Abstract
In 2004 and 2005, an epidemiological survey of Crimean-Congo hemorrhagic fever virus (CCHFV) was conducted in Xinjiang, China. A total of 5,629 serum samples of human and livestock were collected and tested for the CCHFV antibody, and 17,319 ticks were collected for viral identification. Reverse passive hemagglutination inhibition assays showed that the average prevalence of CCHFV antibody was 1.7% for the humans and 12.7% for the livestock. A relatively high antibody prevalence, ranging from 19.1% to 23.4%, was found in the livestock of the northwest, southwest, and northeast parts of the Tarim Basin. When the ticks were pooled to inoculate suckling mice, followed by reverse transcription-PCR (RT-PCR) to detect CCHFV RNA, the average RT-PCR-positive rates for Hyalomma asiaticum kozlovi and H. asiaticum asiaticum were 12.9% and 2.6%, respectively. A significant correlation was found between the antibody prevalence in the livestock and the CCHFV prevalence in H. asiaticum of the same geographic region. No CCHFV RNA was detected in Dermacentor nivenus, Rhipicephalus turanius, or Rhipicephalus sanguineus. A total of 27 partial S segments of CCHFVs were sequenced and used for phylogeny analysis. All but one Chinese isolate grouped into the Asia 1 clade, which contains the strains from Xinjiang and Uzbekistan, while the other strain, Fub90009, grouped with strains from the Middle East.
Crimean-Congo hemorrhagic fever virus (CCHFV) is an RNA virus that belongs to the genus Nairovirus of the family Bunyaviridae. The virus has a tripartite genome composed of a small (S), a medium (M), and a large (L) RNA segment (39). Crimean-Congo hemorrhagic fever (CCHF) is a tick-borne disease with a mortality rate of 10% to 50% (16, 35) and has been reported in more than 30 countries in Africa, Europe, and Asia (16, 20, 26, 36, 38, 43). The potential use of CCHFV as a terrorist agent is a threat to public health (5, 10). Some genera of the family Ixodidae (hard ticks) transmit vectors and reservoirs of this virus (10). Humans can be infected by tick bites and interaction with infected people or animals, which may cause CCHF outbreaks in some regions (1, 9, 17, 19, 24, 27, 33). In China, the first case of CCHF was reported in Bachu county of Xinjiang in 1965 (30), and since then, there have been several outbreaks in that area (3, 4, 21, 23, 37). Several regions in the Tarim Basin, such as the Tarim River and the Yeerqiang River, and the Junggar Basin were identified as natural epidemic foci of CCHF (2, 14). So far, the phylogenetic data for CCHFV in China all relate to the western part of the Tarim Basin (Bachu county and surrounding areas) (22, 28, 37), but the geographic distribution of Hyalomma asiaticum (the local major vector) in Xinjiang appears to occupy a much larger area (40).
In this study, the epidemiology of CCHFV in the Tarim Basin, the Junggar Basin, the Turpan-Hami Basin, and the Ili Valley, which are the habitats of ticks, was studied. A total of 5,629 serum samples from livestock and humans living in these areas were collected and tested for antibodies against CCHFV, and 17,319 ticks belonging to five species/subspecies were collected for viral isolation. Partial sequences of the S segment were amplified and used for phylogenetic analyses. Our results revealed the geographic distribution and phylogeny of CCHFV in Xinjiang, China.
MATERIALS AND METHODS
Investigation areas and sampling.
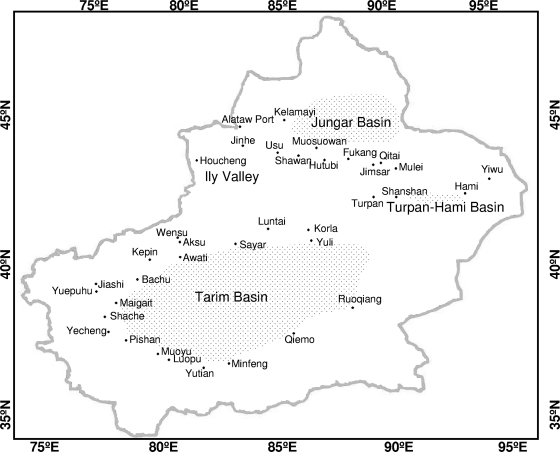
According to the geographic distribution of ticks in Xinjiang, China (40), four geographic areas (the Tarim Basin, the Junggar Basin, the Turpan-Hami Basin, and Ili Valley), including 37 counties (or cities), were investigated from 2004 to 2005 (Fig. 1). The sample collecting spots were virgin droughty deserts which have a distribution of shrubbery, livestock or other herbivores, rodents, and ticks. A total of 3,175 livestock serum samples were collected from animals pastured in the investigation spots, and 2,454 human serum samples were collected from the people who were engaged in stockbreeding or agricultural activities in these spots (see Table 1). A total of 17,319 ticks belonging to five species/subspecies were also collected from these spots.
FIG. 1.
Map of the Xinjiang area showing the locations of the sampling spots for the current study. The small dots indicate the sampling spots, and the names of the sampling counties are shown. The longitudes (E) and latitudes (N) are indicated on the edges of the map.
TABLE 1.
