Abstract
Avian pathogenic Escherichia coli (APEC) is an important respiratory pathogen of poultry. Various virulence factors are responsible for determining the pathogenicity of these strains, and it is commonly believed they are encoded on large plasmids the strains carry. This study examined a series of strains, the pathogenicity of which had previously been determined by aerosol exposure, for possession of large plasmids and found all isolates carried at least one large plasmid, regardless of the level of virulence. Virulence-associated genes carried on these plasmids were also examined, and it was shown that highly virulent strains carried at least four virulence-associated genes on their largest plasmid. Two of the virulence-associated genes were shown to be chromosomally located in a strain of intermediate virulence, while no virulence-associated genes were carried by the low-virulence strain. The organization of the virulence-associated genes was shown to be highly conserved among APEC isolates of high virulence, supporting the concept of a conserved portion of the putative virulence region that contributes to the pathogenicity of APEC strains.
Avian pathogenic Escherichia coli (APEC) strains cause respiratory disease and septicemia in poultry and are economically important worldwide, causing significant mortality (13). The carriage of large plasmids is considered characteristic of APEC isolates (8), and pathogenicity is thought to be determined by virulence-associated factors encoded by them (15). These factors include serum resistance, encoded by the iss gene (14), temperature-sensitive hemagglutination, encoded by tsh (10), adhesins, the production of colicin V (ColV) and the possession of iron-scavenging mechanisms, such as aerobactin production (encoded by the iucABCD operon), and the more recently identified putative iron transport system encoded by the etsABC operon (18).
Another iron acquisition system found in APEC utilizes salmochelin, a catecholate siderophore. The chromosomal iroA gene cluster that encodes this system was first found in Salmonella enterica (2) and is absent from the corresponding region of the E. coli chromosome (32), although it has been found on a transmissible plasmid from a uropathogenic E. coli isolate (34). The iroA gene cluster has been found on multiple APEC virulence plasmids (9, 17, 18, 37), and deletion studies have shown that the iroA gene cluster is required for full virulence (9).
A further iron transport system, designated the sitABCD system, was first identified on a pathogenicity island in Salmonella enterica serovar Typhimurium (39), and it has been shown that sitABCD is required for full virulence of Salmonella serovar Typhimurium (16). Genomic subtraction identified the plasmid-located sitA gene from the sitABCD operon as unique to an APEC strain (32), and the sitA gene was found to be more prevalent in APEC than in commensal E. coli (18, 29, 32).
The sitABCD operon occurs on APEC virulence plasmids (17, 18, 30, 37), but a sitABCD deletion mutant was still pathogenic for birds, suggesting that other iron transport systems are able to compensate for the loss of sitABCD (30).
The carriage of ColV plasmids has previously been thought to be essential for virulence (3, 33, 38). However, other studies have suggested it is not the presence of the ColV gene itself but other genes that these plasmids carry that are responsible for virulence (28, 35). The well-characterized APEC virulence plasmids pAPEC-O2-ColV (18) and pAPEC-1 (9) encode ColV, while carriage of the Australian APEC virulence plasmid pVM01 does not confer production of ColV (12). Despite various ColV statuses, all three of these virulence plasmids are F-type plasmids, and hence this is potentially another way to characterize APEC virulence plasmids.
SopA and SopB, which have similarity to the ParA and ParB proteins of the P1 plasmid, are thought to be essential for F-plasmid partitioning (22, 24). Detection of the genes of the sopABC locus could thus indicate the presence of a putative virulence plasmid.
Strain E3 is an O-nontypeable:H28 APEC field isolate (11) that carries the 151-kb virulence plasmid pVM01 (12), which contains a virulence region with the virulence-associated genes iucA, tsh, iss, iroN, and sitA, as well as hlyF, ompT, and the etsABC operon (37). The arrangement of the virulence-associated genes around pVM01 (37) is similar to that in the plasmids pAPEC-O2-ColV from APEC strain O2 (18), pAPEC-O1-ColBM from APEC strain O1 (17), and pAPEC-1 from APEC strain χ7122 (23). Identifying a specific region that is conserved in highly virulent APEC strains will facilitate diagnosis of colibacillosis by differentiation of pathogenic strains from commensal E. coli and will also enable surveillance for pathogenic isolates in the environment of poultry.
