Abstract
Approximately 84% of legionellosis cases are due to Legionella pneumophila serogroup 1. Moreover, a majority of L. pneumophila serogroup 1 clinical isolates react positively with monoclonal antibody 2 (MAb2) of the international standard panel. Over 94% of the legionellosis outbreaks investigated by the Centers for Disease Control and Prevention are due to this subset of L. pneumophila serogroup 1. To date, there is no complete explanation for the enhanced ability of these strains to cause disease. To better characterize these organisms, we subtyped 100 clinical L. pneumophila serogroup 1 isolates and 50 environmental L. pneumophila serogroup 1 isolates from the United States by (i) reactivity with MAb2, (ii) presence of a lag-1 gene required for the MAb2 epitope, and (iii) sequence-based typing analysis. Our results showed that the MAb2 epitope and lag-1 gene are overrepresented in clinical L. pneumophila serogroup 1 isolates. MAb2 recognized 75% of clinical isolates but only 6% of environmental isolates. Similarly, 75% of clinical isolates but only 8% of environmental isolates harbored lag-1. We identified three distinct lag-1 alleles, referred to as Philadelphia, Arizona, and Lens alleles, among 79 isolates carrying this gene. The Arizona allele is described for the first time in this study. We identified 59 different sequence types (STs), and 34 STs (58%) were unique to the United States. Our results support the hypothesis that a select group of STs may have an enhanced ability to cause legionellosis. Combining sequence typing and lag-1 analysis shows that STs tend to associate with a single lag-1 allele type, suggesting a hierarchy of virulence genotypes. Further analysis of ST and lag-1 profiles may identify genotypes of L. pneumophila serogroup 1 that warrant immediate intervention.
Legionellae are facultative intracellular gram-negative bacteria found within biofilms and freshwater and industrial water systems worldwide (10). They are intracellular parasites of freshwater protozoa and are also able to multiply within mammalian macrophages and epithelial cells (5, 10). Legionellae are transmitted to humans through inhalation of aerosols generated from water containing the bacteria. The bacteria subsequently enter and multiply within alveolar macrophages, initiating the infection. Although over 50 species of legionellae have been identified (http://www.bacterio.cict.fr/l/legionella.html), more than 90% of the isolates associated with Legionnaires' disease are Legionella pneumophila, and up to 84% of these are L. pneumophila serogroup 1 (10, 39).
An international panel of seven monoclonal antibodies (MAbs), including MAb2, was developed in 1986 to discriminate among a fairly heterogeneous group of bacteria that belong to L. pneumophila serogroup 1 (20). An antibody with identity to MAb2 was developed by Helbig et al. and designated MAb3/1 (18). MAb2 and MAb3/1 have been demonstrated to bind to the same L. pneumophila serogroup 1 strains (18, 25). A series of studies indicate that the majority (65% to 100%) of L. pneumophila serogroup 1 clinical isolates, especially ones associated with community-acquired outbreaks and travel-associated infections, react with MAb2 or MAb3/1 (3, 4, 6, 14, 15, 17, 33, 38). In contrast, only 14.9% to 35% of L. pneumophila serogroup 1 environmental isolates not associated with human infection react with MAb2 or MAb3/1 (6, 14, 15, 38). MAb2- or MAb3/1-positive L. pneumophila serogroup 1 isolates have been implicated in approximately 94% of the outbreaks of legionellosis investigated by the Centers for Disease Control and Prevention (B. S. Fields, CDC, personal communication). This is based upon a select number of outbreak investigations, which included matching clinical and environmental isolates from an epidemiologically implicated reservoir.
MAb2 and MAb3/1 recognize an epitope within the 8-O-acetyl group of the legionaminic acid moiety of L. pneumophila serogroup 1 lipopolysaccharide (18, 40, 41). Since this epitope is expressed on the majority of clinical isolates of L. pneumophila, it was designated a virulence-associated epitope by Helbig et al. (18). The lag-1 gene encodes an O-acetyltransferase responsible for the O acetylation of legionaminic acid (41). Although it was shown that a functional lag-1 gene is required for expression of the MAb2 epitope, the molecular basis for the apparently increased virulence of MAb2-positive isolates remains elusive. Lück et al. reported that the wild-type L. pneumophila serogroup 1 strain Corby and its spontaneous lag-1 mutant were indistinguishable in serum resistance and in uptake and intracellular growth in both Acanthamoeba castellanii and macrophages (25). Sequence analysis of the lag-1 locus in several clinical and environmental L. pneumophila serogroup 1 isolates from Europe showed that all MAb2-postitive isolates harbored the lag-1 gene, whereas MAb2-negative isolates either lost lag-1 completely or contained missense mutations or insertions within this gene (2, 26).
Multiple guidelines and official recommendations for preventing the transmission of Legionnaires' disease have been developed by many governments. Many of these guidelines recommend action items based upon certain concentrations of legionellae or upon the number of sites which are positive for the bacteria (1, 11). None of these guidelines discriminates between the large number of Legionella species or serogroups, even though it is established that they differ in their capacities for causing disease (10). Better characterization of the strains which cause the majority of disease should allow more-targeted intervention measures.