Prevalence of the CCHFV antibody in serum samples collected by RPHIA from livestock and humans in different geographic regions in Xinjianga
| Geographic region | Counties | Human sera
|
Livestock sera
|
||||
|---|---|---|---|---|---|---|---|
| No. tested | No. positive | % positive (95% CI) | No. tested | No. positive | % positive (95% CI) | ||
| Tarim Basin | |||||||
| Northwest | Aksu, Awati, Bachu, Jiashi, Yuepuhu | 306 | 8 | 2.6 (2.31-2.89) | 640 | 122 | 19.1 (17.59-20.54) |
| Southwest | Maigait, Shache, Yecheng, Pishan, Luopu, Yutian | 167 | 1 | 0.6 (0.51-0.69) | 363 | 85 | 23.4 (20.99-25.81) |
| Northeast | Sayar, Luntai, Yuli, Ruoqiang | 486 | 2 | 0.4 (0.36-0.44) | 534 | 115 | 21.5 (19.71-23.36) |
| Southeast | Minfeng, Qiemo | 52 | 3 | 5.8 (4.22-7.38) | 256 | 9 | 3.5 (2.08-3.95) |
| Junggar Basin | Jinghe, Kelamayi, Shawan, Muosuowan, Hutubi, Fukang, Mulei | 853 | 24 | 2.8 (2.61-2.99) | 720 | 26 | 3.6 (3.34-3.86) |
| Turpan-Hami Basin | Turpan, Hami, Yiwu | 590 | 5 | 0.8 (0.74-0.86) | 662 | 46 | 6.9 (6.37-7.43) |
| Total | 2,454 | 43 | 1.7 (1.63-1.77) | 3,175 | 403 | 12.7 (12.25-13.13) | |
95% CI, 95% confidence interval.
CCHFV antibody detection in serum samples from livestock and humans.
Reverse passive hemagglutination inhibition assays (RPHIA) were used to detect CCHFV antibodies in the serum samples as described previously (12). The antigen used for RPHIA was prepared with Xinjiang hemorrhagic fever standard strain 66019, which was isolated from patients with hemorrhagic fever in Bachu County, Xinjiang, in 1966 (13). Briefly, the serum samples (25 μl) were serially diluted (1:4 to 1:32) in twofold steps in phosphate-buffered saline containing 1% heat-inactivated normal rabbit serum in V-shaped microplates. The same amount (25 μl) of CCHFV antigen, with a hemagglutination value of 4, was added to each well. After the mixing step, the plates were incubated at 4°C overnight and then transferred to 37°C for 30 min. Twenty-five microliters of 1% sheep erythrocytes coated with a CCHFV monoclonal antibody (43E5) was added to each well. After being shaken, the plates were allowed to stand at 37°C. The hemagglutination pattern of each well was observed after 1 h. Samples with complete hemagglutination inhibition at a dilution of 1:8 or higher were chosen as being positive for the CCHFV antibody.
Virus isolation and identification.
Viral isolation was done by inoculating samples into suckling mice (Kunming white mice) as described previously (12). About 50 ticks of the same species or subspecies collected at the same spot were grouped into one pool for viral isolation. Each pool of ticks was ground with Dulbecco's modified Eagle's medium (Invitrogen Corporation, China) to prepare 10% suspensions. The suspensions were centrifuged at 3,000 rpm for 20 min. The supernatant liquid (0.25 ml) was inoculated into the brain of a newborn (age 24 to 48 h) suckling mouse. The samples that caused typical symptoms at 5 to 9 days postinoculation for at least two passages in suckling mice were regarded as pathogenic. The typical symptoms of the suckling mice included becoming sensitive to stimuli, becoming easily startled, having an arched back, losing balance, and refusing to be nursed. After two to four blind passages in the brains of suckling mice, the brain tissues of the typical symptomatic mice were tested by PCR for partial sequencing of CCHFV S segments.
Sequencing and phylogenetic analyses.
Total RNA from brain tissues of the typical symptomatic mice was extracted by using an SV total RNA isolation system kit (Promega), and normal mouse brain tissues were used as the negative control. About 0.1 g of the homogenized mouse brain was used for RNA extraction. An AMV reverse transcription-PCR (RT-PCR) kit (TaKaRa, Japan) was used for RT-PCR. Specific primer PCM-Tag (5′-CCGAGAATAAAATCGAGGTGAATCTCAAAGAAAT-3′) (22) and random primers (TaKaRa, Japan) were used to amplify cDNA. Primers F2 (5′-TGGACACTTTCACAA ACTC-3′) and R3 (5′-GACAAATTCCCTGCACCA-3′) were used for the first round of nested PCR, while primers F3 (5′-GAGTGTGCCTGGGTTAGTTC-3) and R2 (5′-GACATTACAATTTCACCAGG-3′) were used for the second round nested-PCR for partial sequence of the S segment (31). The PCR products were purified and sequenced by the Shanghai Sangon Company. In addition to samples obtained from 2004 to 2005, seven strains isolated earlier from the same areas were also included in this study to sequence the partial S segments (see Table 3).
TABLE 3.