This study examined six E. coli strains, some of which were isolated from diseased birds and some of which were recovered from healthy birds (11, 36). The pathogenicity of these strains has been determined using aerosol exposure (11, 36), making this the largest known collection of APEC strains fulfilling Koch's postulates. The series of strains includes the highly virulent strains E3, E30, and E956 and the less-virulent strains E133, E1043, and E1292. The presence of the virulence-associated genes iucA, tsh, and iss in these strains has previously been elucidated by PCR amplification (36). However, while previous studies have found many of these virulence factors to be encoded by APEC strains associated with disease (29) and have suggested that they are encoded on virulence plasmids (18), they have not conclusively determined whether they are encoded on virulence plasmids or are chromosomally encoded. Similarly, although previous studies suggest that these virulence-associated genes are consistently present in isolates from diseased birds (1, 6, 18, 21, 26, 29), no study has yet determined if these genes are consistently associated with each other.
The aim of this study was to examine a series of strains of known pathogenicities for the possession of large plasmids and to determine if known virulence-associated genes from the putative virulence region were carried on them. The second objective was to investigate any association between the virulence-associated genes.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study were the field isolates E3, E30, E133, E956, and E1043 (11), E1292 (36), and derivatives of strain E3 with different combinations of plasmids (12). The pathogenicities of the strains were determined previously using aerosol exposure (11, 36). The plasmid profiles, virulence genes, and morbidity rates of birds exposed to the strains are shown in Table 1. All strains were grown in LB broth or on LB agar at 37°C overnight.
TABLE 1.
Plasmid profiles, morbidity rates, and virulence-associated genes of E. coli strains
| Strain | Plasmid(s) | Morbidity (%)a | Virulence-associated genesb |
|---|---|---|---|
| E956 | 01 | 88 | iucA, iss, sitA, iroB, sopAB |
| E3 | pVM01, pVM02, pVM03, pVM04 | 75 | iucA, tsh, iss, sitA, iroB, sopAB |
| E30 | 01, 02 | 59 | iucA, iss, sitA, iroB |
| E1292 | 01, 02 | 31 | iucA*, sitA*, sopAB |
| E133 | 01, 02 | 25 | iss, sitA, iroB, sopAB |
| E1043 | 01, 02 | 9 | None detected |
| E3/2.4 | pVM02, pVM03 | 12 | None detected |
Proportion of aerosol-infected birds that developed air sac lesions as previously determined (12, 36).
Identified by PCR amplification. Virulence-associated genes hybridized to the underlined plasmids except those genes indicated by an asterisk, which did not hybridize to any of the isolated plasmids but did hybridize to digested genomic DNA.
Preparation of plasmid DNA.
Plasmid DNA was prepared using the Qiagen Plasmid Midi kit as recommended by the manufacturer. The modifications recommended to obtain higher yields of low-copy-number plasmids were used. Plasmids were separated by pulsed-field gel electrophoresis (PFGE).
Preparation of genomic DNA.
Genomic DNA from avian E. coli strains was prepared by phenol-chloroform extraction (31).
Restriction endonuclease digestion of plasmid and genomic DNA.
Digestion of plasmid and genomic DNA from avian E. coli strains with HindIII was carried out according to the manufacturer's instructions (New England Biolabs). The digested DNA was separated by PFGE.
PFGE.
PFGE was carried out using a Chef-DR III system (Bio-Rad). Plasmids were separated in 1.0% (wt/vol) DNA-grade agarose (Progen) in 0.5× TBE buffer (1× TBE is 90 mM Tris, 90 mM boric acid, and 2 mM EDTA). A lambda ladder (Bio-Rad) and HindIII-digested lambda DNA were used as molecular weight standards. Electrophoresis was carried out at 6 V/cm with an included angle of 120° for 20 h with a switch time of 1 to 20 s to separate plasmids. Electrophoresis of smaller restriction endonuclease digestion fragments, of both plasmid and genomic DNA, was carried out for 3 h at 9 V/cm with a switch time of 0.1 s. Gels were stained in 1× TBE containing 0.5 mg ethidium bromide ml−1 for 30 min and then destained in distilled water for 30 min. DNA was visualized by UV transillumination.
PCR.
