Abstract
A genotyping study of 285 Hybrid Capture 2 low-risk probe cocktail-positive specimens showed cross-reactivity with several untargeted human papillomavirus genotypes. Cross-reactivity was often clinically beneficial due to the detection of untargeted low-risk genotypes. A total of 8.4% of positive results, usually weak, were due to cross-reactivity with high-risk genotypes. Establishment of a gray zone is recommended.
Low-risk alpha-human papillomaviruses (HPV) have never been at the fore in HPV research, due to their connection with benign neoplasm only. However, interest in these genotypes has increased substantially in recent years, due to the fact that quadrivalent HPV vaccine contains, in addition to a “cervical cancer component” (against HPV16 and HPV18), virus-like particles of the two most important low-risk alpha-HPV genotypes: HPV6 and HPV11. These two closely related HPV genotypes are the etiological agents of at least 90% of genital warts and laryngeal papillomas (1, 8, 10, 20) and at least 12.1% of cervical low-risk squamous intraepithelial lesions (5). In phase III clinical trials, this quadrivalent prophylactic HPV vaccine was shown to be highly effective against genital warts, e.g., reducing the burden of genital warts by 99% (95% confidence interval = 93.7 to 100%) among HPV-naïve vaccinated women aged 16 to 26 years (11). The quadrivalent HPV vaccine is currently licensed in more than 105 countries and has already been included in national vaccination programs in several countries. The widespread use of this vaccine has created an immediate need for a very specific detection tool for low-risk alpha-HPV.
The Hybrid Capture 2 HPV DNA test (hc2), originally developed by Digene Corporation (Gaithersburg, MD) and currently marketed by Qiagen (Hilden, Germany), is the most widely used molecular method for the detection of a subset of clinically important HPV genotypes (14-16, 18). In this assay, exfoliated cells are first treated with alkali-denaturing reagent, and the processed samples are hybridized under high-stringency conditions with two mixtures of unlabeled full-genomic-length RNA probes, one specific for 13 high-risk HPV genotypes (HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, and HPV68) and one for 5 low-risk HPV genotypes (HPV6, HPV11, HPV42, HPV43, and HPV44). Positive specimens are detected by binding the hybridization complexes onto the surface of a microplate well coated with monoclonal antibodies specific to RNA-DNA hybrids. Immobilized hybrids are detected by the addition of an alkaline phosphatase-conjugated antibody to RNA-DNA hybrids, followed by the addition of a chemiluminiscent substrate. The emission of light is measured as relative light units (RLU) in a luminometer. Thus, hc2 does not allow the exact determination of HPV genotype(s) present in a clinical specimen but rather expresses the results of tested high-risk or low-risk HPV genotypes as positive or negative. The hc2 high-risk cocktail is very reliable for the routine detection of clinically important HPV infection and is, at present, the only commercially available HPV DNA assay with sufficient scientific data to support its performance in a clinical setting. However, several studies have shown significant analytical inaccuracy of the high-risk cocktail, mainly due to cross-reactivity with several untargeted HPV genotypes (2-4, 6, 9, 16, 19, 21-26, 27). This phenomenon certainly reduces the analytical specificity of the high-risk cocktail, but cross-reactivity with some HPV genotypes has proven to be clinically beneficial (4, 19).
The U.S. version of hc2, containing the high-risk probe cocktail only, is approved by the U.S. Food and Drug Administration (FDA) for triage (in cases of equivocal cytology results showing the presence of atypical squamous cells of undetermined significance) to determine which patients should be referred to physicians for a colposcopy and as a screening test for use in addition to cytology screening for women 30 years of age and older (15). Although the use of the hc2 low-risk probe cocktail is not recommended in the U.S. due to lack of FDA approval, the “Conformité Européene” (CE)-certified version of hc2, containing both high-risk and low-risk probe cocktails, is currently used in at least 40% of laboratories outside the U.S., mainly for individuals with clinically suspected low-risk HPV infection or as a reflex test for women with atypical squamous cells of undetermined significance who tested negative for high-risk HPVs. In contrast to the established cross-reactivity of the high-risk probe cocktail with several untargeted HPV genotypes, the specificity of the hc2 low-risk cocktail has never been studied in detail. According to the data presented in the hc2 package insert, the only recognized cross-reactivity of the hc2 low-risk cocktail is with HPV13, a genotype commonly detected in lip lesions of certain ethnic groups but never in anogenital lesions (17). In the present study, therefore, we have for the first time systematically examined the analytical specificity of the hc2 low-risk cocktail by determining the exact HPV genotype(s) present in 285 consecutive samples recognized using the hc2 low-risk probe cocktail as HPV DNA positive.