Xinjiang CCHFV strains sequenced in the present study
| Region | Strain | County | Origin | Year | GenBank accession no. |
|---|---|---|---|---|---|
| Northwest Tarim | Aw04318 | Awati | H. asiaticum kozlovi | 2004 | EU715264 |
| Ba66063 | Bachu | H. asiaticum kozlovi | 1966 | EU715257 | |
| Ba68038 | Bachu | Human | 1968 | EU715258 | |
| Hf7403 | Bachu | Human | 1978 | EU715259 | |
| Ba8002 | Bachu | Human | 1980 | EU715261 | |
| Ba8004 | Bachu | Human | 1980 | EU715262 | |
| Ba04005 | Bachu | H. asiaticum kozlovi | 2004 | EU715265 | |
| Ba04203 | Bachu | H. asiaticum kozlovi | 2004 | EU715266 | |
| Ba05108 | Bachu | H. asiaticum kozlovi | 2005 | EU715277 | |
| Southwest Tarim | Lp04224 | Luopu | H. asiaticum kozlovi | 2004 | EU715271 |
| Yt05093 | Yutian | H. asiaticum kozlovi | 2005 | EU715285 | |
| Yt05099 | Yutian | H. asiaticum kozlovi | 2005 | EU715286 | |
| Northeast Tarim | Bz002 | Bazhou | Human | 1979 | EU715260 |
| Lt04035 | Luntai | H. asiaticum kozlovi | 2004 | EU715267 | |
| Lt05146 | Luntai | H. asiaticum kozlovi | 2005 | EU715279 | |
| Yl04032 | Yuli | H. asiaticum kozlovi | 2004 | EU715273 | |
| Yl04033 | Yuli | H. asiaticum kozlovi | 2004 | EU715274 | |
| Yl04038 | Yuli | H. asiaticum kozlovi | 2004 | EU715275 | |
| Yl04041 | Yuli | H. asiaticum kozlovi | 2004 | EU715276 | |
| Yl05034 | Yuli | H. asiaticum kozlovi | 2005 | EU715283 | |
| Yl05035 | Yuli | H. asiaticum kozlovi | 2005 | EU715284 | |
| Rq04219 | Ruoqiang | H. asiaticum kozlovi | 2004 | EU715270 | |
| Rq04244 | Ruoqiang | H. asiaticum kozlovi | 2004 | EU715272 | |
| Southeast Tarim | Qm04222 | Qeimuo | H. asiaticum asiaticum | 2004 | EU715269 |
| Junggar Basin | Fub90009 | Fukang | H. asiaticum asiaticum | 1990 | EU715263 |
| Ml05225 | Mulei | H. asiaticum asiaticum | 2005 | EU715280 | |
| Ml05232 | Mulei | H. asiaticum asiaticum | 2005 | EU715281 |
The phylogeny analysis of CCFHV was conducted using the sequence data in this study as well as 25 other CCHFV S segment sequences available from GenBank (see Table 4). The S partial segment nucleotide sequence alignment was performed by CLUSTALW (version 1.83) (18), and maximum likelihood analysis with PHYLIP (version 3.67) (11) was used to construct phylogenetic trees.
TABLE 4.
The 25 CCHFV isolates obtained from GenBank and used for phylogeny analysis
| Strain | Region | Origin | Year | GenBank accession no. |
|---|---|---|---|---|
| 66019 | Bachu, China | Human | 1966 | AJ010648 |
| 68031 | Bachu, China | Sheep | 1968 | DQ211642 |
| 7001 | Aksu, China | Human | 1970 | AF415236 |
| 75024 | Bachu, China | Human | 1975 | AF362080 |
| 7803 | Bachu, China | Human | 1978 | AF354296 |
| 79121 | Bachu, China | Euchoreutes naso | 1979 | AF358784 |
| 8402 | Bachu, China | H. asiaticum kozlovi | 1984 | AJ010649 |
| 88166 | Bachu, China | Human | 1988 | AY029157 |
| AP92 | Greece | Phipicephalus bursa | 1976 | DQ211638 |
| ArD97268 | Senegal | H. truncatum | 1993 | U15091 |
| ArD15786 | Senegal | Goat | 1973 | U15020 |
| ArTeh1933 | Iran | Alectorobius lahorensis | 1978 | U15022 |
| Baghdad 12 | Iraq | Human | 2000 | AJ538196 |
| Bul/Hu517 | Bulgaria | Human | 1978 | AY277676 |
| HY13 | Bachu, China | H. asiaticum kozlovi | 1968 | AY900145 |
| Hodzha | Uzbekistan | Human | 1967 | AY223475 |
| IbAr10200 | Nigeria | H. excavatum | 1966 | U75674 |
| JD206 | Pakistan | H. anatolicum | 1965 | U88414 |
| Matin | Pakistan | Human | 1976 | AF527810 |
| ROS/TI28044 | Rostov, Russia | H. marginatum | 2000 | AY277672 |
| SPU415/85 | South Africa | Human | 1985 | DQ211648 |
| SPU281/89 | South Africa | Human | 2005 | AY905637 |
| STV/HU29223 | Stavropol, Russia | Human | 2000 | AF481802 |
| Uganda3010 | Uganda | Human | 1956 | U88416 |
| Uzbek/TI10145 | Uzbekistan | H. asiaticum | 2000 | AF481799 |
Statistic analyses.
The correlation of RT-PCR-positive rates for ticks to the CCHFV antibody prevalence in serum samples from different places was estimated by the Pearson product-moment correlation coefficient, and the t test was used to determine the significance of the correlation (34).
RESULTS
CCHFV antibody prevalence in serum samples from livestock and humans in different geographic regions.