The goal of this study was to characterize the prevalences of the MAb2 epitope and the lag-1 gene in the U.S. clinical and environmental isolates to determine if they may serve as markers for enhanced virulence. We further characterized these isolates by using the most recent molecular typing tool for L. pneumophila, sequence-based typing (SBT) analysis. This method is rapid, highly discriminatory, and portable (12, 13, 30). The SBT database, which is available through the website of the European Working Group for Legionella Infections (EWGLI) (www.ewgli.org), allows assignment of the seven-digit allelic profile for alleles in a predetermined order (flaA, pilE, asd, mip, mompS, proA, and neuA) and a corresponding sequence type (ST) to L. pneumophila isolates. SBT is based on the same approach as multilocus sequence typing (28) and, similarly to multilocus sequence typing, may be applied to the study of genetic diversity and clonal expansion of microbial populations (7, 37). Analysis of the phylogenetic relationships between L. pneumophila serogroup 1 STs identified in our study was used to provide insight on how the MAb2 epitope might have emerged and imparted enhanced virulence.
(Some of this work was presented previously as posters at the 107th General Meeting of the American Society for Microbiology, Toronto, Ontario, Canada [23], and the 108th General Meeting of the American Society for Microbiology, Boston, MA [22].)
MATERIALS AND METHODS
L. pneumophila serogroup 1 isolates.
One hundred clinical isolates received through the CDC Data and Specimen Handling system were selected from the CDC reference diagnostics library (Atlanta, GA). No two isolates from a single location and similar time periods were included. The isolates were obtained from 22 states between May 2001 and October 2006. The isolates had no known epidemiologic linkage. Each isolate was from a different patient. Ninety-three of the isolates were from sporadic cases, and seven isolates were from seven outbreaks that took place in different states and at different times, as follows: Pennsylvania, 2002; Vermont, 2002; New Mexico, 2005; North Carolina, 2005; South Dakota, 2005; Connecticut, 2006; and Texas, 2006. A single isolate from each of the seven outbreaks was included in this study. Fifty environmental samples were randomly selected from PathCon Laboratories' (Norcross, GA) culture collection of isolates obtained from water samples collected from buildings with no known association with cases of Legionnaires' disease.
Legionella MAb test.
The isolates were MAb typed by an immunodot method using MAb1 (20), MAb75 (29), and MAb2 (20), as previously described (32). Briefly, a dense suspension of L. pneumophila cultures was prepared in 0.6% formalin-phosphate-buffered saline, immobilized on a nitrocellulose membrane, and washed with phosphate-buffered saline. Then, the antigen was overlaid with the MAbs. A mix of MAb1 and MAb75 was used to confirm that the L. pneumophila samples tested were serogroup 1; MAb2 was used to identify MAb2-positive samples. After the excess MAbs were washed away, a goat anti-mouse antibody labeled with horseradish peroxidase (Bio-Rad, Hercules, CA) was added. The membrane was washed again and placed in peroxidase substrate (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) to allow colorimetric detection of the antigen/antibody reaction.
DNA extraction, PCR amplification, and sequencing.
Genomic DNA was extracted either by using the QIAamp DNA minikit (Qiagen Inc., Valencia, CA) according to manufacturer's guidelines or by emulsifying two colonies of L. pneumophila in 30 μl sterile water and heating for 10 min at 100°C. The PCR was performed on GeneAmp PCR system 9600 (Applied Biosystems, Foster City, CA) using either Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) for amplicons up to 2 kb or Elongase enzyme mix (Invitrogen) for amplicons longer than 2 kb. PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH). DNA sequencing was performed with the ABI BigDye Terminator v1.1 or v3.1 cycle sequencing kit (Applied Biosystems), and the products were analyzed on a model 3130xl ABI Prism genetic analyzer (Applied Biosystems).
Sequence analysis of lag-1 locus in L. pneumophila isolates.