To determine the specificity and accuracy of hc2 in the detection of the five HPV genotypes (HPV6, HPV11, HPV42, HPV43, and HPV44) included in the low-risk probe cocktail, 285 consecutive cervical specimens obtained from the same number of women and recognized as HPV positive using the hc2 low-risk probe cocktail were included in the study. Fifty-six out of 285 samples were positive using both low-risk and high-risk probe cocktails. The specimens included in the study were collected between June 2007 and May 2008, using a DNAPaP cervical sampler and specimen transport medium (STM; Digene Corporation, Gaithersburg, MD), from Croatian and Slovenian women undergoing routine gynecological examination. hc2 testing was performed not later than 5 days after the collection of specimens, strictly following the manufacturer's instructions. According to the manufacturer's interpretation criteria, the specimens with a RLU-per-cutoff (RLU/CO) value higher than 1.0 were considered positive for one or more low-risk HPV genotypes included in the cocktail, and the specimens with a RLU/CO value of <1.0 were considered negative for the five low-risk HPV genotypes tested.
The presence of the five targeted low-risk genotypes, as well as other, untargeted low-risk and high-risk HPV genotypes, in the hc2 low-risk cocktail-positive samples was determined by using seven different PCR-based genotyping methods. To keep the number of tests to a minimum, the seven methods were used consecutively, e.g., if the first genotyping method reliably detected at least one of the five targeted HPV genotypes in the particular hc2 low-risk cocktail-positive sample, this sample was excluded from further testing. All seven genotyping tests were performed on the same sample used for hc2 testing, i.e., 500 μl of STM was removed before the addition of hc2 denaturing reagent solution (first step of hc2 testing) and kept at −70°C until genotyping. After the isolation of DNA from 200 μl of nondenatured STM using a QIAamp DNA mini kit (Qiagen, Hilden, Germany), all 285 samples were first tested using a recently developed real-time PCR (RT-PCR) assay that allows very specific detection and reliable differentiation of HPV6 and HPV11 (13). The sensitivity of the test, assessed by probit analysis at a 95% detection level, is 42.9, 43.4, and 25.3 DNA copies per assay for prototypic and nonprototypic HPV6 variants and HPV11, respectively (13). Sixty-six samples were found to be positive for HPV6, and 6 samples contained HPV11 (Table 1). The remaining 213 samples were then tested using HPV42 genotype-specific RT-PCR, and 105 were found to contain this low-risk HPV. The remaining 108 samples were then tested using HPV43 genotype-specific RT-PCR, followed by testing of all negative samples using HPV44/55 genotype-specific RT-PCR. By using the two RT-PCR tests, the presence of HPV43 and HPV44/55 was identified in 8 and 17 samples, respectively (Table 1). The HPV42, HPV43, and HPV44/55 genotype-specific primers and probes were designed according to the L1 long control region genomic sequences of the prototype isolates (GenBank accession nos. M73236, AJ620205, U31788, and U31791, respectively). As shown in Table 2, two primers and two hybridization (fluorescent resonance energy transfer) probes were selected for each HPV genotype studied. A common probe was designed for HPV44 and HPV55, which has recently been recognized as a subtype of HPV44 and is no longer considered a separate genotype (7). A BLAST search of the GenBank nr database was performed for each sequence in order to verify HPV genotype specificity. The assays were set up on a LightCycler 2.0 real-time system (Roche Diagnostics GmbH, Mannheim, Germany) and performed using a QuantiTect probe PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
TABLE 1.