A total of 2,454 blood samples from humans, and 3,175 serum samples from sheep and camels, were collected from 27 counties (or townships) of the Tarim Basin, the Junggar Basin, and the Turpan-Hami Basin (Table 1). Antibodies against CCHFV were detected by RPHIA in a total of 446 samples (Table 1). The average prevalence of CCHFV antibody in serum samples of humans and livestock was 1.7% and 12.7%, respectively. The CCHFV antibodies in human sera were at low levels (0.4 to 5.8%), with low range variations among different regions. A relatively high prevalence of the CCHFV antibody, ranging from 19.1 to 23.4%, was found in the livestock sera from the northwest, southwest, and northeast parts of the Tarim Basin. In contrast, a relatively low prevalence of the CCHFV antibody, ranging from 3.5 to 6.9%, was found in the livestock sera from the southeast part of the Tarim Basin, the Junggar Basin, and the Turpan-Hami Basin (Table 1). The antibody prevalence in the livestock from the northwest, southwest, and northeast parts of the Tarim Basin is significantly different from that of the southeast part of the Tarim Basin, the Junggar Basin, and the Turpan-Hami Basin (Table 1).
Investigation of CCHFV in different species of ticks in Xinjiang.
From 2004 to 2005, a total of 10,436 H. asiaticum kozlovi specimens were collected from different areas of the Tarim Basin (Table 2). They were grouped into 209 pools and inoculated into sucking mice. Thirty-seven pools induced the typical symptoms in the inoculated suckling mice, of which 27 were positive by RT-PCR for the CCHFV S gene (Table 2). The RT-PCR-positive percentages ranged from 11.1 to 14.3%, with an average of 12.9%. The percentage of tick pools that caused typical symptoms in the inoculated mice (17.7%) was higher than the RT-PCR-positive percentage (12.9%), indicating that some of the CCHFVs might not be amplified by the primers used, or there might be other pathogens in the ticks which induced similar symptoms in the mice.
TABLE 2.
Results of CCHFV detection from H. asiatium collected from different regions in Xinjianga
| Geographic region | County(ies) |
H. asiaticum kozlovi
|
H. asiaticum asiaticum
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of ticks | No. of pools | No. of pools pathogenic to mice | No. of RT-PCR-positive pools | % RT-PCR-positive (95% CI) | No. of ticks | No. of pools | No. of pools pathogenic to mice | No. of RT-PCR-positive pools | % RT-PCR-positive rate (95% CI) | ||
| Tarim Basin | |||||||||||
| Northwest | Aksu, Wensu, Awati, Kepin, Bachu, Yuepuhu | 3,155 | 63 | 12 | 9 | 14.3 (10.8-17.8) | 0 | ||||
| Southwest | Maigait, Shache, Yecheng, Pishan, Moyu, Luopu, Yutian | 1,754 | 36 | 7 | 4 | 11.1 (7.5-14.7) | 0 | ||||
| Northeast | Sayar, Luntai, Korla, Yuli, Ruoqiang | 5,527 | 110 | 18 | 14 | 12.7 (10.3-15.1) | 0 | ||||
| Southeast | Minfeng, Qiemo | 0 | 3,452 | 68 | 4 | 1 | 1.5 (1.1-1.8) | ||||
| Junggar Basin | Alataw Port, Usu, Kelamayi, Muosuowan, Hutubi, Jimsar, Qitai, Mulei | 0 | 1,769 | 36 | 3 | 2 | 5.6 (3.7-7.4) | ||||
| Turpan- Hami Basin | Turpan, Shanshan, Hami, Yiwu | 0 | 590 | 12 | 0 | 0 | 0 | ||||
| Ili Valley | Houcheng | 0 | 50 | 1 | 0 | 0 | 0 | ||||
| Total | 10,436 | 63 | 37 | 27 | 12.9 (11.2-14.7) | 5,861 | 117 | 7 | 3 | 2.6 (2.1-3.0) | |
95% CI, 95% confidence interval.
A total of 5,861 H. asiaticum asiaticum samples were collected from the southeastern part of the Tarim Basin, the Junggar Basin, the Turpan-Hami Basin, and the Ili Valley (Table 2). They were grouped into 114 pools and inoculated into sucking mice. Seven pools induced the typical symptoms in the inoculated suckling mice, of which three tested positive by RT-PCR for CCHFV (Table 2). The average RT-PCR-positive rate for H. asiaticum asiaticum (2.6%) was significantly lower than that for H. asiaticum kozlovi (12.9%) (χ2 = 10.05, P = 0.03).
A total of 1,022 other ticks, including 644 Dermacentor nivenus (12 pools), 327 Rhipicephalus turanius (7 pools), and 51 Rhipicephalus sanguineus (1 pool) specimens collected from the Tarim Basin and the Junggar Basin, were inoculated into suckling mice, but the typical symptoms did not appear in any of the inoculated mice.
Correlation of CCHFV antibody prevalence in livestock with CCHFV RNA prevalence in H. asiaticum ticks.
When the Pearson product-moment correlation coefficient and the t test were used to determine the significance of the correlation (40), a significant correlation between RT-PCR-positive rates in ticks (H. asiaticum kozlovi and H. asiaticum asiaticum) (Table 2) and CCHFV antibody prevalence in serum samples of the livestock from different places (Table 1) was revealed (r = 0.869, P = 0.025).
Phylogenetic analysis of CCHFV in Xinjiang, China.
