Abstract
A total of 101 food-borne and waterborne outbreaks that were caused by norovirus and that resulted in more than 4,100 cases of illness were reported to the Swedish Institute for Infectious Disease Control from January 2002 to December 2006. Sequence and epidemiological data for isolates from 73 outbreaks were analyzed. In contrast to health care-related outbreaks, no clear seasonality could be observed. Sequence analysis showed a high degree of genetic variation among the noroviruses detected. Genogroup II (GII) viruses were detected in 70% of the outbreaks, and of those strains, strains of GII.4 were the most prevalent and were detected in 25% of all outbreaks. The GII.4 variants detected in global outbreaks in health care settings during 2002, 2004, and 2006 were also found in the food-borne outbreaks. GI strains totally dominated as the cause of water-related (drinking and recreational water) outbreaks and were found in 12 of 13 outbreaks. In 14 outbreaks, there were discrepancies among the polymerase and capsid genotype results. In four outbreaks, the polymerase of the recombinant GII.b virus occurred together with the GII.1 or GII.3 capsids, while the GII.7 polymerase occurred together with the GII.6 and GII.7 capsids. Mixed infections were observed in six outbreaks; four of these were due to contaminated water, and two were due to imported frozen berries. Contaminated food and water serve as important reservoirs for noroviruses. The high degree of genetic diversity found among norovirus strains causing food-borne and waterborne infections stresses the importance of the use of broad reaction detection methods when such outbreaks are investigated.
Norovirus (NoV), a member of the Caliciviridae family, is the most common cause of nonbacterial gastroenteritis worldwide and affects humans in all age groups (7, 28). Virus transmission is primarily associated with person-to-person spread or the ingestion of contaminated food or water. Norovirus is highly infectious and causes outbreaks in communities, families, nursing homes, schools, hospitals, and cruise ships (3-6, 8, 11, 15, 19, 21, 23, 28, 41).
NoVs are genetically and antigenically diverse; and they have been classified into five genogroups, three of which (genogroup I [GI], GII, and GIV) are found in humans (1, 10, 16, 17, 43). Within genogroups, strains are further classified into genotypes. NoV has a single-stranded RNA genome of approximately 7,500 nucleotides that contains three open reading frames (ORFs). ORF1 encodes the RNA-dependent RNA polymerase, as well as other proteins; ORF2 encodes the major capsid protein (VP1); and ORF3 encodes a small capsid protein (VP2). Both ORF1 and ORF2 are used for diagnostic PCRs, as well as for genotyping by sequencing.
Exposure to multiple NoV variants, which may occur in contaminated food or water, increases the chance of recombination. The first naturally occurring human NoV recombinant was described by Jiang et al. (13). In Europe, a significant number of the outbreaks detected between 2000 and 2003 were caused by variants with recombinations at the ORF1-ORF2 junction (20, 33).
In the study described here, we investigated gastrointestinal outbreaks of suspected food-borne and waterborne origin reported to the Swedish Institute of Infectious Disease Control (SMI) from January 2002 to December 2006. All outbreaks were primarily analyzed by electron microscopy (EM), and the NoVs detected were further characterized by sequencing.
MATERIALS AND METHODS
Samples and outbreaks.
At SMI, the laboratory diagnosis of human NoV infections is based upon EM. Freshly collected fecal samples from individuals involved in food-borne and waterborne outbreaks suspected of being caused by NoVs reported from 2002 to 2006 were screened for NoV by EM, as described previously (11). Samples were stored at +4°C for further analyses. Outbreaks were reported and handled by the county medical officer responsible for communicable diseases or by the local environmental health protection board. An outbreak was defined as two linked cases with a common food or water intake. At least one sample from each outbreak was characterized by sequencing. No attempts were made to analyze food items for virus contamination.
RNA extraction.
Viral RNA was extracted from a 10% stool suspension in phosphate-buffered saline (pH 7.4) with a QIAamp viral RNA minikit (Qiagen, Hilden, Germany). The extraction was performed according to the manufacturer's instructions. The RNA extracts were used directly for reverse transcription (RT)-PCR or were stored at −70°C.
Polymerase RT-PCR.
NoV-specific primers JV12-JV13 (39), JV12Y-JV13I (38), and JV12BH-Nvp110 (12) were used to amplify 327-bp, 327-bp, and 334-bp fragments of the RNA-dependent RNA polymerase, respectively. RNA was reverse transcribed by using forward primers (primer JV13, JV13I, or Nvp110). RT-PCR was performed as described previously (39).
Capsid RT-PCR.