Summary of genotyping results of 285 consecutive samples recognized as HPV DNA positive using the hc2 low-risk probe cocktail
| HPV genotype detected | No. of positive samples | Detection method(s) | Interpretation |
|---|---|---|---|
| HPV6 | 66 | HPV6/-11-specific RT-PCR (13) | Targeted HPV genotype |
| HPV11 | 6 | HPV6/-11-specific RT-PCR (13) | Targeted HPV genotype |
| HPV42 | 105 | HPV42-specific RT-PCR | Targeted HPV genotype |
| HP43 | 8 | HPV43-specific RT-PCR | Targeted HPV genotype |
| HP44/55 | 17 | HPV44/55-specific RT-PCR | Targeted HPV genotype |
| HPV40 | 4 | INNO-LiPA HPV test | Untargeted low-risk alpha-HPV genotype related to HPV43 (species 8), analytically incorrect but clinically beneficial |
| HPV74 | 13 | INNO-LiPA HPV test | Untargeted low-risk alpha-HPV genotype related to HPV6 and HPV11 (species 10), analytically incorrect but clinically beneficial |
| candHPV91 | 9 | Sequencing of GP5+/GP6+ PCR products | Untargeted low-risk alpha-HPV genotype, related to HPV43 (species 8), analytically incorrect but clinically beneficial |
| HPV61 | 2 | Linear array HPV test | Untargeted low-risk alpha-HPV genotype (species 3), analytically incorrect but clinically beneficial |
| HPV70 | 1 | INNO-LiPA HPV test | Untargeted low-risk alpha-HPV genotype (species 7), analytically incorrect but clinically beneficial |
| candHPV87 | 1 | Sequencing of GP5+/GP6+ PCR products | Untargeted low-risk alpha-HPV genotype (species 3), analytically incorrect but clinically beneficial |
| candHPV89 | 2 | Linear array HPV test | Untargeted low-risk alpha-HPV genotype (species 3), analytically incorrect but clinically beneficial |
| candHPV90 | 4 | Sequencing of GP5+/GP6+ PCR products | Untargeted low-risk alpha-HPV genotype (species 14), analytically incorrect but clinically beneficial |
| HPV16 | 5 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV18 | 3 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV31 | 2 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV45 | 1 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV52 | 1 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV59 | 1 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV68 | 2 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk alpha-HPV genotype |
| HPV51, HPV53, HPV56, HPV58, HPV66, HPV73 | 9 | INNO-LiPA HPV test and linear array HPV test | Untargeted high-risk or probably high-risk alpha-HPV genotypes found only in combination with |
| other high-risk genotypes | |||
| HPV negative | 23 | All seven genotyping methods |
TABLE 2.
Sequences of the primers and probes used in genotype-specific RT-PCRs designed to detect HPV42, HPV43, and HPV44/55
| HPV genotype | Primer/probea | Sequence (5′-3′)b | Nucleotide positionc |
|---|---|---|---|
| HPV42 | 42F | GGTGACTGCCCACCATTAGA(s) | 6374-6393 |
| 42R | CCTCAGCAGACATTTTTAAGTAATCA(a) | 6548-6523 | |
| 42FL | AGTTTTATTCAGGATGGGGATATGGTGG-FL(s) | 6404-6431 | |
| 42LC | LC610-TGTAGGGTTTGGGGCACTAGATTTTGG(s) | 6433-6459 | |
| HPV43 | 43F | AACTTACCCAGTTTCCCTTAGG(s) | 7126-7147 |
| 43R | ACAACCCATACAGGTACAAAACA(a) | 7303-7281 | |
| 43FL | AACTGTAAAGCGTTCTGCACCATCC-FL(s) | 7196-7220 | |
| 43LC | LC640-CCTCTACGTCTGCCCCTGCCT(s) | 7222-7242 | |
| HPV44/55 | 44/55F | GGCCTAGTGAAAACCAGGTATATG(s) | 5701-5724 |
| 44/55R | AGTGTCTTGTTTGCTGGTCGT(a) | 5866-5846 | |
| 44/55FL | CTCCCGCCCCAGTATCCAAAG-FL(s) | 5731-5751 | |
| 44/55LC | LC705-AATAMCTACGGATGCCTATGTCAAACGCAC(s) | 5753-5782 |
HPV genotype-specific forward (F) and reverse (R) primers and hybridization probes (FL and LC; see footnote b) are indicated.