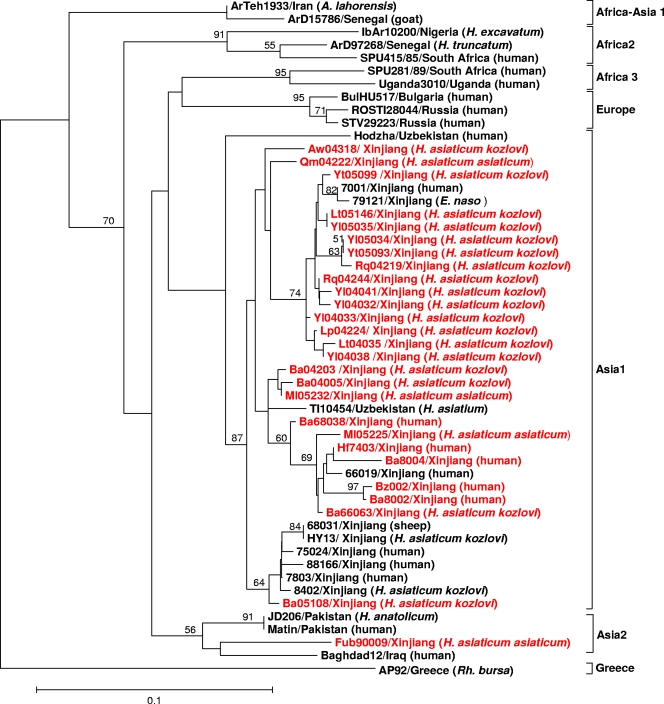
A total of 27 partial S segments (220 bp long, corresponding to nucleotides 329 to 548 of the CCHFV Chinese strain 7001) from this study were analyzed by nested PCR (Table 3). The sequence analyses indicated that the Chinese isolates shared high similarity, with 86 to 99.5% nucleotide identity and 89 to 100% amino acid identity. Figure 2 shows the phylogenetic tree based on the partial sequences of the S segment of the 52 CCHFV sequences (Table 3 and Table 4). Our results showed that CCHFVs could be grouped into seven clades, including (i) Africa-Asia 1 (mainly from West Africa and Iran), (ii) Africa 2 (mainly from South and West Africa), (iii) Africa 3 (mainly from Uganda and West Africa), (iv) Europe (mainly from Eastern Europe, including Russia and Bulgaria), (v) Asia 1 (mainly from China and Uzbekistan), (vi) Asia 2 (mainly from Pakistan and Iraq), and (vii) Greece AP92. This result is very similar to those of previous studies (7, 8, 15, 29, 41, 42).
FIG. 2.
Phylogenetic tree of CCHFVs based on the 220-nucleotide S RNA sequences. The tree was constructed by using the maximum likelihood method with PHYLIP. The sequences obtained from this study are shown in red. The numbers above the branches indicate the bootstrap values in percentages (of 100 replicates). Values lower than 50% are not shown. The scale bar indicates 10% nucleotide sequence divergence.
All but one Chinese isolate were clustered in the Asia 1 clade, which contains the Xinjiang strains from China and the strains from Uzbekistan (Fig. 2). It is interesting that the Fub90009 strain isolated from Fukang County in the Junggar Basin (14) is located in the Asia 2 clade, which contains mainly strains from the Middle East (the Matin strain and JD206 strain from Pakistan and the Baghdad12 strain from Iraq).
DISCUSSION
Geographic distribution of CCHFV in Xinjiang, China.
Since the first report of CCHF in Bachu County in 1965, a greater number of CCHFV samples have been isolated from H. asiaticum, suggesting that it is the major vector and reservoir of CCHFV in this region (2, 14). By combining historical survey data (14, 6, 43) and our current data, it is clear that CCHFV is distributed widely in Xinjiang.
The Tarim Basin (approximately 37° to 42°N, 77° to 89°E), which occupies the center of South Xinjiang, with an area of 560,000 square kilometers, is surrounded by mountains that block the southern warm wet air and the cold air of Siberia, causing extreme drought in the region. The snow from the mountains melts into the rivers and creates oases, but the area where rivers cannot reach remains a barren desert. The relatively isolated oases in the Tarim Basin provide habitats for wild and domestic animals as well as ticks. Our recent study indicated that there are two geographic subspecies of H. asiaticum in the Tarim Basin, H. asiaticum kozlovi and H. asiaticum asiaticum (44). The distribution and epidemiology of CCHFV in the Tarim Basin appeared to be related to the distribution of the two geographic subspecies of H. asiaticum. In the northwestern, southwestern, and northeastern parts of the Tarim Basin, H. asiaticum kozlovi is the main species and accounts for 66.9 to 92% of the tick population (44). The rates of CCHFV antibody-positive serum samples from livestock were 19.1 to 23.4% (Table 1). This result is consistent with the relatively high prevalence of CCHFV (11.1 to 14.3%) in the ticks collected in these regions (Table 2). In the southeastern part of the Tarim Basin, H. asiaticum asiaticum was the main species and accounted for 72.6% of the tick population (44). The CCHFV antibody-positive rate in the serum of livestock in the southeast Tarim Basin was 3.5% (Table 1), which is consistent with the relatively low CCHFV prevalence (1.5%) for the ticks collected in this region (Table 2). Our current results indicate that the correlation of CCHFV antibody prevalence in the livestock with CCHFV RNA prevalence in H. asiaticum ticks of the same geographic region is significant.
The landscape and ecology of the Junggar Basin (approximately 44° to 47°N, 82° to 92°E) are quite different from those of the Tarim Basin. In the Junggar Basin, plants are widely distributed in much greater quantity and at a much higher density than those of the Tarim Basin. Wild animals and ticks are also widely distributed. Like the southeast part of the Tarim, H. asiaticum asiaticum is the main tick species of the Junggar Basin. Likewise, the CCHFV antibody-positive rate in the serum of livestock in the Junggar Basin was 3.6% (Table 1), which is consistent with the relatively low CCHFV prevalence (5.6%) of the ticks collected in this region (Table 2).
