculture method with the same medium, and (iii) culture in modified NNN (Novy-MacNeal-Nicolle) medium with 15% defibrinated rabbit blood. The culture was considered positive if parasites were observed by microscopic analysis with one or more of the three culture techniques.
(iii) LST.
Leishmanin antigen (0.1 ml), prepared from the L. (V.) guyanensis strain LP52 (IPRN/PE/87/Lp52) at the Instituto de Medicina Tropical Alexander von Humboldt in Lima, was injected in the volar surface of the forearm, and induration and erythema were measured after 48 h. The LST result was considered positive if the diameter of the induration was 5 mm or more (31).
Patient classification.
A patient was classified as (i) a confirmed CL patient if positive by smear or culture, (ii) an LST-positive patient if negative by smear and culture but positive by LST, and (iii) an LST-negative patient if negative by smear and culture and LST.
DNA extraction.
DNA of lesion scraping specimens from the recruited patients at San Martín de Pangoa and Kiteni was extracted according to the simplified procedure described by López et al. (17). Briefly, lancets with scraped tissue were immersed in 100 μl of lysis buffer (100 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1,000 mM NaCl) and incubated at 94°C for 30 min. Lysed specimens were directly used in PCR.
kDNA PCR.
A Leishmania (V.) braziliensis complex-specific kDNA PCR was performed on the lesion aspirate and scraping specimens as described by López et al. (17).
Leishmania OligoC-TesT.
The Leishmania OligoC-TesT was performed as described by Deborggraeve et al. (7). Briefly, Leishmania DNA was amplified by PCR and mixed with an equal volume of migration buffer preheated at 55°C. The Leishmania OligoC dipstick was dipped in the solution, and test results were read after 10 min of incubation at 55°C. Two hundred nanograms of L. (V.) guyanensis (strain IPRN/PE/87/Lp52) DNA was used as a positive control for PCR.
RESULTS
Leishmania OligoC-TesT at the leishmaniasis clinic in Lima. (i) Confirmed CL patients (n = 35).
Fifty lesion scrapings from 35 patients were classified in the confirmed-CL category on the basis of the parasitological detection methods (Table 1). Positive LST results were observed for 27 of the 33 patients for which LST results were available, indicating a sensitivity of 81.8%, with a 95% confidence interval (CI) of 65.6% to 91.4%, scored by Wilson's method (32). The DNA extracts from the lesion aspirates were positive by the Leishmania OligoC-TesT for 37 of the 50 specimens, while 47 were positive with the kDNA PCR. Hence, the sensitivities of the Leishmania OligoC-TesT and kDNA PCR for the 50 aspirate specimens are 74% (95% CI, 60.5% to 84.1%) and 94% (95% CI, 83.8% to 98%), respectively. When tested on DNA extracted from lesion scrapings, 46 of the 50 specimens were found to be positive by the Leishmania OligoC-TesT, while 44 were found to be positive with the kDNA PCR, indicating sensitivities of 92% (95% CI, 81.2% to 96.9%) and 88% (95% CI, 76.2% to 94.4%), respectively. Out of the four OligoC-TesT-negative lesion scrapings, three showed the lowest amastigote load (grade 1), while the fourth was negative by smear but positive by culture. Using the McNemar chi-square method, we observed that the kDNA PCR is significantly more sensitive for aspirate specimens (P = 0.001) but that the sensitivities of the two methods did not differ for scrapings (P = 0.317).
TABLE 1.
Diagnostic accuracy of the Leishmania OligoC-TesT and kDNA PCR for 61 lesion specimens from 45 CL-suspected patients at the leishmaniasis clinic in Lima
| Patient group | No. of lesion samples | No. (%) positivea
|
|||
|---|---|---|---|---|---|
| Lesion aspirates
|
Lesion scrapings
|
||||
| OligoC-TesT | kDNA PCR | OligoC-TesT | kDNA PCR | ||
| Confirmed CL | 50 | 37 | 47 | 46 | 44 |
| LST positive | 2 | 0 | 2 | 2 | 1 |
| LST negative | 9 | 4 | 7 | 3 | 4 |
The sensitivities (95% CIs) of the tests were as follows: for lesion aspirates, 74% (60.5% to 84.1%) for the OligoC-TesT and 94% (83.8% to 98%) for kDNA PCR; and for lesion scrapings, 92% (81.2% to 96.9%) for the OligoC-TesT and 88% (76.2% to 84.4%) for kDNA PCR. CIs were scored by Wilson's method.