FL, fluorescein; LC610, LC640, and LC705, LightCycler red dyes; M, A or C; (s), sense orientation; (a), antisense orientation.
As summarized in Table 1, the presence of at least one targeted HPV genotype was detected in 202 (70.9%) of 285 samples recognized as HPV DNA positive using the hc2 low-risk probe cocktail. The remaining 83 HPV6-, HPV11-, HPV42-, HPV43-, HPV44-, and HPV55-negative samples were then tested using the commercially available INNO-LiPA HPV genotyping Extra test (Innogenetics, Gent, Belgium), capable of recognizing 27 different alpha-HPV genotypes (including HPV6, HPV11, HPV43, and HPV44), following the manufacturer's instructions. All remaining HPV-negative samples were additionally tested using the commercially available Linear Array HPV genotyping test (Roche Diagnostics), capable of recognizing 37 different alpha-HPV genotypes (including HPV6, HPV11, and HPV42), following the manufacturer's instructions. Finally, all remaining HPV-negative samples were tested using an in-house GP5+/GP6+ PCR targeting a 150-bp fragment of the L1 gene (12). HPV genotypes were determined by sequencing the GP5+/GP6+ PCR products. By using three additional genotyping methods, the presence of untargeted but low-risk alpha-HPV genotypes phylogenetically related (HPV40, HPV74, and candHPV91) or phylogenetically unrelated (HPV61, HPV70, candHPV87, candHPV89, and candHPV90) to targeted genotypes were detected in a total of 26 and 10 samples, respectively. The detection of these eight untargeted low-risk alpha-HPV genotypes was interpreted as analytically incorrect but clinically beneficial. As summarized in Table 1, the presence of at least one targeted or untargeted low-risk HPV genotype was detected in 238 (83.5%) of 285 samples recognized as HPV DNA positive using the hc2 low-risk probe cocktail. The RLU/CO values measured in these samples ranged between 1.1 and 3,095.7 RLU/CO (median, 17.24 RLU/CO).
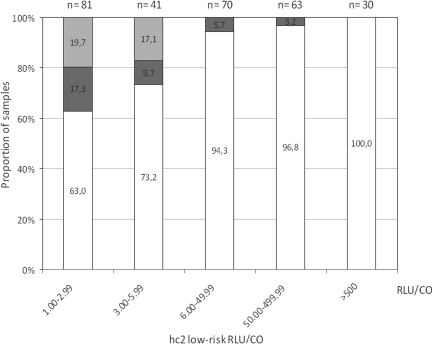
In 24 (8.4%) of 285 samples recognized as HPV positive using the hc2 low-risk probe cocktail, only high-risk or probable high-risk HPV genotypes but no targeted or untargeted low-risk genotypes were detected (Table 1). The unexpected genotyping results were confirmed in all 24 samples, using both the INNO-LiPA HPV genotyping Extra test and the Linear Array HPV genotyping test. Infection with a single high-risk genotype only was thus detected in 15 samples, and 9 samples contained two or more high-risk or probable high-risk HPV genotypes but no targeted or untargeted low-risk genotypes (Table 1). Seven different high-risk alpha-HPV genotypes (HPV16, HPV18, HPV31, HPV45, HPV52, HPV59, and HPV68) were detected as single pathogens and thus interpreted as hc2 low-risk probe cocktail cross-reactive genotypes. Six different high-risk or probable high-risk alpha-HPV genotypes (HPV51, HPV53, HPV56, HPV58, HPV66, and HPV73) were detected only in samples with other high-risk alpha-HPV genotypes and thus interpreted as possible cross-reactive genotypes. As shown in Fig. 1, weak RLU/CO values (below 6 RLU/CO) were observed in the majority of cross-reactive samples. RLU/CO values ranged between 1.1 and 255.6 RLU/CO (median, 2.4). For all 24 cross-reactive samples, the results of testing with hc2 high-risk cocktail were also available. As expected from our genotyping results, all 24 samples clearly tested hc2 high-risk positive, with high-risk RLU/CO values ranging between 21.4 and 3,053.9 (median, 254.1). Thus, similar to what was proposed for the hc2 high-risk probe cocktail (4, 19), the most probable reason for the false-positive hc2 low-risk probe cocktail results, recognized in 24 samples containing high-risk or probable high-risk HPV genotypes only, is the cross-reactivity of long low-risk RNA probes with high-risk HPV DNA present in high concentration in these samples.