The primers lag-F (5′-CTCACAACAAGTCAAGCAAC-3′) and lag-R (5′-AAACCATACCAAAGCAACAT-3′) were used for amplification of an internal lag-1 fragment (base pairs 13 to 597), which contains the region encoding the active center of acylation enzymes. The same primer pair was used to sequence internal lag-1 PCR products from all MAb2-positive isolates. The primers lag_LngD-F (5′-GGAAATTATACCGAACGATGTACTTTGTATTCACC-3′), which anneals to the 3′ region of lpg0773 (L. pneumophila Philadelphia-1 strain), and lag_LngD-R (5′-ATGGCCATAGAAATCTAGTTTGGACCTATTTCA-3′), which anneals to the 5′ region of lpg0779 (L. pneumophila Philadelphia-1 strain), were used to amplify a fragment up to 5.5 kb long that included the lag-1 gene and flanking regions. The size of the amplicons was estimated after separation by agarose gel electrophoresis using a 0.8% E-gel (Invitrogen). PCR products of 2.5 kb from all MAb2-negative clinical isolates and 24/47 MAb2-negative environmental isolates were sequenced using the following primers: lag_LngD-F, lag_LngD-R, Int-ORF3-F (5′-ATGAACAAATATGGTGGTGC-3′), Int-orf3-R (5′-TCCACTTTCAGTAAAATTGCT-3′), orf3UpSeq-F (5′-CTTTTATTGAAACTTTAGAGTCGTT-3′), orf3DnSeq-F (5′-AAATGTAATATTATTTCTGTTTCCT-3′), orf2UpSeq-R (5′-TAACTCTAAATTTGGGGGG-3′), and orf2DnSeq-R (5′-CTCCACCAAACAAATTCC-3′). Five 4-kb PCR products from the isolates with an internal lag-1 sequence homologous to the Lens lag-1 allele were sequenced using primers lag_LngD-F, lag_LngD-R, lag-R, Int-ORF3-F, orf2DnSeq-R, and orf3UpSeq-F. Sequences of six 4-kb PCR products from isolates with an internal lag-1 fragment homologous to the Arizona lag-1 allele were determined using the following primers: lag_LngD-F, lag_LngD-R, lag-F, lag-R, Int-orf3-R, lag_Uint_R (5′-TAGCAAAAGAGCTTTTATGAAAAAAAA-3′), lag_dwn_F (5′-TATGGTTTCTGGGTTCTGGG-3′), lag-low (5′-ATTGAGCCGACGAATTT-3′), and Az_seq30 (5′-CGAAGAAAATTTAGGTAGGCT-3′). Finally, five 5.5-kb PCR products from isolates with an internal lag-1 sequence homologous to the Philadelphia lag-1 allele were sequenced using primers lag_LngD-F, lag_LngD-R, lag-F, lag-R, lag_Uint_R, Int-ORF3-F, orf3UpSeq-F, orf2DnSeq-R, and lag-up (5′-ACCAGGTATGCCTGATAAA-3′).
SBT.
Genotyping was performed using the seven-gene protocol from the EWGLI SBT scheme as previously described (12, 30). Sequences were compared to previously assigned alleles by using the online Legionella SBT Quality Tool (www.hpa-bioinformatics.org.uk/cgi-bin/legionella/sbt/seq_assemble_legionella1.cgi). For each isolate, the combination of seven alleles at each of the loci was defined as a seven-digit allelic profile by using the predetermined order flaA, pilE, asd, mip, mompS, proA, and neuA (e.g., 1-4-3-1-1-1-1) and an ST represented by a number (e.g., ST1). New alleles and STs encountered for the first time in this study were submitted to the EWGLI SBT database (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php).
Phylogenetic analysis.
The examination of relationships between STs within clonal complexes was conducted using the eBURST v3 website (http://eburst.mlst.net). The stringent group definition, according to which a clonal complex consists of STs that share 6/7 alleles with at least one other member of the group and are all believed to be descended from the same founding genotype (the primary founder), was used (8). Comparative eBURST analysis was used to compare 150 L. pneumophila serogroup 1 isolates from the United States with 2,054 L. pneumophila serogroup 1 isolates submitted to the EWGLI database from Europe, Asia, and North America as of 11 December 2008. The SBT profiles of 2,054 L. pneumophila serogroup 1 isolates deposited in EWGLI SBT database were available to the database password holders. The evolutionary relationship between STs was also inferred using the neighbor-joining method (31) with concatenated SBT alleles. The evolutionary distances were computed using the Jukes-Cantor method (21), and the optimal tree was drawn. The bootstrap test (with 1,000 bootstrap replicates) was used to examine the confidence in the tree (9). The analyses were conducted with the MEGA4 software (34).
Statistical analysis.
Proportions of MAb2-positive and lag-1-positive isolates as well as the STs and lag-1 alleles were compared using the chi-square test in the SAS Proc Freq program (SAS v9.2; SAS Institute, Cary, NC). Diversity was estimated by calculating Hunter and Gaston's modification of Simpson's index of diversity (19) as previously described (15). The implicit normal distributions for indexes of diversity (IODs) for clinical and environmental isolates were compared using the t test (SAS Proc Ttest).
Nucleotide sequence accession numbers.
The sequences of DNA fragment encompassing the lag-1, open reading frame 2 (ORF2), and ORF3 genes (27) of Arizona isolate D5597 and the lag-1 of MAb2-negative isolate E73 were submitted to GenBank under accession numbers EU868813 and FJ899835, respectively.
RESULTS
Reactivity with MAb2.
MAb2 recognized 75/100 (75%) clinical isolates and 3/50 (6%) environmental isolates, with a P value of <0.005 (chi-square test on proportions) (Tables 1 and 2). All seven clinical isolates associated with seven distinct legionellosis outbreaks were MAb2 positive (Table 1).
TABLE 1.