Two sets of primers, primers G1-SKF and G1-SKR and primers COG2F and G2-SKR (18), were used to amplify NoV GI and GII fragments of the capsid gene and generated 330-bp and 387-bp products, respectively. Random hexamers were used for cDNA synthesis. RT and PCR were performed as described by Yan et al. (42). All PCR products were run on 2% agarose gels.
Sequencing.
The PCR products were purified on spin columns (Jet Quick; Saween Werner, Limhamn, Sweden), according to the manufacturer's instructions. Both strands of the PCR fragments were sequenced by using a BigDye Terminator cycle sequencing kit (Applied Biosystems) and were run on an automated sequencer (ABI Prism 3100 DNA sequencer; Applied Biosystems). The primers used for sequencing were identical to those used in the PCRs.
Sequence analysis and alignment.
The sequences were edited by the Seqman II module in the Lasergene software package (DNAStar, Inc., Madison, WI). All sequences (RNA polymerase, capsid GI, and capsid GII sequences separately) were aligned; and dendrograms, supplemented with the sequences of NoV reference strains from the sequence database of the European Network (www.rivm.nl/bnwww), as well as NoV strains from the GenBank database, were generated by the unweighted pair group method with arithmetic averages method with BioNumerics software (version 4.61; Applied Mathematics, Belgium). Bootstrap values for branches of >70% are given at appropriate nodes; 500 simulations were run.
Nucleotide sequence accession numbers.
The nucleotide sequences have been submitted to GenBank and assigned accession numbers EU007698 to EU007817.
RESULTS
Food-borne and waterborne outbreaks of NoV gastroenteritis in Sweden from 2002 to 2006.
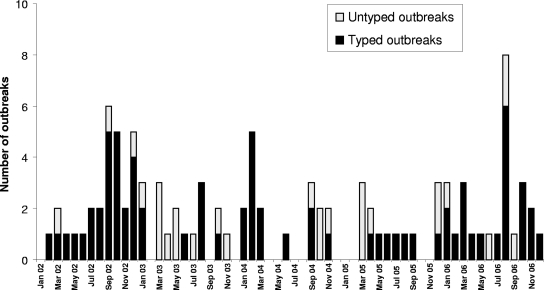
Between January 2002 and December 2006, 101 suspected food-borne and waterborne outbreaks caused by NoV were reported to SMI and together comprised more than 4,100 cases. Fecal material was not available from all outbreaks, but it proved possible to obtain genotyping results for NoV strains from 73 outbreaks (Table 1). More outbreaks were reported in 2002 (n = 28) and 2006 (n = 26) than in 2003 (n = 17), 2004 (n = 17), and 2005 (n = 13). The outbreaks occurred throughout the year, and there was no clear peak in winter (Fig. 1). Of the 73 outbreaks studied, 52% were related to food intake at restaurants or cafés, while water (drinking or recreational) was the suspected cause in 18% of the outbreaks.
TABLE 1.
Food-borne and waterborne outbreaks genotyped by sequencing of polymerase and capsid gene fragmentsa
| Outbreak no. | Month | Setting | No. of people affected | Genotype by sequencing of:
|
|
|---|---|---|---|---|---|
| Polymerase gene | Capsid gene | ||||
| SE/2002/02 | 3 | Health resort | 25 | ND | GII.6 |
| SE/2002/03 | 2 | Drinking water | 400 | GI.3 | GI.3 |
| SE/2002/05 | 7 | Drinking water | 74 | ND | GI.8/GII.4 |
| SE/2002/06 | 8 | Restaurant | 60 | GI.3 | GI.3 |
| SE/2002/07 | 8 | Drinking water | 30 | GI.3 | GI.3 |
| SE/2002/08 | 9 | Restaurant | 70 | GII.4 | GII.4 |
| SE/2002/09 | 9 | Private home | 22 | GI.3 | GI.3 |
| SE/2002/11 | 9 | Restaurant | 35 | GII.b | GII.1 |
| SE/2002/12 | 10 | Restaurant | 40 | ND | GII.4 |
| SE/2002/13 | 10 | Restaurant | 15 | GII.2 | ND |
| SE/2002/14 | 5 | Drinking water | 119 | GII.2 | GII.2 |
| SE/2002/15 | 11 | Drinking water | 50 | GI.4 | GI.4 |