FIG. 1.
The relationship between the proportion of hc2 false reactivity and hc2 RLU/CO values. The light gray part of each bar represents the percentage of hc2 false-positive results due to the absence of HPV DNA, the dark gray part the percentage of hc2 false-positive results due to cross-reactivity with untargeted high-risk HPV genotypes, and the white part the percentage of hc2 true-positive results.
Finally, in 23 (8.0%) of 285 samples recognized as HPV positive using the hc2 low-risk probe cocktail, no HPV DNA was detected using all seven genotyping methods (Table 1). Focusing on five hc2-targeted HPV genotypes, the presence of HPV6 and HPV11 was excluded using four different and very sensitive PCR-based methods and the presence of HPV42, HPV43, and HPV44 using three different PCR-based methods each. In addition, all 23 samples tested positive using primers KM29/RS42 targeting 536 bp of the ubiquitous human beta-globin gene (12), indicating an adequate quality of DNA and the absence of PCR inhibitors. As shown in Fig. 1, low-risk hc2 RLU/CO values measured in these samples were weak and ranged between 1.1 and 5.6 RLU/CO (median, 2.1). In addition to weak hc2 low-risk probe cocktail positivity, in 15 of 23 samples, weak hc2 high-risk probe cocktail positivity was also found, ranging between 1.4 and 7.7 RLU/CO (median, 4.3). Another eight samples tested hc2 high-risk probe cocktail negative. Although the possibility remains of the presence in the tested samples of HPV genotypes not covered by the seven genotyping methods but recognized by hc2 due to cross-reactivity with untargeted genotypes, this seems highly unlikely. Due to weak hc2 signals in these specimens and the recently published observation that in at least 4.8% of hc2 high-risk probe cocktail-positive samples, no HPV could be identified using two very sensitive PCR assays (4), we strongly believe that in the 23 hc2 low-risk probe cocktail-positive samples in which no HPV DNA was detected using all seven genotyping methods, no HPV DNA is present or it is present in clinically irrelevant quantities.
In conclusion, in the present genotyping study, which was performed on 285 hc2 low-risk HPV-positive cervical specimens, we found that the hc2 low-risk probe cocktail, similarly to the hc2 high-risk cocktail, cross-reacts with several untargeted HPV genotypes. The broader-than-assigned HPV genotype detection range of the hc2 low-risk probe cocktail is clinically beneficial in the majority of cases, due to the detection of phylogenetically related and unrelated low-risk HPV genotypes. However, 8.4% of all hc2 low-risk probe cocktail-positive results, usually with weak signal strength, were due to cross-reactivity with untargeted high-risk genotypes. We thus suggest a more cautious interpretation of all samples with weak low-risk hc2 signal strength until the clinical consequences of cross-reactivity are finally determined. On the basis of our results, and similar to a recent proposal for the high-risk probe cocktail (24), we suggest the following strategy. If a tested sample has an hc2 low-risk RLU/CO value of less than 6 and, at the same time, an hc2 high-risk RLU/CO value above 20, a high possibility of cross-reactivity with untargeted high-risk genotypes should be considered. If a tested sample has an hc2 low-risk RLU/CO value of less than 6 and, at the same time, a weak hc2 high-risk RLU/CO value or if it is hc2 high-risk negative, a high possibility of HPV DNA false positivity due to unresolved reasons should be considered.