STs of the 100 L. pneumophila serogroup 1 clinical samples isolated in the United States
| STa | SBT profile (flaA-pilE-asd-mip-mompS-proA-neuA) | No. of isolates with indicated result
|
State of origin (no. of isolates or outbreak name)b | |||
|---|---|---|---|---|---|---|
| MAb2
|
lag-1
|
|||||
| Positive | Negative | Positive | Negative | |||
| ST1 | 1-4-3-1-1-1-1 | 4 | 15 | 4 | 15 | AZ (5), GA, HI, IL (6), NH, OH (4), SD |
| ST18 | 2-10-9-13-2-5-6 | 1 | 0 | 1 | 0 | CT |
| ST22 | 2-3-6-10-2-1-6 | 0 | 2 | 0 | 2 | GA, NH |
| ST36 | 3-4-1-1-14-9-1 | 4 | 0 | 4 | 0 | NC, NJ, TX (TX06-1), VA (NC05-1) |
| ST37 | 3-4-1-1-14-9-11 | 6 | 0 | 6 | 0 | AZ (3), IL, ME, SD |
| ST42 | 4-7-11-3-11-12-9 | 4 | 0 | 4 | 0 | AZ, IN, OH, SD (SD05-1) |
| ST44 | 4-8-11-10-10-12-2 | 1 | 0 | 1 | 0 | AZ |
| ST59 | 7-6-17-3-13-11-11 | 0 | 2 | 0 | 2 | OH, WI |
| ST62 | 8-10-3-15-18-1-6 | 5 | 0 | 5 | 0 | IL (2), IN, ME, OH |
| ST89 | 4-10-11-15-29-1-6 | 2 | 0 | 2 | 0 | AZ (2) |
| ST94 | 12-8-11-5-20-12-2 | 3 | 0 | 3 | 0 | OH (2), PA |
| ST109 | 5-1-22-15-6-10-6 | 2 | 0 | 2 | 0 | CO, KY |
| ST154 | 11-14-16-16-15-13-2 | 0 | 1 | 0 | 1 | IL |
| ST205 | 5-10-22-15-6-1-6 | 3 | 0 | 3 | 0 | CO, OH (2) |
| ST213 | 2-19-5-10-18-1-2 | 8 | 0 | 8 | 0 | CT (CT06-1), IL, OH (2), RI (4) |
| ST222 | 2-19-5-10-18-1-10 | 8 | 0 | 8 | 0 | CT, IL (4), OH (2), VT (VT02-1) |
| ST224 | 4-8-11-16-42-12-2 | 1 | 0 | 1 | 0 | OH |
| ST251*# | 21*-14-29*-35*-15-29*-10 | 1 | 0 | 1 | 0 | OH |
| ST252* | 22*-4-3-1-1-30*-1 | 0 | 1 | 0 | 1 | NM |
| ST253*# | 2-10-5-10-38*-5-10 | 1 | 0 | 1 | 0 | NM |
| ST256* | 6-10-14-5-39*-14-9 | 1 | 0 | 1 | 0 | IL |
| ST258*# | 4-8-11-16-43*-12-2 | 1 | 0 | 1 | 0 | OH |
| ST259* | 21*-27*-28*-2-15-29*-6 | 2 | 0 | 2 | 0 | CO, PA (PA02-1) |
| ST260*# | 12-8-11-23-29-26-2 | 0 | 1 | 0 | 1 | AZ |
| ST266* | 8-10-3-10-18-1-6 | 1 | 0 | 1 | 0 | IN |
| ST267*# | 3-4-1-1-8-9-1 | 0 | 1 | 0 | 1 | OH |
| ST268*# | 6-10-14-3-21-4-2 | 1 | 0 | 1 | 0 | GA |
| ST269*# | 7-10-17-3-13-11-11 | 0 | 1 | 0 | 1 | GA |
| ST270*# | 5-1-22-16-6-10-2 | 1 | 0 | 1 | 0 | OH |
| ST271*# | 6-10-19-5-19-4-9 | 2 | 0 | 2 | 0 | AZ, PA |
| ST272*# | 8-10-3-15-18-1-20 | 1 | 0 | 1 | 0 | RI |
| ST273*# | 6-10-19-3-19-4-2 | 0 | 1 | 0 | 1 | AZ |
| ST274*# | 4-7-11-15-11-12-9 | 1 | 0 | 1 | 0 | OH |
| ST275*# | 2-10-3-5-19-4-9 | 1 | 0 | 1 | 0 | NM (NM05-1) |
| ST276*# | 2-19-11-10-18-12-2 | 1 | 0 | 1 | 0 | CT |
| ST278* | 6-10-15-28-4-14-11 | 1 | 0 | 1 | 0 | SD |
| ST288*# | 12-8-11-16-29-12-2 | 2 | 0 | 2 | 0 | OH (2) |
| ST289*# | 2-19-11-10-18-1-2 | 2 | 0 | 2 | 0 | IN, NY |
| ST290*# | 4-8-11-10-29-12-10 | 1 | 0 | 1 | 0 | CO |
| ST291*# | 6-10-14-10-13-3-9 | 1 | 0 | 1 | 0 | AZ |
| ST294*# | 8-3-3-15-21-1-6 | 1 | 0 | 1 | 0 | AZ |
STs found more than twice among 150 L. pneumophila serogroup 1 isolates are shown in boldface. *, ST/SBT allele identified for the first time in this study; #, ST unique to the United States as of 10 June 2009.