The Turipan-Hami Basin (approximately 42° to 42°40′N, 88° to 94°E) located in east Xinjiang. Although CCHFV-positive antibodies were identified from the serum samples from human and livestock in this region (Table 1), we were unable to isolate CCHFV from the ticks (Table 2). Whether this region is a natural focus of CCHFV remains to be further investigated.
In this study, only one sample site (Houcheng) belongs to the Ily Valley, and no human or livestock sera were collected there. Therefore, the data are too preliminary to draw any conclusions about the region.
Historically, CCHF cases have been reported from around the Bachu region (Bachu, Awati, Jiashi, Kepin, Kuche, and Maigait) in Xinjiang (3, 4, 21, 23, 30, 32, 37), and CCHFVs have been isolated mainly from that region (14, 22, 28, 37). Our results indicate that CCHFV is likely to be distributed over a much larger area in Xinjiang. The isolates Ml05225 and Ml05232 (Table 3), collected from Mulei (43°55′N, 90°25′E) in this study, to our knowledge, are the easternmost isolates to be reported so far.
No significant correlation between CCHFV antibody prevalence in humans with CCHFV prevalence in the ticks of the same geographic region was identified. This may be due to the different lifestyles of the people and/or to differences in our sample population. For example, the human samples of the southeast Tarim were mainly from herders or butchers, while human samples from other areas include not only herders or butchers but also people involved in other agriculture activities. This may partially explain why the CCHFV antibody prevalence was relatively high in samples from southeast Tarim compared to those from other regions (Table 1).
Phylogenetic analyses of CCHFVs in Xinjiang, China.
There have been several genetic analyses of CCHFVs isolated from Xinjiang (22, 25, 28, 37); however, all the reported Xinjiang strains were isolated from the area around Bachu County (at the western part of the Tarim Basin) from the 1960s to the 1980s. In this study, 27 CCHFV strains isolated from different geographic regions (the Tarim Basin and the Junggar Basin) in Xinjiang were partially sequenced (Table 3). Together with the previously published CCHFV data (Table 4), a phylogenetic tree with more information on Chinese isolates is presented (Fig. 2). The phylogeny analysis reveals that all but one Chinese isolate (Fub90009) belong to one clade, Asia 1 (Fig. 2), regardless of their origin (different tick species, humans, or animals). The Xinjiang isolates grouped with the isolates from Uzbekistan, indicating that the virus circulating in Xinjiang and Uzbekistan are genetically related, consistent with previous reports (37, 41). However, the Fub90009 strain, which was isolated from the Junggar Basin, was grouped into another clade (Asia 2) with Iraq, Pakistan, and other Central Asian strains (Fig. 2). Xinjiang has a geographical location and environmental climate similar to those of Central Asian countries. The migration of virus-infected birds or tick-infected birds may contribute to the spread of CCHFV (16, 39). In addition, frequent livestock trading and economic and cultural exchanges could also cause the spread of CCHFV. Our results indicate that the CCHFVs in Xinjiang may have multiple origins.
It has been shown that although phylogenetic analyses based on S and L segments generate similar results, M segments often show greater diversity, which could lead to a better-resolved phylogenetic tree (7). Morikawa et al. (25) previously sequenced M segments of 7 CCHFV isolates around Bachu County, and their results indicated that there is a multisource virus population in that region. At the moment, we are focusing on sequencing more M segments to further reveal the evolutionary history of CCHFV in Xinjiang.
Acknowledgments
This work was supported by a grant from the National Key Technologies R&D Program of China during the 10th Five-Year Plan Period (2003BA712A08-03), a project from the Chinese Academy of Science (KSCX2-YW-N-065), a project from the National Natural Science Foundation of China (30860255), a project from MOST (2007FY210700), and a joint project from the State Key Laboratory of Virology.
We thank Yue Jiang for help with the phylogenetic analyses and Xiulian Sun for help with the statistic analyses.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Altaf, A., S. Luby, A. J. Ahmed, N. Zaidi, A. J. Khan, S. Mirza, J. McCormick, and S. Fisher-Hoch. 1998. Outbreak of Crimean-Congo haemorrhagic fever in Quetta, Pakistan: contact tracing and risk assessment. Trop. Med. Int. Health 3878-882. [DOI] [PubMed] [Google Scholar]
- 2.Chai, J. 2004. The preliminary investigation report on geographical distribution of Xinjiang hemorrhagic fever in upper and middle reach of Tarim River in Xinjiang in 1968. Endemic Dis. Bull. 19(Suppl.)34-36. [Google Scholar]
- 3.Chai, J., Y. Han, C. Feng, Y. Zhang, Y. Nuer, Y. Liu, Y. Zhang, Y. Qiao, and H. Li. 2004. Report of investigation on hemorrhagic fever in Bachu County, Xinjiang in 1966. I. Clinical observation on 5 cases with hemorrhagic fever. Endemic Dis. Bull. 19(Suppl.)1-5. [Google Scholar]
- 4.Chai, J., Y. Nuer, C. Feng, G. Li, and J. Liu. 2004. Hemorrhagic fever in Bachu: two case reports. Endemic Dis. Bull. 19(Suppl.)23-25. [Google Scholar]