| SE/2002/16 | 9 | Drinking water | ND | GI.4 | ND |
| SE/2002/17 | 4 | Restaurant | 41 | GII.b | ND |
| SE/2002/18 | 9 | Restaurant | 20 | GI.3 | GI.8 |
| SE/2002/19 | 10 | Restaurant | ND | ND | GII.2 |
| SE/2002/21 | 10 | Drinking water | 110 | GI.3 | GII.2 |
| SE/2002/22 | 12 | Restaurant | ND | GII.4 | GII.4 |
| SE/2002/23 | 12 | Restaurant | ND | GI.6 | GI.8 |
| SE/2002/24 | 12 | Restaurant | ND | GII.4 | GII.4 |
| SE/2002/25 | 10 | Drinking water | ND | GI.3 | GI.3 |
| SE/2002/26 | 6 | Restaurant | ND | GII.7 | GII.6 |
| SE/2002/27 | 12 | Restaurant | ND | GI.3 | GI.3 |
| SE/2002/28 | 11 | Unknown | ND | GII.4 | ND |
| SE/2002/29 | 7 | Private home | ND | GII.4 | GII.4 |
| SE/2003/08 | 6 | Restaurant | 54 | GII.4 | GII.4 |
| SE/2003/09 | 8 | Catering | 63 | GII.4 | GII.4 |
| SE/2003/10 | 8 | Restaurant | 350 | GII.7 | GII.6 |
| SE/2003/11 | 8 | Catering | 13 | GII.4 | GII.4 |
| SE/2003/12 | 10 | Restaurant | 50 | GI.3 | GI.14 |
| SE/2003/15 | 1 | Restaurant | 103 | GI.3 | GI.3 |
| SE/2003/16 | 1 | Restaurant | 10 | GII.4 | GII.4 |
| SE/2004/01 | 1 | Restaurant | 20 | GI.6 | GI.8 |
| SE/2004/03 | 1 | Restaurant | 74 | GII.7 | GII.6 |
| SE/2004/04 | 2 | Restaurant | 120 | GII.b | GII.3 |
| SE/2004/05 | 2 | Catering | 32 | GII.7 | GII.6 |
| SE/2004/06 | 2 | Restaurant | 6 | GII.7 | GII.7 |
| SE/2004/07 | 2 | Restaurant | 6 | GII.7 | GII.7 |
| SE/2004/08 | 2 | Private home | 35 | ND | GII.7 |
| SE/2004/09 | 3 | Café | 19 | GII.7 | GII.7 |
| SE/2004/10 | 3 | Catering | 14 | GII.b | GII.3 |
| SE/2004/12 | 6 | Restaurant | 35 | GII.b | GII.3 |
| SE/2004/13 | 9 | Restaurant | 16 | GI.6 | GI.8 |
| SE/2004/15 | 9 | Restaurant | 5 | GII.4 | GII.4 |
| SE/2004/18 | 11 | Restaurant | 200 | GII.4 | GII.4 |
| SE/2005/04 | 5 | Catering | 20 | GII.2 | GII.2 |
| SE/2005/05 | 4 | Private home | 16 | GII.4 | GII.4 |
| SE/2005/06 | 6 | Restaurant | 18 | GII.8 | GII.8 |
| SE/2005/07 | 7 | Recreational water | 35 | GI.5 | GI.5/GII.6 |
| SE/2005/08 | 9 | Private home | ND | GII.7 | GII.7 |
| SE/2005/09 | 8 | Private home | ND | ND | GII.6 |
| SE/2005/13 | 12 | Private home | 20 | ND | GII.4 |
| SE/2006/02 | 1 | Restaurant | 48 | ND | GI.8 |
| SE/2006/03 | 1 | Restaurant | 35 | ND | GII.7 |
| SE/2006/04 | 2 | Private home | 15 | ND | GII.7 |
| SE/2006/05 | 3 | Catering | 9 | ND | GII.7 |
| SE/2006/06 | 3 | Catering | 9 | ND | GII.7 |
| SE/2006/07 | 3 | Restaurant | 15 | ND | GIIx |
| SE/2006/08 | 4 | Restaurant | 10 | ND | GII.7 |
| SE/2006/09 | 5 | Restaurant | 30 | ND | GII.2 |
| SE/2006/10 | 8 | Private home | ND | ND | GII.5/GII.8 |
| SE/2006/11 | 7 | Recreational water | 150 | ND | GI.4 |
| SE/2006/13 | 8 | School | 12 | ND | GII.2/GII.8 |
| SE/2006/15 | 8 | Catering | 9 | ND | GII.8 |
| SE/2006/17 | 10 | Nursing home | 50 | ND | GII.4 |
| SE/2006/18 | 10 | Restaurant | 24 | ND | GII.4 |
| SE/2006/19 | 11 | Catering | 55 | ND | GII.4 |
| SE/2006/21 | 12 | Restaurant | 30 | ND | GI.14 |
| SE/2006/22 | 8 | Recreational water | 22 | ND | GI.1/GI.2 |
| SE/2006/23 | 8 | Recreational water | 100 | ND | GI.1/GI.4 |
| SE/2006/24 | 8 | Drinking water | 19 | ND | GI.14 |
| SE/2006/25 | 10 | Restaurant | 9 | ND | GII.3 |
| SE/2006/26 | 11 | Restaurant | 21 | ND | GII.6 |
The data are for 73 outbreaks. The number of people affected is given when known, and the total number of people affected was 3,117. ND, no data.