The virulence-associated genes sitA, iroB, and sopAB were amplified by PCR using the primers listed in Table 2. Amplification was performed using 1.25 U of Taq DNA polymerase (Promega) in a 25-μl reaction mix containing 1× Mg-free buffer, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, and 1 μM of both the forward and reverse primers. PCRs were inoculated with colonies picked from freshly streaked plates with a straight wire. The reaction mixtures were incubated at 95°C for 4 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 7 min, using a Bio-Rad iCycler PCR thermocycler. PCR products were separated in 2% (wt/vol) agarose gels containing 0.5× TPE (1× TPE is 30 mM NaH2PO4·2H2O, 36 mM Tris, and 1 mM EDTA) buffer and 0.1 μg of ethidium bromide ml−1 by electrophoresis at 4.5 V/cm for 1 to 2 h. Hyperladder IV (Bioline) was used as a molecular weight marker.
TABLE 2.
Primers used for PCR amplification of genes
| Primer | Gene specificity (expected size [bp]) | Sequences (5′ → 3′) | Accession no.a |
|---|---|---|---|
| SitA F | sitA (252) | TGCCTCGCCCTCACCTGCTC | AY126440 |
| Sita R | AGCGTTGGAACCACAATTCCA | ||
| IroB F | iroB (272) | CGGCCTGACCTCATCATCTA | AY545598 |
| IroB R | ATACGGGACGTACTGCATCG | ||
| SopAB F | sopAB (589) | ATATGCTTCGTGATCTGCTCA | NC004998 |
| SopAB R | CGGTAAGCAGTTCCTGGTCAC |
GenBank accession number for sequence used for primer design.
Southern blot hybridization.
Following PFGE of plasmids or digested plasmid and genomic DNA, DNA was transferred from the agarose gels to a nylon membrane (Hybond-N+; Amersham) using the method described by Sambrook et al. (31). Hybridization probes were constructed as described previously (36). Briefly, 25 ng of gel-purified PCR product derived from the virulence-associated genes (iss, iucA, tsh, sitA, iroB, or sopAB) in pVM01::TnphoA DNA was labeled with [α-32P]dATP (Perkin Elmer) using a random priming DNA labeling kit (Roche). Prehybridization and hybridization were carried out in Church buffer (0.5 M Na2HPO4 [pH 7.4], 7% sodium dodecyl sulfate, 1 mM EDTA, 1% bovine serum albumin) (4) overnight at 58°C. Membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate at 58°C three times for 20 min each and exposed to Kodak BioMax MS film at −80°C.
Long-range PCR amplification.
Regions between the virulence-associated genes were amplified using PCR and the primers listed in Table 3. For regions less than 5 kb in size, amplification was performed using 0.5 U Platinum Taq high-fidelity DNA polymerase (Invitrogen) in a 50-μl reaction mixture containing 1× high-fidelity PCR buffer, 2 mM MgSO4, 100 μM of each deoxynucleoside triphosphate, and 500 nM of each primer. The PCR mixture was incubated in a Bio-Rad iCycler PCR thermocycler at 94°C for 2 min, followed by 25 cycles at 94°C for 30 s, 55°C for 30 s, and 68°C for 45 s, and then finally at 68°C for 7 min. The Expand long-template PCR system (Roche) was used to amplify regions predicted to be larger than 5 kb. PCR amplifications were performed according to the manufacturer's instructions using a Bio-Rad iCycler PCR thermocycler. Long-range PCR products were visualized by agarose gel electrophoresis.
TABLE 3.
Primers used for PCR amplification of regions between virulence-associated genes
| Gene specificity (expected size [kb]) | Primer | Sequence (5′ → 3′) | PCR-positive strainsa |
|---|---|---|---|
| iucA → sitD (2.8) | iuc300r | ATAATAAGCCGGGAGAGAGTG | E3, E956 |
| shigsit3700f | TTCTGGGGTTACCTCTGGCT | ||
| sitA → repA (3.5) | shigsitAr | AGCGTTGGAACCACAATTCCA | E3, E133, E956 |
| sitA 1 fwd | ATCATTATCCATATCCAGGCG | ||
| etsC → iss (8.3) | sitA 27 rev | AGAGTGGCAGTTGCGTGAGGC | E3, E133 |
| iss 1 rev | TCCTGTAATAAGCATTGCCAG | ||
| iss → iroB (4.3) | iss fwd | GTGGCGAAAACTAGTAAAACAGC | E3, E30, E133, E956 |
| iroB rev | ATACGGGACGTACTGCATCG | ||
| iroN → cvaA (8) | iroN3 fwd | AGGCGCTATTGCCGGTAAGAT | E3, E30, E133, E956 |
| cvaA rev | TGAGTCTGAATCTGACTCTCC |
PCR products amplified from underlined strains differed in size from that expected for pVM01 and obtained from E3.