- 5.Charrel, R. N., H. Attoui, A. M. Butenko, J. C. Clegg, V. Deubel, T. V. Frolova, E. A. Gould, T. S. Gristsun, F. X. Heinz, M. Labuda, V. A. Lashkevich, V. Loktev, A. Lundkvist, D. V. Lvov, C. W. Mandl, M. Niedrig, A. Papa, V. S. Petrov, A. Plyusnin, S. Randolph, J. Süss, V. I. Zlobin, and X. de Lamballerie. 2004. Tick-borne virus diseases of human interest in Europe. Clin. Microbiol. Infect. 101040-1055. [DOI] [PubMed] [Google Scholar]
- 6.Dai, X., A. Muhtar, C. H. Feng, R. Sun, X. P. Tai, X. H. Wang, K. Burenmind, W. W. Meng, R. Azat, and Y. J. Zhang. 2006. Geography and host distribution of Crimean-Congo hemorrhagic fever in the Tarim Basin. Zhonghua Liu Xing Bing Xue Za Zhi 271048-1052. [PubMed] [Google Scholar]
- 7.Deyde, V. M., M. L. Khristova, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 2006. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J. Virol. 178834-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten, C., D. Minnak, P. Emmerich, H. Schmitz, and T. Reinicke. 2002. Crimean-Congo hemorrhagic fever in Kosovo. J. Clin. Microbiol. 401122-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engin, A., A. Yildirim, T. Kunt, M. Bakir, I. Dokmetas, and L. Ozdemir. 2008. Clinical investigation of the transient evoked otoacoustic emission test in Crimean-Congo hemorrhagic fever. Int. J. Infect. Dis. 12162-165. [DOI] [PubMed] [Google Scholar]
- 10.Ergonul, O. 2006. Crimean-Congo hemorrhagic fever. Lancet Infect. Dis. 6203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 12.Feng, C. 2004. Laboratory assays applied for diagnosis of Xinjiang hemorrhagic fever. Endemic Dis. Bull. 19(Suppl.)101-116. [Google Scholar]
- 13.Feng, C., J. Chai, Y. Han, and H. Li. 2004. Report of investigation on hemorrhagic fever in Bachu County, Xinjiang in 1966. II. A preliminary study on etiology of hemorrhagic fever in Bachu. Endemic Dis. Bull. 19(Suppl.)6-14. [Google Scholar]
- 14.Feng, C. H., X. Bai, H. Liu, F. Li., and Y. Gu. 1991. Discovery of natural foci of Xinjiang hemorrhagic fever in the southern margin area of Junggar Basin, Xinjiang. Endemic Dis. Bull. 652-55. [Google Scholar]
- 15.Hewson, R., J. Chamberlain, V. Mioulet, G. Lloyd, B. Jamil, R. Hasan, A. Gmyi, L. Gmyi, S. E. Smirnova, A. Lukashev, G. Karganova, and C. Clegg. 2004. Crimean-Congo haemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 102185-189. [DOI] [PubMed] [Google Scholar]
- 16.Hoogstraal, H. 1979. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J. Med. Entomol. 15307-417. [DOI] [PubMed] [Google Scholar]
- 17.Jauréguiberry, S., P. Tattevin, A. Tarantola, F. Legay, A. Tall, P. Nabeth, H. Zeller, and C. Miachelet. 2005. Imported Crimean-Congon hemorrhagic fever. J. Clin. Microbiol. 434905-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeanmougin, F. J., D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23403-405. [DOI] [PubMed] [Google Scholar]
- 19.Joubert, J. R., J. B. King, D. J. Rossouw, and R. Cooper. 1985. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part III. Clinical pathology and pathogenesis. S. Afr. Med. J. 68722-728. [PubMed] [Google Scholar]
- 20.Khan, A. S., G. O. Maupin, P. E. Rollin, A. M. Noor, H. H. Shurie, A. G. Shalabi, S. Wasef, Y. M. Haddad, R. Sadek, K. Ijaz, C. J. Peters, and T. G. Ksiazek. 1997. An outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates, 1994-1995. Am. J. Trop. Med. Hyg. 57519-525. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., J. Chai, C. Xiao, and W. Li. 2004. Epidemiological analysis of 140 cases with Xinjiang hemorrhagic fever. Endemic Dis. Bull. 19(Suppl.)47-49. [Google Scholar]
- 22.Ma, B., C. Hang, and A. Papa. 2001. Sequencing and comparative analysis of the complete glycoprotein gene of three Crimean-Congo hemorrhagic fever virus Chinese isolates. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 15105-111. [PubMed] [Google Scholar]
- 23.Ma, B., and H. Wang. 2000. Tianshan hacker: Crimean-Congo hemorrhagic fever virus. Foreign Med. Sci. 78-10. [Google Scholar]
- 24.Midilli, K., A. Gargili, O. Ergonul, G. Sengoz, R. Ozturrk, M. Bakar, and F. Jongejan. 2007. Imported Crimean-Congo hemorrhagic fever cases in Istanbul. BMC Infect. Dis. 541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morikawa, S., T. Qing, Z. Xinqin, M. Saijo, and I. Kurane. 2002. Genetic diversity of the M RNA segment among Crimean-Congo hemorrhagic fever virus isolates in China. Virology 296159-164. [DOI] [PubMed] [Google Scholar]
- 26.Morikawa, S., M. Saijio, and I. Kurane. 2007. Recent progress in molecular biology of Crimean-Congo hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 30375-389. [DOI] [PubMed] [Google Scholar]
- 27.Papa, A., I. Christova, E. Papadimitriou, and A. Antoniadis. 2004. Crimean-Congo hemorrhagic fever in Bulgaria. Emerg. Infect. Dis. 101465-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papa, A., B. J. Ma, S. Kouidou, Q. Tang, C. S. Hang, and A. Antoniadis. 2002. Genetic characterization of the M RNA segment of Crimean Congo hemorrhagic fever virus strains, China. Emerg. Infect. Dis. 850-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrova, I. D., S. V. Seregin, V. S. Petrov, O. I. Vyshemirskii, I. I. Kuzina, D. K. Lvov, E. I. Samokhvalov, G. I. Tyummikov, V. V. Gutorov, L. N. Iashina, and S. V. Netesov. 2003. Genetic characteristics of the S-segment of RNA from two strains of the Crimean-Congo hemorrhagic fever virus isolated in the south of Russia and in Uzbekistan. Vopr. Virusol. 488-11. (In Russian.) [PubMed] [Google Scholar]
- 30.Region HaEPSoXUA. 1975. Compilation of information on Xinjiang haemorrhagic fever. Health and Epidemic Prevention Station of Xinjiang Uygur Autonomous Region, Urumqi, China.