Outbreaks with which the listed STs were associated are indicated in parentheses. PA02-1, Pennsylvania, 2002; VT02-1, Vermont, 2002; NM05-1, New Mexico, 2005; NC05-1, North Carolina, 2005; SD05-1, South Dakota, 2005; CT06-1, Connecticut, 2006; TX06-1, Texas, 2006.
TABLE 2.
STs of the 50 environmental samples isolated in the United States
| STa | SBT profile (flaA- pilE-asd-mip-mompS- proA-neuA) | No. of isolates with indicated result
|
State of origin (no. of isolates) | |||
|---|---|---|---|---|---|---|
| MAb2
|
lag-1
|
|||||
| Positive | Negative | Positive | Negative | |||
| ST1 | 1-4-3-1-1-1-1 | 1 | 19 | 1 | 19 | CA (2), MA, MD (3), MI (2), MN, NJ (3), NY (3), OH, PA, TN, TX (2) |
| ST7 | 1-4-3-1-1-1-6 | 0 | 1 | 0 | 1 | IL |
| ST8 | 1-4-3-1-1-1-9 | 0 | 8 | 0 | 8 | CA, DE, MD (2), OH (2), TN, TX |
| ST36 | 3-4-1-1-14-9-1 | 1 | 0 | 1 | 0 | MN |
| ST154 | 11-14-16-16-15-13-2 | 0 | 2 | 0 | 2 | FL, RI |
| ST257*# | 12-8-11-5-40*-12-22* | 1 | 0 | 1 | 0 | NH |
| ST261*# | 7-28*-17-3-13-11-3 | 0 | 1 | 0 | 1 | IL |
| ST262*# | 7-6-30*-2-12-11-11 | 0 | 1 | 0 | 1 | TX |
| ST263*# | 7-10-31*-3-13-11-11 | 0 | 1 | 0 | 1 | MS |
| ST264*# | 22*-6-3-1-1-30*-2 | 0 | 1 | 0 | 1 | LA |
| ST265*# | 8-10-3-15-18-1-21* | 0 | 1 | 0 | 1 | GA |
| ST279*# | 6-10-1-28-4-14-11 | 0 | 1 | 0 | 1 | MN |
| ST280*# | 2-10-19-28-21-14-6 | 0 | 1 | 0 | 1 | TX |
| ST281*# | 7-6-17-28-4-14-11 | 0 | 1 | 1 | 0 | MS |
| ST282*# | 6-23-17-28-21-4-11 | 0 | 2 | 0 | 2 | TX (2) |
| ST283*# | 6-23-19-28-19-4-11 | 0 | 1 | 0 | 1 | CA |
| ST284*# | 1-6-3-10-1-1-11 | 0 | 1 | 0 | 1 | CA |
| ST285*# | 2-10-14-10-19-4-3 | 0 | 1 | 0 | 1 | WY |
| ST286*# | 1-4-3-1-14-1-1 | 0 | 1 | 0 | 1 | MA |
| ST287*# | 1-12-1-1-1-9-2 | 0 | 1 | 0 | 1 | MI |
| ST296* | 1-4-3-1-1-1-11 | 0 | 2 | 0 | 2 | CA, MA |
STs found more than twice among 150 L. pneumophila serogroup 1 isolates are shown in boldface. *, ST/SBT allele identified for the first time in this study; #, ST unique to the United States as of 10 June 2009.
lag-1 locus.