(ii) LST-positive patients (n = 2).
The lesion aspirates from the two LST-positive patients were positive by kDNA PCR, but those from both were negative by the Leishmania OligoC-TesT. Upon testing with lesion scrapings, a positive kDNA PCR result was observed in one of the two cases, and positive Leishmania OligoC-TesT results were observed in both cases.
(iii) LST-negative patients (n = 7).
Nine lesion scrapings from seven LST-negative patients were negative by smear and culture. Seven aspirate specimens were positive by kDNA PCR, and four were positive by the Leishmania OligoC-TesT. Four scrapings were positive by kDNA PCR and three by the Leishmania OligoC-TesT.
Leishmania OligoC-TesT at the peripheral health centers.
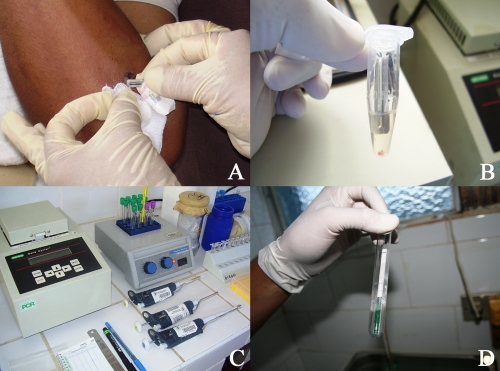
The Leishmania OligoC-TesT was evaluated for 17 CL-suspected patients at two peripheral health centers in areas where CL is highly endemic (Fig. 1). Lesion scrapings from 2 of the 4 suspected patients at San Martín de Pangoa and from 8 of the 13 suspected patients at Kiteni showed parasites during microscopic analysis of lesion tissue smears and/or in vitro culture of parasites in modified NNN medium with 15% defibrinated rabbit blood. Scrapings from both confirmed CL patients at San Martín de Pangoa were positive by the Leishmania OligoC-TesT, while those from the two nonconfirmed patients were negative by the Leishmania OligoC-TesT. The latter were additionally tested with kDNA PCR, and negative test results were observed as well. Lesion scrapings from seven of the eight confirmed CL patients and two of the five nonconfirmed suspected patients at Kiteni showed positive Leishmania OligoC-TesT results. No kDNA PCR was performed at Kiteni.
FIG. 1.
(A) Lesion scraping procedure using a sterile lancet; (B) lancet with scraped lesion material in buffer prior to tissue lysis; (C) equipment used in the assay; (D) macroscopic visualization of results.
DISCUSSION
Molecular approaches have shown great potential for diagnosis of leishmaniasis, follow-up after treatment, disease control, and epidemiology (1, 8, 18, 19, 24, 29). Recently, the Leishmania OligoC-TesT was introduced as an innovative PCR format for simplified and standardized molecular diagnosis of Leishmania infections (7). We have demonstrated that the Leishmania OligoC-TesT offers performance comparable to that of conventional kDNA PCR for lesion scrapings in cases of suspected and confirmed CL in Peru.
