A novel H1N1 subtype influenza A virus, derived by reassortment between two known circulating swine influenza strains, has now been declared a human pandemic influenza A virus (8). The pandemic virus, centered initially in Mexico and the United States, has now spread to 74 countries with the World Health Organization reporting 29,669 cases and 145 deaths as of 15 June 2009 (2, 7). The potential severity of this pandemic virus is currently unknown. Because the novel virus began circulating at the end of the 2008-2009 influenza season in the northern hemisphere, distinguishing it from other influenza A viruses, especially the circulating human H1N1 viruses, is crucial not only for pandemic planning, containment, and surveillance but also for treatment. The current human H1N1 viruses are now predominantly resistant to the neuraminidase inhibitor oseltamivir (4), while the novel swine origin H1N1 viruses are resistant to the adamantanes (2). To this end, we developed and evaluated several reverse transcription (RT)-PCR-based assays to distinguish the human lineage H1N1 and the novel swine origin virus and report here specific RT-PCR assays for human and swine origin H1N1 and also an RT-PCR assay with one primer set that can amplify a portion of the H1 subtype hemagglutinin (HA) gene segment from avian, swine, and human origin influenza A viruses and, in a sensitive, quantitative real-time assay, distinguish both the current human H1N1 virus and the novel H1N1 virus by using specific probes in a one-tube format. Each of these real-time assays uses primers designed to match highly conserved regions of the H1 HA gene.
Influenza A virus RNA was isolated from MDCK-grown viral stocks by using the QIAamp Viral RNA extraction kit (Qiagen Inc., Valencia, CA). The viruses used in the analysis are listed in the legend to Fig. 1. H1 HA sequences were aligned by using the MegAlign program (DNAStar Inc., Madison, WI) for design of H1 HA primers and specific probes (Table 1). First-strand cDNA was produced by using random primers with 4 μl viral RNA lysate in a 20-μl reaction mixture containing 200 U Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and following the manufacturer's instructions.
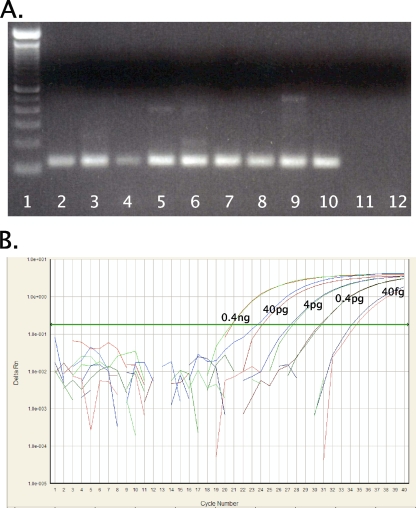
FIG. 1.
RT-PCR for H1 subtype HAs. (A) RT-PCR amplification with the universal H1 HA primer set (set 3). cDNAs from swine, human, and avian origin H1 subtype influenza A viruses were amplified. The final reaction mixture contained 1× PCR Gold buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mixture, 4 U AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA), and 0.6 μM primers. Thermocycling was performed in a DNAEngine (Bio-Rad, Hercules, CA) under the following cycling conditions: 10 min at 95°C, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. PCR products were electrophoresed on a 2% Tris-acetate-EDTA agarose gel, stained with ethidium bromide, and photographed under UV transillumination. Lanes (left to right): 1, 100-bp DNA molecular weight ladder; 2, A/swine/Ohio/23/1935(H1N1), 3, A/swine/Jamesburg/1942 (H1N1), 4, A/swine/Wisconsin/1/1967 (H1N1); 5, A/Maryland/NIH-37/2009 (H1N1); 6, A/Maryland/NIH-39/2009 (H1N1); 7, A/mallard/Ohio/171/1990 (H1N1); 8, A/green-winged teal/Ohio/72/1999(H1N1); 9, A/green-winged teal/Ohio/430/1987 (H1N1); 10, A/California/04/2009 (H1N1); 11, A/New York/470/2004 (H3N2); 12, water. (B) Amplification curve for influenza A virus H1 HA real-time RT-PCR assay. Shown are the 10−1 to 10−5 dilutions of A/California/04/2009 (H1N1) cDNA (final concentrations, 0.4 ng to 40 fg viral RNA per reaction mixture) with the FAM (novel swine origin) probe. Reactions were run in duplicate. The final reaction mixture contained 1× ABI master mix (Applied Biosystems), 0.9 μM primers, and 0.25 μM of each probe under the following cycling conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
TABLE 1.