FIG. 1.
Number of food-borne and waterborne outbreaks caused by NoV reported to SMI each month, with and without genotyping results. It proved possible to genotype samples from 73 of 101 NoV-positive outbreaks.
Molecular epidemiology of NoVs.
The strains from 73 outbreaks were genotyped by sequencing fragments of the polymerase region (ORF1) and/or the capsid region (ORF2) of the NoV genome. A variety of strains were found. The results are summarized in Table 1, which lists the genotypes detected, together with the setting, the month of the outbreak, and the number of people affected, when they were known. GII strains were the most frequently detected (70%), while approximately 30% of the strains belonged to GI. Throughout the whole study period, the proportion of GI strains ranged from 14 to 52% per year. Strains of genotype GII.4, as determined by sequencing of both the capsid and the polymerase regions, were the most prevalent and were detected in 25% of all outbreaks during the study period.
Genotyping of strains from 45 outbreaks (from 2002 to 2005) by sequencing of the polymerase gene.
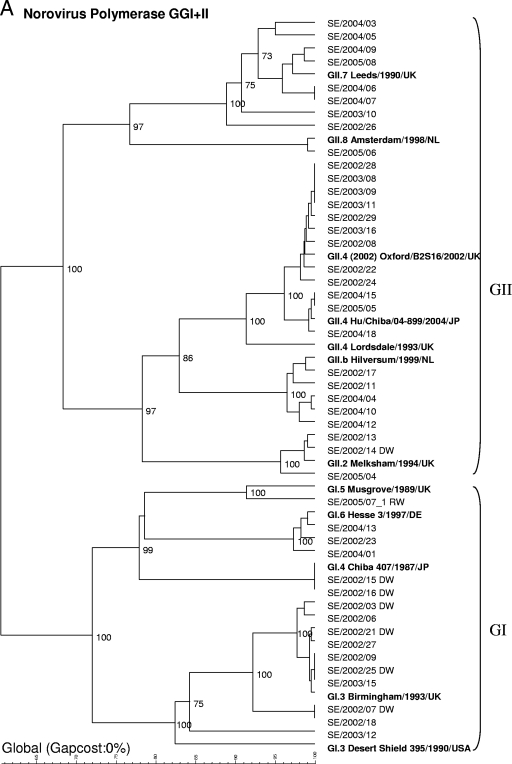
On the basis of sequencing of the polymerase gene (234 nucleotides), nine different genotypes were found (Fig. 2A): GII.4 (n = 12 strains), GI.3 (n = 10), GII.7 (n = 8), GII.b (n = 5), GI.6 (n = 3), GII.2 (n = 3), GI.4 (n = 2), GI.5 (n = 1), and GII.8 (n = 1). The dominant strain in 2002 was GI.3 (which was detected in 8/21 outbreaks), that in 2003 was GII.4 (4/7 outbreaks), and that in 2004 was GII.7 (5/11 outbreaks). In 2005, strains of five different genotypes were detected in five outbreaks studied. Different clusters were observed within the group of GII.4 strains. The strains occurring in 2002 and 2003 were of a variant that caused large outbreaks all over Europe during this period (26), whereas the strains observed in 2004 and 2005 clustered with a lineage that appeared in Europe for the first time in 2004 (22).
FIG. 2.
Phylogenetic dendrograms based on 234-nucleotide fragments of the polymerase gene for GI and GII NoV strains (A) or 291-nucleotide fragments (GI) (B) or 282-nucleotide fragments (GII) (C) of the capsid gene supplemented with reference strains. Waterborne outbreaks are marked DW for drinking water and RW for recreational water. When two genotypes were detected within one outbreak, they are given the suffixes _1 and _2.
Genotyping of strains from 69 outbreaks (from 2002 to 2006) by sequencing of the capsid gene.