On lesion aspirate specimens, we observed lower sensitivity for the Leishmania OligoC-TesT than for kDNA PCR (74% versus 92%). This could be explained by the lower number of copies of the 18S rRNA gene (10 to 100 copies) than of the minicircles (10,000 copies) in the kDNA. However, when lesion scrapings from the same lesions were tested, the sensitivity of the Leishmania OligoC-TesT was found to be 92% and the sensitivity of the kDNA PCR was found to be 88%. The same trend was observed with the two LST-positive patients, where lesion scrapings from both were negative by the Leishmania OligoC-TesT with aspirates but positive with lesion scrapings. In contrast to lesion aspirates, scrapings may contain fewer PCR-inhibiting factors but also fewer parasites. The Leishmania OligoC-TesT is probably more liable to inhibition than the kDNA PCR, giving rise to a higher sensitivity for lesion scrapings in spite of the lower parasite load. Moreover, our results confirm the findings of García et al. (13), who reported on the use of lesion scrapings for parasite species identification by conventional PCR. Hence, this favors the use of lesion scrapings over aspirates and biopsy specimens, since scrapings are much less invasive. The positive PCR results in the LST-negative-specimen group are probably from patients with low-level cell-mediated immunity responses to the infection or from recent skin lesions. The LST is a more sensitive tool in the setting of chronic infection (31). Contamination during PCR is unlikely since negative controls remained negative and since positive results were observed with two PCRs targeting two different DNA sequences. The high sensitivity of the LST for the confirmed CL group (81.8%) supports the valuable contribution of LST to CL diagnosis.
The results obtained with the Leishmania OligoC-TesT at the peripheral health centers in the Peruvian jungle show that the Leishmania OligoC-TesT can be conducted in low-level-equipped laboratories. Both health centers are located in high-CL-endemicity regions characterized by impoverished populations and limited access to appropriate health care. The results obtained by the test for the two CL-suspected patients at the health center in San Martin de Pangoa were perfectly concordant with the results obtained by conventional diagnosis and were confirmed by kDNA PCR. At the health center in Kiteni, the Leishmania OligoC-TesT results were in agreement with conventional diagnosis for 10 of the 13 CL-suspected patients. One patient showed parasites during parasitological detection but a negative result with the Leishmania OligoC-TesT.
This study demonstrates the high accuracy of the Leishmania OligoC-TesT for noninvasive lesion scrapings. A major advantage of the test compared to parasite culture and LST is the time reduction. OligoC-TesT results are available in 5 h after the initial specimen is obtained, while culture may require up to 2 to 4 weeks and LST 2 days. Diagnosis within 1 day is important since patients often live far from the health centers and extra costs due to transport or lodging should be avoided. Furthermore, the OligoC-TesT can be produced in a kit format including a quality-controlled PCR mixture and dipsticks. This will likely enhance the integration of the PCR technique in leishmaniasis reference centers as well as in peripheral health centers in areas of endemicity. The development of a PCR dipstick which is able to discriminate among different New World Leishmania species would be most welcome (16), as disease progression and treatment response are species specific (1, 19).
Acknowledgments
This study received financial support from the Directorate-General for Development Cooperation of the Belgian Government (framework agreement 03, project 95502) and from the European Community through the project “Control strategies for visceral leishmaniasis (VL) and mucocutaneous leishmaniasis (MCL) in South America: applications of molecular epidemiology” (contract INCO-CT2005-015407).