Restriction endonuclease digestion of long-range PCR products.
Restriction endonuclease digestion of long-range PCR products with HindIII was carried out according to the manufacturer's instructions (New England Biolabs). The digested DNA was separated by agarose gel electrophoresis.
RESULTS
PCR amplification of virulence genes.
The presence of the virulence-associated genes iucA, tsh, and iss in the strains has been elucidated previously by PCR amplification (36). Each of the strains in the series was examined for the presence of the virulence-associated genes sitA, iroB, and sopAB by PCR amplification. The sitA gene from the sitABCD operon was amplified from strains E3, E30, E133, E956, and E1292. The iroB gene of the iroA gene cluster was amplified from strains E3, E30, E133, and E956, whereas sopAB was amplified from strains E3, E133, E956, and E1292. The virulence-associated genes sitA, iroB, and sopAB were amplified from strain E3 and from derivatives that carried pVM01 but not from the E3 derivatives that lacked pVM01. This indicated that the sitA, iroB, and sopAB genes were carried on the virulence plasmid pVM01 and not the chromosome of strain E3.
Isolation of plasmids from strains.
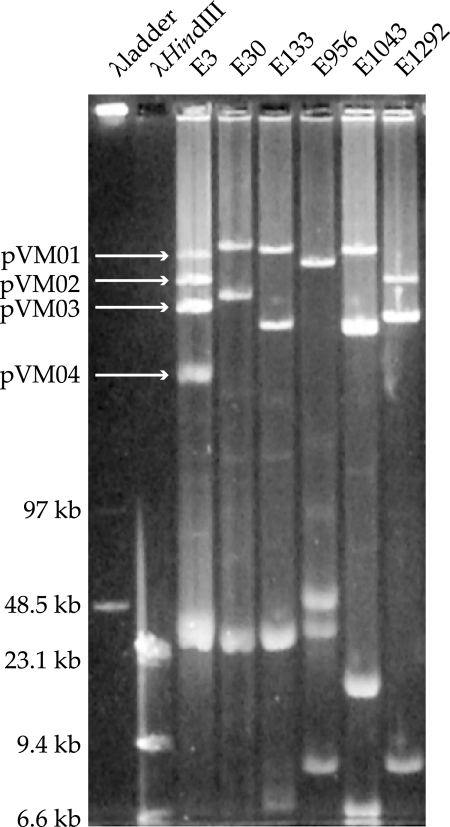
Plasmid DNA was prepared from each of the strains and separated by PFGE. Two large plasmids were identified in all of the strains except E956, which carried only one large plasmid. These large plasmids were all of sizes similar to those of the three largest plasmids of strain E3 (pVM01, pVM02, and pVM03) (Fig. 1).
FIG. 1.
Plasmids purified from six strains of known pathogenicity. All strains contained at least one large plasmid, irrespective of virulence. The four large plasmids known to be carried by strain E3 are indicated (pVM01 to pVM04). Smaller plasmids (<48.5 kb) can also be seen in all isolates.
Hybridization of virulence-associated genes to plasmids of strains.
A Southern blot of the separated plasmids (Fig. 1) was probed with each of the six virulence-associated genes to determine whether they were carried by any of the plasmids. Since pVM01 was known to carry all six virulence-associated genes, it was used as an internal control.
As expected, all six virulence-associated genes hybridized to the largest plasmid, pVM01, in strain E3. None of these genes hybridized to any of the other plasmids of strain E3. The virulence-associated genes iucA, iss, sitA, and iroB hybridized to the largest plasmid of strain E30, while iss, sitA, iroB, and sopAB hybridized to the largest plasmid of strain E133. All the virulence-associated genes except tsh hybridized to the largest plasmid of strain E956, but none of the virulence-associated genes hybridized to plasmids from strain E1043. Only the sopAB probe hybridized to a plasmid from strain E1292, and this was to the second-largest plasmid. PCR amplification has shown that E1292 contained the virulence-associated genes iucA (36) and sitA, but these genes did not hybridize to either of its plasmids. This suggested that these virulence-associated genes may be chromosomally located in strain E1292.
Chromosomally located virulence genes.