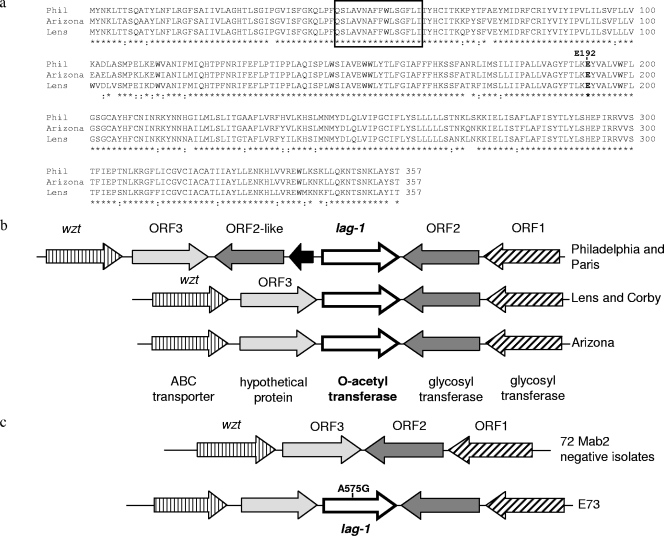
PCR amplification and sequencing of the lag-1 locus in all 150 L. pneumophila serogroup 1 isolates showed that the lag-1 gene was present in every MAb2-positive isolate (Tables 1 and 2). In contrast, this gene was missing in 72/73 MAb2-negative isolates (Tables 1 and 2). Seventy-five percent of clinical isolates and 8% of environmental isolates harbored the gene (Tables 1 and 2), with a P value of <0.0001 (chi-square test on proportions). Sequence analysis of a 510-bp internal DNA fragment of lag-1 in all 79 lag-1-positive isolates and of the entire 1,074-bp gene in 16 representative isolates revealed three alleles of lag-1, which we named after their representative strains: Philadelphia-1 (Philadelphia), Lens, and Arizona. Specifically, 16 isolates had lag-1 sequences that were 100% identical to the lag-1 sequences of both the Philadelphia-1 and Paris strains (GenBank accession numbers NC_002942 and NC_006368, respectively). Thirty-nine isolates had lag-1 sequences that were 100% identical to the lag-1 sequence of Lens and Corby strains (GenBank accession numbers NC_006369 and NC_009494, respectively). The Arizona allele was identified for the first time in this study; 24 isolates had lag-1 sequences that were 100% identical to the lag-1 sequence of the representative L. pneumophila serogroup 1 isolate D5597, which originated from Arizona. Multiple sequence alignment of the three different lag-1 alleles indicated that there were 94%, 90%, and 89% sequence identities between Philadelphia/Arizona, Philadelphia/Lens, and Arizona/Lens alleles, respectively (data not shown). The amino acid sequences of O-acetyltransferases encoded by lag-1 also differed between strains (Fig. 1a), with 94%, 92%, and 91% identities for Philadelphia/Arizona, Philadelphia/Lens, and Arizona/Lens O-acetyltransferase proteins, respectively. However, a highly conserved amino acid motif, V-X-X-F-F-X-I/V/L-S-G-F/W/Y, which composes the active center of acylation enzymes (25), appeared identically in all lag-1-encoded proteins (Fig. 1a).
FIG. 1.
Compositions and sequences of lag-1 loci differ in Philadelphia-1, Lens, and Arizona strains. (a) Sequences of the O-acetyltransferase encoded by the Philadelphia (Phil), Lens, and Arizona alleles of lag-1 were aligned using CLUSTAL_W. Identical residues are denoted below the alignment by asterisks, and positions of strongly conserved residues are indicated by colons. The highly conserved amino acid motif composing the active center of acylation enzymes is enclosed within the rectangle. Glutamate 192, which was mutated to glycine in the MAb2-negative environmental isolate E73, is in boldface. (b) A schematic representation of the lag-1 loci from sequenced and annotated genomes of L. pneumophila serogroup 1 MAb2-positive strains Philadelphia-1, Paris, Lens, and Corby is shown. The reference loci are compared to the lag-1 locus identified in a group of L. pneumophila serogroup 1 MAb2-positive clinical isolates, the representative member of which originated in Arizona. The designations of ORF1 to ORF3 are according to Lüneberg et al. (27). (c) The majority of L. pneumophila serogroup 1 MAb2-negative isolates analyzed in our study lack the lag-1 gene, with ORF2 and ORF3 adjacent to each other and highly homologous to their respective loci in Philadelphia-1 strain. In a single isolate, E73, the lag-1 was present in a configuration similar to the Arizona type but contained a single-nucleotide substitution, A575G.
Comparison of the lag-1 region in the published genomes of Corby, Lens, Paris, and Philadelphia-1 strains with the lag-1 locus in Arizona isolate D5597 revealed differences in the compositions of genes adjacent to lag-1. In Lens and Corby strains as well as in isolates with the Arizona type lag-1 allele, the lag-1 gene was flanked by ORF3 and ORF2 of the L. pneumophila lipopolysaccharide biosynthesis cluster described by Lüneberg et al. (27) (Fig. 1b). ORF2 encodes a glycosyl transferase, whereas ORF3 encodes a hypothetical protein unique to Legionella. As shown in Fig. 1b, the lag-1 locus in Philadelphia-1 and Paris strains contained additional genes, namely, a small transposase-encoding gene and an ORF2-like gene, which shared 96% identity with Philadelphia-1 ORF2 and was predicted to encode a glycosyl transferase.
In the 72 MAb2-negative L. pneumophila serogroup 1 isolates that did not carry the lag-1 gene, ORF2 and ORF3 were adjacent to each other (Fig. 1c) and highly homologous to their respective loci in the Philadelphia-1 strain. Environmental isolate E73 was the only MAb2-negative isolate that carried a full-length lag-1 gene of the Arizona allele and had an Arizona-like composition of the lag-1 locus (Fig. 1c). Analysis of the E73 lag-1 sequence (GenBank accession number FJ899835) revealed a single-nucleotide substitution at position 575 that resulted in an amino acid exchange from polar glutamate to neutral glycine (Fig. 1a). The affected glutamate 192 is conserved in lag-1-encoded O-acetyltransferases from Arizona, Philadelphia-1, and Lens strains.
SBT analysis.