Acknowledgments
This study was in part supported by grants 143-1080116-0097 and 143-0000000-0117 from the Croatian Ministry of Science, Education, and Sports; by a grant from the Slovenian Research Agency, project number Z3-0220-0381-08; and by Slovenian-Croatian Bilateral Project BI-HR/09-10-017.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Aubin, F., J. L. Prétet, A. C. Jacquard, M. Saunier, X. Carcopino, F. Jaroud, P. Pradat, B. Soubeyrand, Y. Leocmach, C. Mougin, D. Riethmuller, and EDiTH Study Group. 2008. Human papillomavirus genotype distribution in external acuminata condylomata: a large French national study (EDiTH IV). Clin. Infect. Dis. 47610-615. [DOI] [PubMed] [Google Scholar]
- 2.Castle, P. E., P. E. Gravitt, D. Solomon, C. M. Wheeler, and M. Schiffman. 2008. Comparison of linear array and line blot assay for detection of human papillomavirus and diagnosis of cervical precancer and cancer in the Atypical Squamous Cell of Undetermined Significance and Low-Grade Squamous Intraepithelial Lesion Triage Study. J. Clin. Microbiol. 46109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castle, P. E., M. Schiffman, R. D. Burk, S. Wacholder, A. Hildesheim, R. Herrero, M. C. Bratti, M. E. Sherman, and A. Lorincz. 2002. Restricted cross-reactivity of Hybrid Capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol. Biomarkers Prev. 111394-1399. [PubMed] [Google Scholar]
- 4.Castle, P. E., D. Solomon, C. M. Wheeler, P. E. Gravitt, S. Wacholder, and M. Schiffman. 2008. Human papillomavirus genotype specificity of Hybrid Capture 2. J. Clin. Microbiol. 462595-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clifford, G. M., R. K. Rana, S. Franceschi, J. S. Smith, G. Gough, and J. M. Pimenta. 2005. Human papillomavirus genotype distribution in low-grade cervical lesions: comparison by geographic region and with cervical cancer. Cancer Epidemiol. Biomarkers Prev. 141157-1164. [DOI] [PubMed] [Google Scholar]
- 6.de Cremoux, P., J. Coste, X. Sastre-Garau, M. Thioux, C. Bouillac, S. Labbe, I. Cartier, M. Ziol, A. Dosda, C. Le Gales, V. Molinie, M. C. Vacher-Lavenu, B. Cochand-Priollet, P. Vielh, H. Magdelenat, and French Society of Clinical Cytology Study Group. 2003. Efficiency of the Hybrid Capture 2 HPV DNA test in cervical cancer screening. A study by the French Society of Clinical Cytology. Am. J. Clin. Pathol. 120492-499. [DOI] [PubMed] [Google Scholar]
- 7.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 8.Dianzani, C., S. Calvieri, A. Pierangeli, and A. M. Degener. 2004. Identification of human papillomaviruses in male dysplastic genital lesions. New Microbiol. 2765-69. [PubMed] [Google Scholar]
- 9.Federschneider, J. M., L. Yuan, J. Brodsky, G. Breslin, R. A. Betensky, and C. P. Crum. 2004. The borderline or weakly positive Hybrid Capture II HPV test: a statistical and comparative (PCR) analysis. Am. J. Obstet. Gynecol. 191757-761. [DOI] [PubMed] [Google Scholar]
- 10.Gale, N., M. Poljak, V. Kambič, D. Ferluga, and J. Fischinger. 1994. Laryngeal papillomatosis: molecular, histopathologic, and clinical evaluation. Virchows. Arch. 425291-295. [DOI] [PubMed] [Google Scholar]
- 11.Garland, S. M., M. Hernandez-Avila, C. M. Wheeler, G. Perez, D. M. Harper, S. Leodolter, G. W. Tang, D. G. Ferris, M. Steben, J. Bryan, F. J. Taddeo, R. Railkar, M. T. Esser, H. L. Sings, M. Nelson, J. Boslego, C. Sattler, E. Barr, L. A. Koutsky, and Females United to Unilaterally Reduce Endo/Ectocervical Disease (FUTURE) I Investigators. 2007. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N. Engl. J. Med. 3561928-1943. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, M. V., P. J. F. Snijders, A. J. C. van den Brule, T. J. M. Helmerhorst, C. J. L. M. Meijer, and J. M. M. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocjan, B. J., K. Seme, and M. Poljak. 2008. Detection and differentiation of human papillomavirus genotypes HPV-6 and HPV11 by FRET-based real-time PCR. J. Virol. Methods 153245-249. [DOI] [PubMed] [Google Scholar]