To establish whether the virulence-associated genes iucA and sitA were chromosomally located in E1292, genomic DNA preparations from the six strains, along with plasmid DNA, were digested with HindIII and the resultant fragments were separated by PFGE. Southern blot hybridization showed that the iucA and sitA probes hybridized to the genomic DNA of E1292 but not to the digested plasmid DNA of the same strain, proving that these genes were carried on the chromosome in strain E1292.
Arrangement of virulence-associated genes.
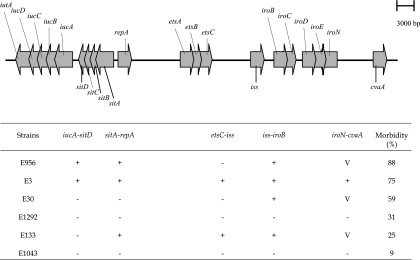
The arrangement of the virulence genes in pAPEC-O2-ColV (18), pAPEC-O1-ColBM (17), pAPEC-1 (23), and pVM01 (37) appears to be quite similar, suggesting that the arrangement of the virulence genes in virulence plasmids of APEC may be somewhat conserved. This was examined in the strains using PCR to amplify regions between the virulence-associated genes. Strain E3, carrying pVM01, was used as a control for all PCRs. The orientation and location of the genes in pVM01 and the expected size of the PCR products are shown in Table 3.
The sizes of PCR products amplified from strain E3 were as predicted. The 2.8-kb iucA-sitD PCR product was amplified only from strains E3 and E956. The 8.3-kb etsC-iss PCR product was amplified only from E3 and E133. The 3.5-kb sitA-repA PCR product was amplified from strains E3, E133, and E956. The 4.3-kb iss-iroB PCR product was amplified from E3 and from three other strains, E30, E133, and E956. However, PCR products of various and different sizes from those of products from E3 were amplified from strains E30, E133, and E956 in the iroN-cvaA PCR assay. Products of 4.5 kb were amplified from both E30 and E956, while two PCR products were amplified from E133 using this assay, one of approximately 9 kb and the other of 3 kb. The product from E3 was 8 kb. These results are summarized in Fig. 2.
FIG. 2.
Schematic diagram of the virulence-associated genes as found in pVM01, drawn to scale. The table lists the strains of known pathogenicity in order of virulence. +, amplification of a PCR product; −, no amplification; V, amplification of a PCR product differing in size from that predicted from the sequence of pVM01 and seen in E3.
Restriction endonuclease digestion of PCR products.
PCR products were subjected to restriction endonuclease digestion with HindIII, and the digested DNA was separated by continuous-field agarose gel electrophoresis. PCR products of the same size from different strains produced fragments of the same size after digestion with HindIII. However, the iroN-cvaA PCR products, which varied in size, resulted in fragments of differing sizes after digestion. Digestion of the iroN-cvaA PCR product from E3 resulted in three fragments of 1,231 bp, 1,743 bp, and 5,051 bp, as predicted from the pVM01 DNA sequence. Digestion of the PCR products amplified from E30 and E956 resulted in two fragments, one of 3,500 bp and the other of 1,231 bp, similar in size to one of those obtained from E3. The products from E133 yielded three restriction endonuclease cleavage products, the largest being 6,500 bp and the smallest 1,743 bp, similar to one of those generated from E3, and the third 3,500 bp, similar to that obtained from products from E30 and E956.
DISCUSSION
The genes iss and tsh and the aerobactin operon have been associated with virulence in APEC for many years (10, 14), and it has also been suggested that virulence of APEC is mediated by the large plasmids they carry (8, 12, 15). Recent studies have investigated the genes carried on these virulence plasmids (17, 18, 36). This is the first study to look beyond PCR amplification to identify genes carried by both virulent and avirulent strains of E. coli isolated from birds.
All the strains except the nonpathogenic strain E1043 were found to carry the virulence-associated gene sitA, suggesting that the sitABCD operon may play a key role in virulence. The gene iroB was detected in E3, E30, E133, and E956. These were the same intermediately or highly virulent strains that were found to carry the iss gene, suggesting that iroB may also play a significant role in virulence. The sopAB genes were amplified only from strains E3, E133, E956, and E1292, suggesting that strains E30 and E1043 did not carry an F-type plasmid.
All six strains, which were isolated from both diseased and healthy birds and ranged from nonpathogenic to highly pathogenic, were found to carry at least one large plasmid. This is consistent with the published observations of Doetkott et al. (8), who found that possession of large plasmids did not appear to correlate with capacity to cause disease.