SBT analysis assigned the 150 L. pneumophila serogroup 1 isolates in this study to 59 different STs, 40 (68%) of which were identified for the first time. In addition, 34 STs (58%) were unique to the United States (Tables 1 and 2). We also identified 18 new alleles of SBT genes: 2 of flaA, 2 of pilE, 4 of asd, 1 of mip, 4 of mompS, 2 of proA, and 3 of neuA (Tables 1 and 2). Three STs (ST205, ST213, and ST222) were found only in the United States and Canada. Of the 11 STs that were seen more than twice, 2 STs (ST213 and ST222) were previously identified in Canada, and the other 9 STs occurred throughout the world (Tables 1 and 2). The majority of isolates that belonged to the 11 most prevalent STs were clinical isolates, including 5 isolates collected during outbreak investigations (Table 1). The exceptions were ST8, found exclusively among environmental isolates, and ST1, ST36, and ST154, which were identified in both clinical and environmental isolates.
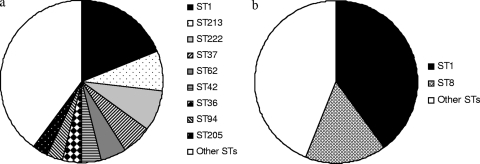
Clinical isolates showed more diversity than environmental isolates. Among 100 clinical isolates, 41 STs were identified, 24 of which were represented by single isolates (IOD, 0.946). The three most frequent STs (ST1, ST213, and ST222) accounted for 35% of the total clinical isolates (Fig. 2a and Table 1). Among 50 environmental isolates, 21 STs were found, 16 of which were represented by single isolates (IOD, 0.822). The two most frequent STs (ST1 and ST8) accounted for 56% of the total environmental isolates (Fig. 2b and Table 2). The IODs of the clinical and environmental isolates differed significantly (P value of <0.0001 by t test for comparison of normally distributed IODs). This difference was mainly caused by the proportion of isolates belonging to ST1, since 40% of environmental isolates but only 19% of clinical isolates belonged to this ST.
FIG. 2.
Diversities of clinical and environmental isolates based on ST analysis. Frequencies of STs among clinical (a) and environmental (b) isolates. Each fill pattern represents an ST comprised of three or more isolates. STs representing two or less isolates are included as “other STs”.
eBURST analysis.
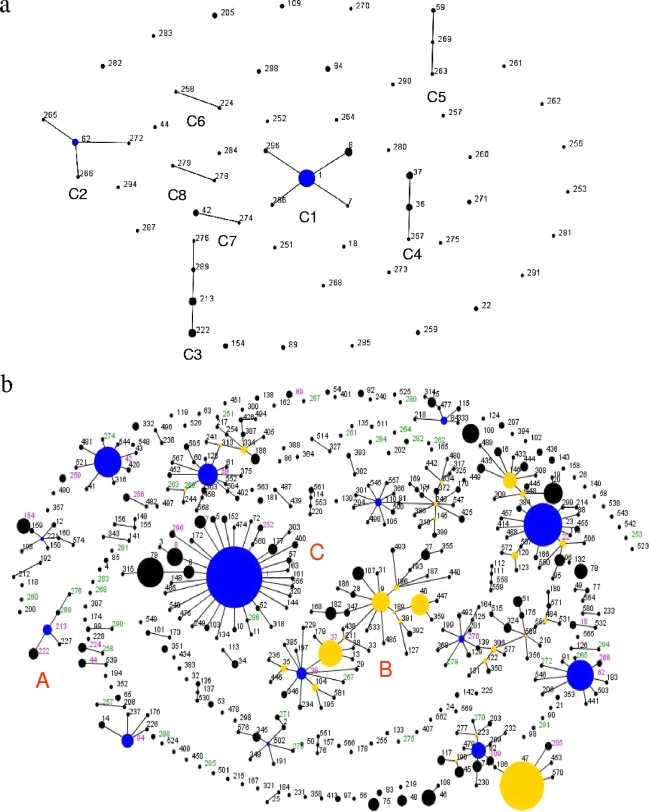
We examined relationships between STs identified in our study by using eBURST analysis (8). As shown in Fig. 3a, 25 STs from our study were predicted to form eight clonal complexes, whereas 34 STs differ from every other ST in two or more genes and were thus identified as singletons. Clonal complex C1 (Fig. 3a) consisted of the highest number of STs (ST1, ST8, ST296, ST7, and ST286), and 51/150 (34%) isolates belonged to this complex.
FIG. 3.
Phylogenetic relationships between L. pneumophila serogroup 1 STs identified by eBURST analysis. Blue circles indicate the predicted primary founder of a clonal complex. Yellow circles denote a subgroup founder (an ST that has at least two descendant single locus variants). The area of each circle represents the prevalence of the ST in the input data. Only single-locus variant links are shown. (a) A population snapshot of 59 STs identified in our study: 25 STs were grouped into eight clonal complexes (C1 to C8), and 34 STs were singletons. (b) Comparative analysis between the U.S. and EWGLI STs. The population snapshot contained 38 clonal complexes and 115 singletons. STs in green font were first identified in our study and are unique to the United States. STs in black font were listed in the EWGLI database but not found among our samples. STs in magenta font were found in both the EWGLI database and our sample pool.