RT-PCR primers and probes designed for this study
| Set and oligonucleotide | Sequence | Locationa |
|---|---|---|
| 1 | ||
| Novel swine origin H1 forward primer | 5′-TGTGAATCACTCTCCACAGCAAGC-3′ | 250-273 |
| Novel swine origin H1 probe | 5′-(FAM)CAGACAATGGAACGTGTTACCCAGGA(TAMRA)-3′b | 305-330 |
| Novel swine origin H1 reverse primer | 5′-ATTGGGCCATGAACTTGTCTTGGG-3′ | 403-426 |
| 2 | ||
| Human H1 forward primer | 5′-AAAGAAAGCTCATGGCCCAACCAC-3′ | 406-429 |
| Human H1 probe | 5′-(FAM)ACCGTAACCGGAGTGTCAGCATCAT(TAMRA)-3′ | 431-457 |
| Human H1 reverse primer | 5′-AAAGAAAGCTCATGGCCCAACCAC-3′ | 521-544 |
| 3 | ||
| H1 universal forward primer | 5′-ATTGCCGGTTTCATTGAAGG-3′ | 1048-1067 |
| Novel swine origin H1 probe | 5′-(FAM)ATGAGCAGGGGTCAGGATATGCAGCCGACC(TAMRA)-3′ | 1115-1144 |
| Human H1 probe | 5′-(VIC)ATGAGCAAGGATCTGGCTATGCTGCAGATC(TAMRA)-3′ | 1115-1144 |
| H1 universal reverse primer | 5′-ATGGCATTYTGTGTGCTYTT-3′ | 1147-1166 |
Oligonucleotides are numbered as aligned to the HA sequence of A/California/04/2009 (H1N1), GenBank accession no. GQ117044.
TAMRA, 6-carboxytetramethylrhodamine.
A conventional RT-PCR was performed with 2 μl cDNA in a 50-μl reaction mixture (the legend to Fig. 1 contains reaction details). PCR products were analyzed with a 2% Tris-acetate-EDTA agarose gel. A real-time RT-PCR was performed with a 7500 Real Time PCR system (ABI, Foster City, CA) with 1 μl cDNA in a 25-μl reaction mixture (the legend to Fig. 1 contains reaction details). A control real-time RT-PCR for the influenza A virus matrix gene was performed as described previously (6).
All influenza A virus cDNA samples were positive in the control matrix gene RT-PCR assay. The universal H1 primer set (set 3) was able to amplify cDNAs from H1 subtype influenza A viruses of avian, classical swine, human, and novel swine origins. Template cDNA from a human H3N2 virus was negative (Fig. 1A). The analytical sensitivities of the novel H1 TaqMan assays were determined. RT-PCRs were set up with template cDNA equivalent to 4 ng of viral RNA, followed by five 10-fold serial dilutions to 40 fg of viral RNA equivalent. In all three sets (Table 1), consistent amplification and detection were observed across 6 orders of magnitude (Fig. 1B). The estimated limit of influenza A virus detection was 40 fg viral RNA (∼5,200 viral genome copies) with the three TaqMan assays, similar to results obtained with other quantitative influenza A virus real-time assays (3). The specific real-time assays for both the novel swine origin H1 (set 1) and human (seasonal) H1 (set 2) sequences each demonstrated high specificity and did not result in cycle threshold (CT) values of ≤40 in the absence of a novel swine origin H1N1 or human H1N1 template, respectively. With the universal primer set (set 3), templates from H1 subtype viruses of avian, classical swine, human, and novel swine origins were all amplified (Fig. 1A); however, when run as a TaqMan assay, the human H1N1 template was only positive with the human H1 probe, while the novel swine origin H1N1 template was only positive with the novel swine H1 probe (Fig. 1B), demonstrating the specificity of the assay. Other real-time assays for the novel virus have been developed (1, 5). Because of rapid mutation in the novel swine origin H1N1 H1 HA gene, the TaqMan assay widely distributed for rapid testing (1) now has several mismatches in the reverse primer sequence, which may decrease its sensitivity. The advantage of the one-tube assay with two probes labeled with 6-carboxyfluorescein (FAM) and VIC reported here is that samples can be specifically identified as being either human H1N1 or novel swine origin H1N1 in one sensitive real-time RT-PCR.
Acknowledgments
This work was supported by the intramural program of the NIAID and the NIH.
We thank Ruben Donis, Richard Webby, and Matthew Memoli for providing influenza viral samples.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Centers for Disease Control and Prevention. 12 May 2009, posting date. H1N1 Flu (Swine Flu): Resources for Laboratories. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/h1n1flu/lab/.
- 2.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 22 May 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed]
- 3.Krafft, A. E., K. L. Russell, A. W. Hawksworth, S. McCall, M. Irvine, L. T. Daum, J. L. Connoly, A. H. Reid, J. C. Gaydos, and J. K. Taubenberger. 2005. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J. Clin. Microbiol. 431768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Layne, S. P., A. S. Monto, and J. K. Taubenberger. 2009. Pandemic influenza: an inconvenient mutation. Science 3231560-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon, L. L., K. H. Chan, G. J. Smith, C. S. Leung, Y. Guan, K. Y. Yuen, and J. S. Peiris. 21 May 2009. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin. Chem. doi: 10.1373/clinchem.2009.130229. [DOI] [PMC free article] [PubMed]
- 6.Runstadler, J. A., G. M. Happ, R. D. Slemons, Z. M. Sheng, N. Gundlach, M. Petrula, D. Senne, J. Nolting, D. L. Evers, A. Modrell, H. Huson, S. Hills, T. Rothe, T. Marr, and J. K. Taubenberger. 2007. Using RRT-PCR analysis and virus isolation to determine the prevalence of avian influenza virus infections in ducks at Minto Flats State Game Refuge, Alaska, during August 2005. Arch. Virol. 1521901-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. 12 June 2009, posting date. Influenza A (H1N1)—update 48. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/don/2009_06_12/en/index.html.
- 8.World Health Organization. 11 June 2009, posting date. World now at the start of 2009 influenza pandemic. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html.