All the virulence-associated genes were found to be plasmid borne, with the exception of the iucA and sitA genes in E1292. It is possible that the chromosomally located genes iucA and sitA could be located on a pathogenicity island in strain E1292. Pathogenicity islands have been found in the APEC strain Ec222 (25) and APEC O1 (20), and genes encoding the synthesis and transport of aerobactin have previously been found on an island in a strain of Shigella boydii (27). A previous study found genes for aerobactin production to be chromosomally located in 3 E. coli isolates and plasmid carried in another 20 isolates (5), while in APEC strain O1, the sitABCD operon occurs on pAPEC-O1-ColBM and also in its chromosome (17).
The most prevalent virulence genes, iucA, sitA, iss, and iroB, were carried on the largest plasmid in the three strains of highest virulence. This was also seen in strain E133, except that this lower-virulence strain did not contain the iucA gene. Since the avirulent strain E1043 did not carry any of these virulence-associated genes, the findings reinforce previous suggestions of the contribution of these virulence-associated genes to the pathogenicity of APEC strains (32, 36).
Isolates that contained sopAB also carried the virulence-associated gene sitA, but not all isolates carrying sitA were found to carry sopAB. F-type plasmids, as determined by possession of sopAB, that also bear other virulence-associated genes were carried by the two most virulent APEC strains.
The next most virulent strain, E30, had most of the virulence-associated genes but did not contain sopAB and thus presumably did not carry an F-type plasmid. This suggests that identification of sopAB alone is not a reliable indicator of carriage of a virulence plasmid and hence it should not be considered a virulence-associated gene.
All the strains previously found to carry the virulence-associated gene iss were also found to contain iroB, suggesting these two putative virulence factors may be closely associated with each other. Further examination of this association by amplification of the region between iss and iroB generated a 4.3-kb product from strain E3. The same-size product was consistently amplified from all strains carrying these two genes, providing further evidence that the genes are closely associated with each other.
APEC virulence plasmids appear to be similar throughout the world, with similar gene arrangements found in the putative virulence region of three APEC plasmids from the United States (17, 18, 23) and in the Australian APEC plasmid pVM01 (37). Comparing the putative virulence regions in six APEC strains of known pathogenicity by PCR amplification showed that the arrangements of many of these genes are conserved in different strains.
The 3.5-kb sitA-repA PCR product amplified from pVM01 was the only region examined in this study that would not be amplified from pAPEC-O2-ColV. In this plasmid (18), the IS1-flanked region containing the aerobactin and sitABCD operons was in the opposite orientation to that seen in pVM01, and thus, the gene adjacent to repA is iutA. However, as observed in pVM01 (37), sitA lies adjacent to repA in the other fully sequenced APEC virulence plasmid, pAPEC-O1-ColBM (17). The presence of inverted IS1 elements bracketing this region in both pAPEC-O2-ColV and pAPEC-O1-ColBM offers an explanation for the differing orientations of this region within the virulence plasmids.
APEC strain E30 carried both iucA and sitA on its largest plasmid, but no PCR product was amplified using the iucA-sitD primer pair. This could be explained if the sitABCD operon is not complete or if the two operons are in an orientation different from that seen in pVM01, although the aerobactin and sitABCD operons are consistently found in the same orientation as in the sequenced pVM01 (37) in both pAPEC-O1-ColBM (17) and pAPEC-O2-ColV (18). Similarly, although it contained sitA, the sitA-repA PCR product could not be amplified from E30. Since sopAB was not detected in E30, its largest plasmid may not be an F-type plasmid, explaining the absence of the RepFIB replicon gene.
Both E30 and E956 carried the iss gene on their largest plasmid, but the etsC-iss product could not be amplified from either strain. This could be accounted for if both strains lack the putative ABC transport system encoded by etsABC or if they are found on the plasmids in an orientation different from that seen in pVM01 and pAPEC-O2-ColV. This is seen in pAPEC-O1-ColBM, in which the etsABC genes are adjacent to the RepFIB gene repA (17).
Apart from the 8.3-kb etsC-iss PCR product, all the regions amplified from E3 were also amplified from the highly virulent E956 strain, indicating that the arrangement of virulence-associated genes on the largest plasmid of E956 is very similar to that on pVM01. Similarly, the arrangement of the virulence-associated genes in strain E133 was found to be very similar to that on pVM01, except that strain E133 lacked the virulence-associated gene iucA, which may account for its lower pathogenicity. This may be indicative of the evolution of the virulence region found on APEC plasmids by addition or deletion of specific segments bracketed by IS elements.