Since January 2006, only sequencing of the capsid region (291 or 282 nucleotides) has routinely been used for genotyping, since the assay of the capsid region has been found to be more sensitive and the typing results are more easily interpreted. Sequencing of the capsid region of strains from 69 outbreaks (in 6 outbreaks, two strains were found) detected the following strain types (Fig. 2B and C): GII.4 (n = 17), GII.7 (n = 10), GII.6 (n = 8), GI.3 (n = 7), GI.8 (n = 6), GII.2 (n = 6), GII.3 (n = 4), GII.8 (n = 4), GI.4 (n = 3), GI.14 (n = 3), GI.1 (n = 2), GI.2 (n = 1), GI.5 (n = 1), GII.1 (n = 1), and GII.5 (n = 1). In addition, one strain (GIIx) did not cluster with any of the known reference strains. The closest match of the strain (recovered on 7 July 2008) with sequences in the GenBank database was Hu/GII/Leverkusen267/2005/DE (GenBank accession number EU424333) and Hu/Minase/1/05/JP (GenBank accession number AB234184), and the latter strain was therefore added as a reference in the dendrogram.
In 2002, the most prevalent strains were GI.3 (6/21 outbreaks) and GII.4 (6/21), in 2003 the most prevalent strains were GII.4 (4/7), in 2004 the most prevalent strains were GII.7 (4/13), in 2005 the most prevalent strains were GII.4 (2/7) and GII.6 (2/7), and in 2006 the most prevalent strains were GII.7 (5/21). Sequencing of the capsid region also revealed different GII.4 clusters, although they were not as clearly defined as they were in the analysis of the polymerase gene region. Primarily, one cluster was found for the GII.4 strains recovered in 2002 and 2003, another clustered the strains recovered in 2004 and 2005, and two separate clusters were found among the strains recovered in 2006. The clusters detected in 2006 were recently described by Kroneman et al. (22) and named GII.4 2006a and GII.4 2006b by the European Network (http://www.rivm.nl/bnwww).
Correlation between genotyping results by sequencing of polymerase and capsid gene regions.
From 2002 to 2005, sequencing of both the polymerase and the capsid gene regions was performed for 41 of 52 outbreaks. The same genotype was identified by sequencing of both the polymerase and the capsid gene regions in most of the outbreaks (Table 1). However, there were five outbreaks in 2002, two in 2003, and seven in 2004 in which different genotypes were identified by sequencing of the polymerase region (pol) and the capsid region (cap). The GII.bpol variant was associated with GII.1cap or GII.3cap; GII.7pol was found together with GII.7cap as well as with GII.6cap; and GI.3pol strains were observed together with GI.3cap, GI.8cap, GI.14cap, and GII.2cap, while a GI.6pol strain was found together with GI.8cap.
Mixed NoV infections.
Sequencing of the capsid gene region detected six outbreaks caused by strains of more than one genotype: one outbreak in 2002 (caused by GI.8 and GII.4 strains), one in 2005 (caused by GI.5 and GII.6 strains), and four in 2006 (caused by GII.5 and GII.8 strains, GII.2 and GII.8 strains, GI.1 and GI.2 strains, and GI.1 and GI.4 strains). Strains of the different genotypes were found in fecal samples from different patients, and recombinations are unlikely. Four of the outbreaks were due to contaminated drinking or recreational water, and two were presumed to have been caused by contaminated berries imported from China. For outbreaks in which several samples were analyzed, the sequences of strains of one genotype were usually identical or differed by only 1 or 2 nucleotides. A common feature for most outbreaks linked to contaminated water was the detection of GI strains, which were found in all but one case.
DISCUSSION
The genetic diversity of NoV infections in Swedish food-borne and waterborne outbreaks during the period from 2002 to 2006 was investigated by sequencing fragments of the polymerase and/or capsid gene. A high degree of genetic diversity was found among the NoV strains. Strains of 9 polymerase genotypes (4 GI and 5 GII) and 16 capsid genotypes (7 GI and 9 GII) and a great variety of subgroups were recovered from the 73 outbreaks from which strains were analyzed.
No obvious seasonality was observed among the outbreaks presented here, as opposed to the typical winter peak in NoV infections seen in health care settings (14, 27, 29, 31). The relatively large number of outbreaks reported in July 2006 was related to the co-occurrence of waterborne outbreaks and contaminated frozen berries. The lack of seasonality is not easy to understand, but the international food trade and the use of frozen food items could be possible explanations. NoV has so far been detected in only a few products, as we still lack standardized techniques for the detection of NoV in food items (2, 6). In Sweden, NoV has been demonstrated in drinking water and raspberries (24, 32).