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Arévalo, J., L. Ramirez, V. Adaui, M. Zimic, G. Tulliano, C. Miranda-Verástegui, M. Lazo, R. Loayza-Muro, S. De Doncker, A. Maurer, F. Chappuis, J. C. Dujardin, and A. Llanos-Cuentas. 2007. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J. Infect. Dis. 1951846-1851. [DOI] [PubMed] [Google Scholar]
- 2.Bensoussan, E., A. Nasereddin, F. Jonas, L. F. Schnur, and C. L. Jaffe. 2006. Comparison of PCR assays for diagnostics of cutaneous leishmaniasis. J. Clin. Microbiol. 441435-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boggild, A. K., C. Miranda-Verástegui, D. Espinosa, J. Arévalo, V. Adaui, G. Tulliano, A. Llanos-Cuentas, and D. E. Low. 2007. Evaluation of a microculture method for the isolation of Leishmania parasites from cutaneous lesions in Peru. J. Clin. Microbiol. 453680-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvopina, M., R. X. Armijos, and Y. Hashiguchi. 2004. Epidemiology of leishmaniasis in Ecuador: current status of knowledge. Mem. Inst. Oswaldo Cruz 99663-672. [DOI] [PubMed] [Google Scholar]
- 5.Chulay, J. D., and A. D. Bryceson. 1983. Quantitation of amastigotes of Leishmania donovani in smears of splenic aspirates from patients with visceral leishmaniasis. Am. J. Trop. Med. Hyg. 32475-479. [DOI] [PubMed] [Google Scholar]
- 6.Deborggraeve, S., F. Claes, T. Laurent, P. Mertens, T. Leclipteux, J. C. Dujardin, P. Herdewijn, and P. Büscher. 2006. Molecular dipstick test for diagnosis of sleeping sickness. J. Clin. Microbiol. 442884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deborggraeve, S., T. Laurent, D. Espinosa, G. Van der Auwera, M. Mbuchi, M. Wasunna, S. El-Safi, A. A. Al-Basheer, J. Arévalo, C. Miranda-Verástegui, T. Leclipteux, P. Mertens, J. C. Dujardin, P. Herdewijn, and P. Büscher. 2008. A simplified and standardized polymerase chain reaction format for the diagnosis of leishmaniasis. J. Infect. Dis. 1981565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Oliveira, C. I., A. Báfica, F. Oliveira, C. B. F. Favali, T. Correa, L. A. R. Freitas, E. Nascimento, J. M. Costa, and A. Barral. 2003. Clinical utility of polymerase chain reaction-based detection of Leishmania in the diagnosis of American cutaneous leishmaniasis. Clin. Infect. Dis. 37e149-e153. [DOI] [PubMed] [Google Scholar]
- 9.Desjeux, P. 2004. Disease watch focus: leishmaniasis. Nat. Rev. Microbiol. 2692-693. [DOI] [PubMed] [Google Scholar]
- 10.Desjeux, P. 2001. The increase of risk factors for leishmaniasis worldwide. Trans. R. Soc. Trop. Med. Hyg. 95239-243. [DOI] [PubMed] [Google Scholar]
- 11.Dirección General de Epidemiología. 2002. Anuario del sistema de vigilancia epidemiológica. Ministerio de Salud, Lima, Peru.
- 12.Faber, W. R., L. Oskam, T. Van Gool, N. C. M. Kroon, K. J. Knegt-Junk, H. Hofwegen, A. C. van der Wal, and P. A. Kager. 2003. Value of diagnostic techniques for cutaneous leishmaniasis. J. Am. Acad. Dermatol. 4970-74. [DOI] [PubMed] [Google Scholar]
- 13.García, A. L., R. Parrado, S. De Doncker, H. Bermudez, and J. C. Dujardin. 2007. American tegumentary leishmaniasis: direct species identification of Leishmania in non-invasive clinical samples. Trans. R. Soc. Trop. Med. Hyg. 101368-371. [DOI] [PubMed] [Google Scholar]
- 14.Hepburn, N. C. 2003. Cutaneous leishmaniasis: an overview. J. Postgrad. Med. 4950-54. [DOI] [PubMed] [Google Scholar]
- 15.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 3541191-1199. [DOI] [PubMed] [Google Scholar]
- 16.Laurent, T., G. Van der Auwera, M. Hide, P. Mertens, W. Quispe-Tintaya, S. Deborggraeve, S. De Doncker, T. Leclipteux, A. L. Bañuls, P. Büscher, and J. C. Dujardin. 2009. Identification of Old World Leishmania spp. by specific polymerase chain reaction amplification of cysteine proteinase B genes and rapid dipstick detection. Diagn. Microbiol. Infect. Dis. 63173-181. [DOI] [PubMed] [Google Scholar]