Although the variable portion of the putative virulence region surrounding the tsh gene is found in pAPEC-O2-ColV, pAPEC-O1-ColBM, pAPEC-1, and pVM01, tsh was not found in the other five strains (36).
The 8-kb PCR product amplified from E3 in the iroN-cvaA PCR was not obtained from any of the other strains, in direct contrast to a previous suggestion that the putative virulence region is conserved through to the cvaB gene (18), suggesting it may be conserved only as far as the iroA gene cluster. However, the products from the three most virulent strains yielded a restriction endonuclease cleavage fragment of approximately 1,231 bp. This suggests that although the sequence between iroN and cvaA varied between the strains, possession of this 1,231-bp fragment may be associated with virulence. The corresponding nucleotide sequence was identified in pVM01 and was found to encode the end of the IroN protein and a hypothetical protein (CAC43413). Interestingly, this hypothetical protein is also found in uropathogenic E. coli strain 536 (7), providing further evidence of the similarity between virulence determinants of APEC and uropathogenic E. coli.
Further evidence that the virulence-associated regions are conserved between APEC isolates was provided by the restriction endonuclease digestion patterns of the PCR products. With the exception of the iroN-cvaA PCR products, which varied in size, the PCR products, ranging from 2.8 to 8.3 kb, from six field isolates were found to have the same restriction endonuclease cleavage sites.
Furthermore, a recent study by Johnson et al. has shown they can predict virulence of APEC strains by the presence of five genes, iroN, ompT, hlyF, iss, and iutA (19), all of which are present in the conserved virulence region, thereby validating the current findings.
The purported conserved virulence region may be similar to pathogenicity islands, and mobility of this region would be consistent with its presence not only in F-type plasmids but in other types of plasmids, as was seen in strain E30, and in the chromosome, as seen in strain E1292.
Thus, it appears that in highly virulent APEC strains, the majority of the virulence-associated genes are present and are carried on a single plasmid and that in strains of lower virulence some of the virulence-associated genes may be present and may be either plasmid or chromosomally carried, while in avirulent strains the virulence-associated genes are absent. When present on these large virulence plasmids, the virulence-associated genes seem to be closely associated with each other and the sequences surrounding them appear to be well conserved in different APEC strains, even those from geographically distinct regions. The findings of this study support the concept of the “conserved portion of the putative virulence region” derived from gene prevalence studies of APEC isolates (18) and suggest a common ancestor for these virulence plasmids in APEC.
Acknowledgments
This work was funded by the Australian Poultry CRC.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Altekruse, S. F., F. Elvinger, C. DebRoy, F. W. Pierson, J. D. Eifert, and N. Sriranganathan. 2002. Pathogenic and fecal Escherichia coli strains from turkeys in a commercial operation. Avian Dis. 46562-569. [DOI] [PubMed] [Google Scholar]
- 2.Baumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 1801446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, J. E., M. Blanco, A. Mora, and J. Blanco. 1997. Production of toxins (enterotoxins, verotoxins, and necrotoxins) and colicins by Escherichia coli strains isolated from septicemic and healthy chickens: relationship with in vivo pathogenicity. J. Clin. Microbiol. 352953-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 811991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna, B., L. Ranucci, P. A. Fradiani, M. Casalino, A. Calconi, and M. Nicoletti. 1992. Organization of aerobactin, hemolysin, and antibacterial resistance genes in lactose-negative Escherichia coli strains of serotype O4 isolated from children with diarrhea. Infect. Immun. 605224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delicato, E. R., B. G. de Brito, L. C. Gaziri, and M. C. Vidotto. 2003. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 9497-103. [DOI] [PubMed] [Google Scholar]
- 7.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 706365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doetkott, D. M., L. K. Nolan, C. W. Giddings, and D. L. Berryhill. 1996. Large plasmids of avian Escherichia coli isolates. Avian Dis. 40927-930. [PubMed] [Google Scholar]
- 9.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 684145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginns, C., G. Browning, M. Benham, and K. Whithear. 1998. Development and application of an aerosol challenge method for reproduction of avian colibacillosis. Avian Pathol. 27505-511. [DOI] [PubMed] [Google Scholar]