In Sweden, as in the rest of Europe, increased numbers of cases of NoV infections were seen in health care settings in the autumns of 2002, 2004, and 2006. On all occasions, the increase was correlated to the emergence of new GII.4 variants (14, 22, 26, 37, 40). The increased activity caused by GII.4 variants was not reflected as clearly in the outbreaks related to food and water, but the genotype was common throughout the whole study period. However, in this study, several of the outbreaks caused by GII.4 strains occurred in private homes or health care settings in which the epidemiological links to food were rather weak. Person-to-person spread could be an alternative explanation in these cases.
The genetic drift within the NoV GII.4 genotype has caused the global spread of new variants. The increased numbers of outbreaks due to these new variants have been studied on several continents (9, 35, 36, 40). Whether genetic drift also occurs within other genotypes is less well known. However, it is worth noting that in some of the outbreaks described in this study, the causative NoV strain showed sequence data similar or identical to those for the reference strains described 10 or 15 years ago. This observation indicates a high degree of genetic stability or unknown NoV reservoirs, at least for some genotypes. The relatively rare appearance of these strains might be explained by some kind of persistent population immunity or an inherited low level of susceptibility to certain strains.
A rather large proportion of GI strains was observed during the study period, and these were mostly observed in 2002 and 2006. In total, 13 waterborne outbreaks were analyzed, with the majority involving GI strains. The abundance of GI viruses in water-related outbreaks has also been observed in Finland (30). The outbreaks occurred during both winter and summer, which could imply that GI NoV strains are more stable in water than GII strains. The GI strains are usually immunogenetically distinct from the dominant GII.4 strains found in the winter, as found in a serological enzyme-linked immunosorbent assay investigation with a whole range of virus-like particles (10). Population immunity only to GII strains would therefore make individuals relatively more susceptible to rare GI strains between the winter seasons, as in the waterborne outbreaks. In 2002, strains from eight outbreaks involving drinking water were analyzed. Most of these occurred after heavy rains, when wastewater entered the drinking water system. Four water-related outbreaks occurred after periods of hot weather during late summer in 2005 and 2006, when large numbers of people gathered at lakes and were exposed to polluted recreational lake water. More than 200 people fell ill under similar circumstances in 2004, when GI.1 viruses were recovered from affected swimmers who had visited one of two lakes (34). The detection of GI strains in outbreaks connected to recreational water could indicate that surface water is an important reservoir for these viruses.
Most outbreaks were reported from restaurants, while some were waterborne or occurred in private homes. The number of food-borne and waterborne outbreaks is probably heavily underestimated, since most cases are never reported or patients do not seek health care. Suspected food items in this study were first and foremost raspberries; but layer cake, Christmas smorgasbord, cold buffets, and water were also implicated. An epidemiological link could sometimes be established, but in most cases the food item was unknown.
It has previously been shown that food-borne and waterborne outbreaks are often caused by multiple NoV strains (16). In this study, infections caused by strains of two genotypes were found in at least six outbreaks, four of which were due to contaminated water (drinking and recreational water). In outbreaks in which data for the sequences of both the polymerase and capsid gene regions were available, discrepancies were occasionally found. These differences could result from mixed infections, but they could also reflect inconsistencies in genotype assignment (e.g., GI.6 and GI.8 strains) or recombination at the ORF1-ORF2 junction (e.g., GII.b strains). The capsid region of GI.6 and GI.8 strains has been described as GI.6, on the basis of a shorter fragment spanning the capsid N/S domain gene (16), and as GI.8 (43), on the basis of analysis of the entire capsid gene. GII.bpol has been described in combination with the GII.1, GII.2, GII.3, and GII.4 capsids in Hungary (33). In this study the variant was found with GII.1 and GII.3 capsids. GII.b has previously been described in Sweden in contaminated drinking water (32), as well as in health care settings, mainly in young children (25).
We used direct sequencing of PCR products and made no attempts at cloning. By this approach, it is likely that some outbreaks caused by multiple strains may have been overlooked. Only the predominant strains from single patients and outbreaks in which only a few samples have been examined can be sequenced. Strains of different genotypes were usually found in fecal samples from different patients, as was the case for two outbreaks caused by contaminated water (outbreaks SE/2002/21 and SE/2005/07), in which GI and GII strains were found. Despite these drawbacks, we believe that the high degree of genetic variation found among strains from Swedish food-borne and waterborne outbreaks is well documented.