- 31.Rodriguez, L. L., G. O. Maupin, T. G. Ksiazek, P. E. Rollin, A. S. Khan, T. F. Schwarz, R. S. Lofts, J. F. Smith, A. M. Norr, C. J. Peters, and S. T. Nichol. 1997. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 57512-518. [DOI] [PubMed] [Google Scholar]
- 32.Saijo, M., Q. Tang, B. Shimayi, L. Han, Y. Zhang, M. Asiguma, T. Dong, A. Maeda, and I. Kurane. 2004. Possible horizontal transmission of Crimean-Congo hemorrhagic fever virus from a mother to her child. Jpn. J. Infect. Dis. 5755-57. [PubMed] [Google Scholar]
- 33.Schwarz, T. F., H. Nsanze, M. Longson, H. Nitschko, S. Gilch, H. Shurie, A. Ameen, A. R. Zahir, U. G. Acharya, and G. Jager. 1996. Polymerase chain reaction for diagnosis and identification of distinct variants of Crimean-Congo hemorrhagic fever virus in the United Arab Emirates. Am. J. Trop. Med. Hyg. 5590-96. [DOI] [PubMed] [Google Scholar]
- 34.SPSS, Inc. 2003. SPSS 12.0 for Windows user's guide. SPSS, Inc., Chicago, IL.
- 35.Swanepoel, R., A. J. Shepherd, P. A. Leman, S. P. Shepherd, G. M. McGillivray, M. J. Erasmus, L. A. Searle, and D. E. Gill. 1987. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am. J. Trop. Med. Hyg. 36120-132. [DOI] [PubMed] [Google Scholar]
- 36.Swanepoel, R., J. K. Struthers, A. J. Shepherd, G. M. McGillivray, M. J. Nel, and P. G. Jupp. 1983. Crimean-congo hemorrhagic fever in South Africa. Am. J. Trop. Med. Hyg. 321407-1415. [DOI] [PubMed] [Google Scholar]
- 37.Tang, Q., X. Q. Zhao, H. Y. Wang, B. Simayi, Y. Z. Zhang, M. Saijo, M. Shigeru, G. D. Liang, and I. Kurane. 2005. Molecular epidemiology of Xinjiang hemorrhagic fever viruses. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 19312-318. [PubMed] [Google Scholar]
- 38.Tikriti, S. K., F. K. Hassan, I. M. Moslih, F. Jurji, M. I. Mahmud, and H. H. Tantawi. 1981. Congo/Crimean hemorrhagic fever in Iraq: a seroepidemiological survey. J. Trop. Med. Hyg. 84117-120. [PubMed] [Google Scholar]
- 39.Whitehouse, C. A. 2004. Crimean-Congo hemorrhagic fever. Antivir. Res. 64145-160. [DOI] [PubMed] [Google Scholar]
- 40.Xin, Y., Y. Rui, and G. Zheng-Da. 1997. The tick fauna of Xinjiang, p. 3-8. Xinjiang Scientific, Technological and Medical Publishing House, Urumqi, China.
- 41.Yashina, L., I. Petrova, S. Seregin, O. Vyshemirskii, D. Lvov, V. Aristova, J. Kuhn, S. Morzunnov, V. Gutorov, I. Kuzina, G. Tyunnikov, S. Netesov, and V. Petrov. 2003. Genetic variability of Crimean-Congon hemorrhagic fever virus in Russia and Central Asia. J. Gen. Virol. 841199-1206. [DOI] [PubMed] [Google Scholar]
- 42.Yashina, L., O. Vyshemirskii, S. Seregin, I. Petrova, E. Samokhvalov, D. Lvov, V. Gutorov, I. Kuzina, G. Tyunnikov, Y. W. Tang, S. Netesov, and V. Petrov. 2003. Genetic analysis of Crimean-Congon hemorrhagic fever virus in Russia. J. Clin. Microbiol. 41860-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen, Y. C., L. X. Kong, L. Lee, Y. Q. Zhang, F. Li, B. J. Cai, and S. Y. Gao. 1985. Characteristics of Crimean-Congo hemorrhagic fever virus (Xinjiang strain) in China. Am. J. Trop. Med. Hyg. 341179-1182. [PubMed] [Google Scholar]
- 44.Zhang, Y.-J., H.-L. Cao, X. Dai, M. Azaz, W. Jiang, A. Abulikm, B. Li, M. Ablimiti, G. Lei, A. Rezwan, X.-H. Liang, H.-B. Liu, X. Yu, and C.-H. Feng. 2006. Classification and diversity of tick community in Tarim Basin. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 246-11. [PubMed] [Google Scholar]