- 14.Lörincz, A. T. 1996. Hybrid Capture method for detection of human papillomavirus DNA in clinical specimens: a tool for clinical management of equivocal Pap smears and for population screening. J. Obstet. Gynaecol. Res. 22629-636. [DOI] [PubMed] [Google Scholar]
- 15.Meijer, C. J., J. Berkhof, P. E. Castle, A. T. Hesselink, E. L. Franco, G. Ronco, M. Arbyn, F. X. Bosch, J. Cuzick, J. Dillner, D. A. Heideman, and P. J. Snijders. 2009. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int. J. Cancer. 124516-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyton, C. L., M. Schiffman, A. T. Lorincz, W. C. Hunt, I. Mielzynska, C. Bratti, S. Eaton, A. Hildesheim, L. A. Morera, A. C. Rodriguez, R. Herrero, M. E. Sherman, and C. M. Wheeler. 1998. Comparison of PCR- and Hybrid Capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J. Clin. Microbiol. 363248-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfister, H., I. Hettich, U. Runne, L. Gissmann, and G. N. Chilf. 1983. Characterization of human papillomavirus type 13 from focal epithelial hyperplasia Heck lesions. J. Virol. 47363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poljak, M., A. Brenčič, K. Seme, A. Vince, and I. J. Marin. 1999. Comparative evaluation of first- and second-generation Digene Hybrid Capture assays for detection of human papillomaviruses associated with high or intermediate risk for cervical cancer. J. Clin. Microbiol. 37796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poljak, M., I. J. Marin, K. Seme, and A. Vince. 2002. Hybrid Capture II HPV test detects at least 15 human papillomavirus genotypes not included in its current high risk cocktail. J. Clin. Virol. 25(Suppl. 3)S89-S97. [DOI] [PubMed] [Google Scholar]
- 20.Potočnik, M., B. J. Kocjan, K. Seme, and M. Poljak. 2007. Distribution of human papillomavirus (HPV) genotypes in genital warts from males in Slovenia. Acta Dermatovenerol. Alp. Panonica Adriat. 1691-98. [PubMed] [Google Scholar]
- 21.Safaeian, M., R. Herrero, A. Hildesheim, W. Quint, E. Freer, L. J. Van Doorn, C. Porras, S. Silva, P. Gonzalez, M. C. Bratti, A. C. Rodriguez, and P. Castle. 2007. Comparison of the SPF10-LiPA system to the Hybrid Capture 2 Assay for detection of carcinogenic human papillomavirus genotypes among 5,683 young women in Guanacaste, Costa Rica. J. Clin. Microbiol. 451447-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiffman, M., C. M. Wheeler, A. Dasgupta, D. Solomon, and P. E. Castle. 2005. A comparison of a prototype PCR assay and Hybrid Capture 2 for detection of carcinogenic human papillomavirus DNA in women with equivocal or mildly abnormal Papanicolaou smears. Am. J. Clin. Pathol. 124722-732. [DOI] [PubMed] [Google Scholar]
- 23.Schneede, P., P. Hillemanns, F. Ziller, A. Hofstetter, E. Stockfleth, R. Arndt, and T. Meyer. 2001. Evaluation of HPV testing by Hybrid Capture II for routine gynecologic screening. Acta Obstet. Gynecol. Scand. 80750-752. [DOI] [PubMed] [Google Scholar]
- 24.Seme, K., K. Fujs, B. J. Kocjan, and M. Poljak. 2006. Resolving repeatedly borderline results of Hybrid Capture 2 HPV DNA test using polymerase chain reaction and genotyping. J. Virol. Methods 134252-256. [DOI] [PubMed] [Google Scholar]
- 25.Solomon, D., M. Schiffman, R. Tarone, and the ALTS Study Group. 2000. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J. Natl. Cancer Inst. 92397-402. [DOI] [PubMed] [Google Scholar]
- 26.Terry, G., L. Ho, P. Londesborough, J. Cuzick, I. Mielzynska-Lohnas, and A. Lorincz. 2001. Detection of high-risk HPV types by the Hybrid Capture 2 test. J. Med. Virol. 65155-162. [PubMed] [Google Scholar]
- 27.Yamazaki, H., T. Sasagawa, W. Basha, T. Segawa, and M. Inoue. 2001. Hybrid Capture-II and LCR-E7 PCR assays for HPV typing in cervical cytologic samples. Int. J. Cancer. 94222-227. [DOI] [PubMed] [Google Scholar]