One of the main advantages of genotyping is the possibility of linking outbreaks. In this study, similar GII.8 strains were detected in patient samples from two outbreaks involving imported raspberries from China (outbreaks SE/2006/13 and SE/2006/15). An identical GII.8 strain, also associated with a food-borne outbreak (outbreak SE/2006/10), was found at about the same time, but it could not be verified whether the Chinese raspberries were the cause of that outbreak. Three outbreaks caused by GII.7 strains (outbreaks SE/2004/06 to SE/2004/08) occurred closely in time and within a small geographical area, but no epidemiological link could be confirmed. Furthermore, identical GI.1 strains were found in two outbreaks related to recreational water. They occurred at about the same time, but they were geographically separated.
This study showed that a diversity of NoV strains caused food-borne and waterborne infections in Sweden from 2002 to 2006. This is in contrast to the situation in institutional settings in Sweden during the same period, when single strains usually dominated during 1 year (14). The high degree of genetic diversity found among strains responsible for food-borne and waterborne infections stresses the importance of the use of broad reaction detection methods when these outbreaks are studied.
Acknowledgments
This work was supported in part by the European Union grant Prevention of Emerging (Food-Borne) Enteric Viral Infections: Diagnosis, Viability Testing, Networking and Epidemiology (grant DIVINE-NET, 2003213).
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2)S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Baert, L., M. Uyttendaele, and J. Debevere. 2008. Evaluation of viral extraction methods on a broad range of ready-to-eat foods with conventional and real-time RT-PCR for norovirus GII detection. Int. J. Food Microbiol. 123101-108. [DOI] [PubMed] [Google Scholar]
- 3.Berg, D. E., M. A. Kohn, T. A. Farley, and L. M. McFarland. 2000. Multi-state outbreaks of acute gastroenteritis traced to fecal-contaminated oysters harvested in Louisiana. J. Infect. Dis. 181(Suppl. 2)S381-S386. [DOI] [PubMed] [Google Scholar]
- 4.Boccia, D., A. E. Tozzi, B. Cotter, C. Rizzo, T. Russo, G. Buttinelli, A. Caprioli, M. L. Marziano, and F. M. Ruggeri. 2002. Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg. Infect. Dis. 8563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 434659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 1811467-1470. [DOI] [PubMed] [Google Scholar]
- 7.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19467-474. [DOI] [PubMed] [Google Scholar]
- 8.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1861-7. [DOI] [PubMed] [Google Scholar]
- 9.Gallimore, C. I., M. Iturriza-Gomara, J. Xerry, J. Adigwe, and J. J. Gray. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 1521295-1303. [DOI] [PubMed] [Google Scholar]
- 10.Hansman, G. S., K. Natori, H. Shirato-Horikoshi, S. Ogawa, T. Oka, K. Katayama, T. Tanaka, T. Miyoshi, K. Sakae, S. Kobayashi, M. Shinohara, K. Uchida, N. Sakurai, K. Shinozaki, M. Okada, Y. Seto, K. Kamata, N. Nagata, K. Tanaka, T. Miyamura, and N. Takeda. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87909-919. [DOI] [PubMed] [Google Scholar]
- 11.Hedlund, K. O., E. Rubilar-Abreu, and L. Svensson. 2000. Epidemiology of calicivirus infections in Sweden, 1994-1998. J. Infect. Dis. 181(Suppl. 2)S275-S280. [DOI] [PubMed] [Google Scholar]
- 12.Hoebe, C. J., H. Vennema, A. M. de Roda Husman, and Y. T. van Duynhoven. 2004. Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. J. Infect. Dis. 189699-705. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 1442377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen, K., K. Mannerqvist, A. Allard, Y. Andersson, L. G. Burman, L. Dillner, K. O. Hedlund, K. Jonsson, U. Kumlin, T. Leitner, M. Lysén, M. Thorhagen, A. Tiveljung-Lindell, C. Wahlström, B. Zweygberg-Wirgart, and A. Widell. 2008. Norovirus strains belonging to the GII.4 genotype dominate as a cause of nosocomial outbreaks of viral gastroenteritis in Sweden 1997-2005. Arrival of new variants is associated with large nation-wide epidemics. J. Clin. Virol. 42129-134. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, P. J., M. Torvén, A. C. Hammarlund, U. Björne, K. O. Hedlund, and L. Svensson. 2002. Food-borne outbreak of gastroenteritis associated with genogroup I calicivirus. J. Clin. Microbiol. 40794-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 422988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299225-239. [DOI] [PubMed] [Google Scholar]