- 12.Ginns, C. A., M. L. Benham, L. M. Adams, K. G. Whithear, K. A. Bettelheim, B. S. Crabb, and G. F. Browning. 2000. Colonization of the respiratory tract by a virulent strain of avian Escherichia coli requires carriage of a conjugative plasmid. Infect. Immun. 681535-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, W. G. 1994. Diseases due to Escherichia coli in poultry, p. 237-259. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 14.Horne, S. M., S. J. Pfaff-McDonough, C. W. Giddings, and L. K. Nolan. 2000. Cloning and sequencing of the iss gene from a virulent avian Escherichia coli. Avian Dis. 44179-184. [PubMed] [Google Scholar]
- 15.Ike, K., K. Kawahara, H. Danbara, and K. Kume. 1992. Serum resistance and aerobactin iron uptake in avian Escherichia coli mediated by conjugative 100-megadalton plasmid. J. Vet. Med. Sci. 541091-1098. [DOI] [PubMed] [Google Scholar]
- 16.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 351146-1155. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 1885975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, T. J., Y. Wannemuehler, C. Doetkott, S. J. Johnson, S. C. Rosenberger, and L. K. Nolan. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli (APEC) virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 463987-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kariyawasam, S., T. J. Johnson, and L. K. Nolan. 2006. The pap operon of avian pathogenic Escherichia coli strain O1:K1 is located on a novel pathogenicity island. Infect. Immun. 74744-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knobl, T., M. R. Baccaro, A. M. Moreno, T. A. Gomes, M. A. Vieira, C. S. Ferreira, and A. J. Ferreira. 2001. Virulence properties of Escherichia coli isolated from ostriches with respiratory disease. Vet. Microbiol. 8371-80. [DOI] [PubMed] [Google Scholar]
- 22.Libante, V., L. Thion, and D. Lane. 2001. Role of the ATP-binding site of SopA protein in partition of the F plasmid. J. Mol. Biol. 314387-399. [DOI] [PubMed] [Google Scholar]
- 23.Mellata, M., J. W. Touchman, and R. Curtiss. 2009. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli chi7122 (O78:K80:H9). PLoS ONE 4e4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori, H., Y. Mori, C. Ichinose, H. Niki, T. Ogura, A. Kato, and S. Hiraga. 1989. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 26415535-15541. [PubMed] [Google Scholar]
- 25.Parreira, V. R., and C. L. Gyles. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 715087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaff-McDonough, S. J., S. M. Horne, C. W. Giddings, J. O. Ebert, C. Doetkott, M. H. Smith, and L. K. Nolan. 2000. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Dis. 4423-33. [PubMed] [Google Scholar]
- 27.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 1834176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quackenbush, R. L., and S. Falkow. 1979. Relationship between colicin V activity and virulence in Escherichia coli. Infect. Immun. 24562-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36241-256. [DOI] [PubMed] [Google Scholar]
- 30.Sabri, M., S. Leveille, and C. M. Dozois. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152745-758. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a Laboratory manual, 2nd ed., vol. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Schouler, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 1502973-2984. [DOI] [PubMed] [Google Scholar]
- 33.Smith, H. W., and M. B. Huggins. 1976. Further observations on the association of the colicin V plasmid of Escherichia coli with pathogenicity and with survival in the alimentary tract. J. Gen. Microbiol. 92335-350. [DOI] [PubMed] [Google Scholar]
- 34.Sorsa, L. J., S. Dufke, J. Heesemann, and S. Schubert. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 713285-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart, S. J., K. T. Greenwood, and R. K. Luke. 1980. Hydroxamate-mediated transport of iron controlled by ColV plasmids. J. Bacteriol. 14335-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tivendale, K. A., J. L. Allen, C. A. Ginns, B. S. Crabb, and G. F. Browning. 2004. Association of iss and iucA, but not tsh, with plasmid-mediated virulence of avian pathogenic Escherichia coli. Infect. Immun. 726554-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tivendale, K. A., A. H. Noormohammadi, J. L. Allen, and G. F. Browning. 2009. The conserved portion of the putative virulence region contributes to virulence of avian pathogenic Escherichia coli. Microbiology 155450-460. [DOI] [PubMed] [Google Scholar]
- 38.Vidotto, M. C., E. E. Muller, J. C. de Freitas, A. A. Alfieri, I. G. Guimaraes, and D. S. Santos. 1990. Virulence factors of avian Escherichia coli. Avian Dis. 34531-538. [PubMed] [Google Scholar]
- 39.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 671974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]