- 17.López, M., C. Orrego, M. Cangalaya, R. Inga, and J. Arévalo. 1993. Diagnosis of Leishmania via the polymerase chain reaction a simplified procedure for field work. Am. J. Trop. Med. Hyg. 49348-356. [DOI] [PubMed] [Google Scholar]
- 18.Lucas, C. M., E. D. Franke, M. I. Cachay, A. Tejada, M. E. Cruz, R. D. Kreutzer, D. C. Barker, S. H. McCann, and D. M. Watts. 1998. Geographic distribution and clinical description of leishmaniasis cases in Peru. Am. J. Trop. Med. Hyg. 59312-317. [DOI] [PubMed] [Google Scholar]
- 19.Llanos-Cuentas, A., G. Tulliano, R. Araujo-Castillo, C. Miranda-Verastegui, G. Santamaria-Castrellon, L. Ramírez, M. Lazo, S. De Doncker, M. Boelaert, J. Robays, J. C. Dujardin, J. Arévalo, and F. Chappuis. 2008. Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Peru. Clin. Infect. Dis. 46223-231. [DOI] [PubMed] [Google Scholar]
- 20.Marfurt, J., A. Nasereddin, I. Niederwieser, C. L. Jaffe, H. P. Beck, and I. Felger. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 413147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques, M. J., A. C. Volpini, G. L. L. Machado-Coelho, J. Machado-Pinto, C. A. da Costa, W. Mayrink, O. Genaro, and A. J. Romanha. 2006. Comparison of polymerase chain reaction with other laboratory methods for diagnosis of American cutaneous leishmaniasis: diagnosis of cutaneous leishmaniasis by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 5437-43. [DOI] [PubMed] [Google Scholar]
- 22.Ministerio de Salud del Perú. 2000. Leishmaniasis. Módulos técnicos serie documentos monográficos. Ministerio de Salud, Lima, Peru.
- 23.Murray, H. W., J. D. Berman, C. R. Davies, and N. Saravia. 2005. Advances in leishmaniasis. Lancet 3661561-1577. [DOI] [PubMed] [Google Scholar]
- 24.Reithinger, R., and J. C. Dujardin. 2007. Molecular diagnosis of leishmaniasis: current status and future applications. J. Clin. Microbiol. 4521-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Barraquer, I., R. Góngora, M. Prager, R. Pacheco, L. M. Montero, A. Navas, C. Ferro, M. C. Miranda, and N. G. Saravia. 2008. Etiologic agent of an epidemic of cutaneous leishmaniasis in Tolima, Colombia. Am. J. Trop. Med. Hyg. 78276-282. [PubMed] [Google Scholar]
- 26.Segura, E. L., N. Juan, A. L. M. Piquin, C. A. Cuba Cuba, L. Abramo Orrego, D. McMahon-Pratt, E. E. Montamat, H. Momen, and G. Grimaldi, Jr. 2000. Molecular and biologic characterization of Leishmania parasites implicated in an epidemic outbreak in northwestern Argentina. Parasitol. Res. 86504-508. [DOI] [PubMed] [Google Scholar]
- 27.Singh, S. J. 2003. Recent advances in the diagnosis of leishmaniasis. J. Postgrad. Med. 4955-60. [DOI] [PubMed] [Google Scholar]
- 28.van der Meide, W. F., G. J. Schoone, W. R. Faber, J. E. Zeegelaar, H. J. C. de Vries, Y. Özbel, R. F. M. Lai A Fat, L. I. A. R. C. Coelho, M. Kassi, and H. D. F. H. Schallig. 2005. Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples. J. Clin. Microbiol. 435560-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Meide, W. F., I. Peekel, P. P. A. M. van Thiel, H. D. F. H. Schallig, H. J. C. de Vries, J. E. Zeegelaar, and W. R. Faber. 2008. Treatment assessment by monitoring parasite load in skin biopsies from patients with cutaneous leishmaniasis, using quantitative nucleic acid sequence-based amplification. Clin. Exp. Dermatol. 33394-399. [DOI] [PubMed] [Google Scholar]
- 30.Weigle, K. A., L. A. Labrada, C. Lozano, C. Santrich, and D. C. Barker. 2002. PCR-based diagnosis of acute and chronic cutaneous leishmaniasis caused by Leishmania (Viannia). J. Clin. Microbiol. 40601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weigle, K. A., L. Valderrama, A. L. Arias, C. Santrich, and N. Saravia. 1991. Leishmanin skin test standarization and evaluation of safety, dose, storage, longevity of reaction and sensitization. Am. J. Trop. Med. Hyg. 44260-271. [DOI] [PubMed] [Google Scholar]
- 32.Wilson, E. B. 1927. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 22209-212. [Google Scholar]