- 18.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100107-114. [DOI] [PubMed] [Google Scholar]
- 19.Koopmans, M., and E. Duizer. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 9023-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koopmans, M., H. Vennema, H. Heersma, E. van Strien, Y. van Duynhoven, D. Brown, M. Reacher, and B. Lopman. 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerg. Infect. Dis. 91136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koopmans, M., C. H. von Bonsdorff, J. Vinjé, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. E. Böttiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro Surveill. 11E061214 1. [DOI] [PubMed] [Google Scholar]
- 23.Le Guyader, F. S., F. Bon, D. DeMedici, S. Parnaudeau, A. Bertone, S. Crudeli, A. Doyle, M. Zidane, E. Suffredini, E. Kohli, F. Maddalo, M. Monini, A. Gallay, M. Pommepuy, P. Pothier, and F. M. Ruggeri. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J. Clin. Microbiol. 443878-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Guyader, F. S., C. Mittelholzer, L. Haugarreau, K. O. Hedlund, R. Alsterlund, M. Pommepuy, and L. Svensson. 2004. Detection of noroviruses in raspberries associated with a gastroenteritis outbreak. Int. J. Food Microbiol. 97179-186. [DOI] [PubMed] [Google Scholar]
- 25.Lindell, A. T., L. Grillner, L. Svensson, and B. Z. Wirgart. 2005. Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in children. J. Clin. Microbiol. 431086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Böttiger, K. O. Hedlund, M. Torvén, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szücs, B. Melegh, L. Svensson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363682-688. [DOI] [PubMed] [Google Scholar]
- 27.Lopman, B. A., G. K. Adak, M. H. Reacher, and D. W. Brown. 2003. Two epidemiologic patterns of norovirus outbreaks: surveillance in England and Wales, 1992-2000. Emerg. Infect. Dis. 971-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24137-160. [DOI] [PubMed] [Google Scholar]
- 29.Lopman, B. A., M. Reacher, C. Gallimore, G. K. Adak, J. J. Gray, and D. W. Brown. 2003. A summertime peak of “winter vomiting disease”: surveillance of noroviruses in England and Wales, 1995 to 2002. BMC Public Health 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maunula, L., I. T. Miettinen, and C. H. von Bonsdorff. 2005. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 111716-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2)S284-S287. [DOI] [PubMed] [Google Scholar]
- 32.Nygård, K., M. Torvén, C. Ancker, S. B. Knauth, K. O. Hedlund, J. Giesecke, Y. Andersson, and L. Svensson. 2003. Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg. Infect. Dis. 91548-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter, G., H. Vennema, M. Koopmans, and G. Szücs. 2006. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J. Clin. Virol. 3584-88. [DOI] [PubMed] [Google Scholar]
- 34.Sartorius, B., Y. Andersson, I. Velicko, B. de Jong, M. Löfdahl, K. O. Hedlund, G. Allestam, C. Wangsell, O. Bergstedt, P. Horal, P. Ulleryd, and A. Söderström. 2007. Outbreak of norovirus in Vastra Gotaland associated with recreational activities at two lakes during August 2004. Scand. J. Infect. Dis. 39323-331. [DOI] [PubMed] [Google Scholar]
- 35.Siebenga, J. J., H. Vennema, E. Duizer, and M. P. Koopmans. 2007. Gastroenteritis caused by norovirus GGII.4, The Netherlands, 1994-2005. Emerg. Infect. Dis. 13144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 819932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vainio, K., and M. Myrmel. 2006. Molecular epidemiology of norovirus outbreaks in Norway during 2000 to 2005 and comparison of four norovirus real-time reverse transcriptase PCR assays. J. Clin. Microbiol. 443695-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25233-235. [DOI] [PubMed] [Google Scholar]
- 39.Vinjé, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 174610-615. [DOI] [PubMed] [Google Scholar]
- 40.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 19027-36. [DOI] [PubMed] [Google Scholar]
- 41.Widdowson, M. A., A. Sulka, S. N. Bulens, R. S. Beard, S. S. Chaves, R. Hammond, E. D. Salehi, E. Swanson, J. Totaro, R. Woron, P. S. Mead, J. S. Bresee, S. S. Monroe, and R. I. Glass. 2005. Norovirus and foodborne disease, United States, 1991-2000. Emerg. Infect. Dis. 1195-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan, H., F. Yagyu, S. Okitsu, O. Nishio, and H. Ushijima. 2003. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J. Virol. Methods 11437-44. [DOI] [PubMed] [Google Scholar]
- 43.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]