Figure 3b displays an eBURST-generated “population snapshot” of all L. pneumophila serogroup 1 STs listed in the EWGLI database and from our study. Out of 34 STs unique to the United States, 16 STs belonged to already established clonal complexes, but 17 STs did not relate to any other ST and remained assigned as singletons. L. pneumophila serogroup 1 isolates from clonal complex A, consisting of ST213, ST222, ST227, ST289, and ST276, were isolated from either the northeastern United States or Ontario, Canada, or both (Table 1) (36).
As shown in Fig. 3b, clonal complex B was the largest clonal complex, containing 45 STs. ST36 was predicted to be a primary founder, since it had 10 single-locus variants, representing the highest number in the group. Two of the four ST36 clinical isolates from our study were implicated in two separate outbreaks (Table 1). The Philadelphia-1 strain, isolated from the seminal 1976 outbreak, belonged to ST37, which shares 6/7 alleles with ST36.
MAb2 reactivity and lag-1 distribution within the ST1 clonal complex.
The primary founder of clonal complex C (Fig. 3b) was the most common sequence type, ST1. In our study, ST1 was represented by equal numbers of clinical and environmental isolates, the majority of which did not react with MAb2 (Tables 1 and 2). Similar trends were observed for isolates listed in the EWGLI database belonging to ST1 and related STs. Specifically, in clonal complex C, there were 162 MAb2-negative ST1 isolates and only 57 MAb2-positive ones. Of those that were MAb2 negative, 77% were environmental isolates (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php). Sequencing analysis of the lag-1 locus in MAb2-negative isolates that belonged to ST1 showed that they carried ORF2 and ORF3, which were highly homologous to the respective loci in the Philadelphia-1 strain and that they completely lacked the lag-1 gene (Fig. 1c). On the other hand, ST1 MAb2-positive isolates had a classical Philadelphia-1 locus with highly homologous ORF2 and ORF2-like sequences flanking the lag-1 gene (Fig. 1b).
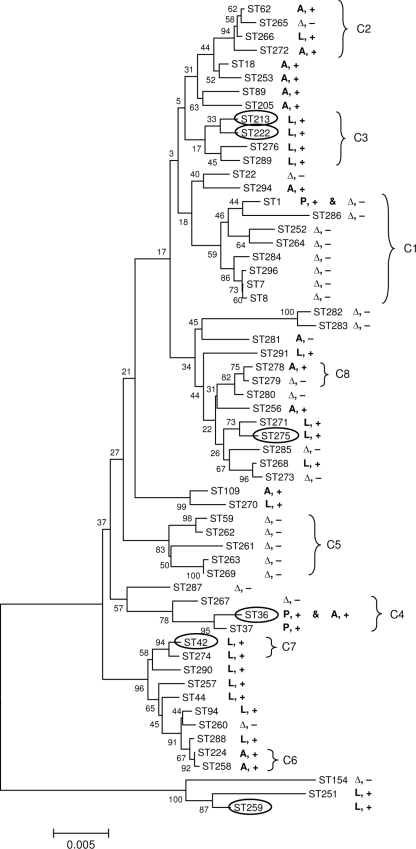
Neighbor-joining tree analysis.
The neighbor-joining method (31) was used to infer the phylogenetic relationship between STs, based on concatenated sequences of SBT alleles (Fig. 4). We found an incomplete correlation between the groups delineated by the neighbor-joining analysis and the clonal complexes determined by eBURST. However, 94% and 78% of 1,000 bootstrap trees supported the delineation of clonal complexes C2 and C4, respectively, from other STs (Fig. 3a and 4). Analysis of the distribution of lag-1 allele types and reactivity with MAb2 showed a strong correlation between sequence-based type and lag-1 allele (Fig. 4), with a P value of <0.0015 (chi-square test on proportions); there was a single lag-1 allele type for each ST, with the exception of ST36. However, there were no separate lineages for MAb2-positive and MAb2-negative strains and no indication that a MAb2-positive strain emerged within a MAb2-negative lineage.
FIG. 4.
Correlations between STs, presence of lag-1, and reactivity with MAb2. The evolutionary relationships between STs were inferred using the neighbor-joining method. The optimal tree is shown. The percentages of replicate trees in which the associated STs clustered together in the bootstrap test are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are in the units of the number of base substitutions per site. The allele type of the lag-1 gene of the strains within each ST is indicated as A (Arizona), L (Lens), or P (Philadelphia). Δ, lag-1 gene was missing; +, reactivity with MAb2; −, failure to react with MAb2. STs of strains isolated from outbreaks are in ovals. Correlation with clonal complexes (C1 to C8) identified by eBURST analysis is indicated by brackets.
DISCUSSION
MAb2-positive L. pneumophila serogroup 1 strains are responsible for the majority of outbreaks of Legionnaires' disease reported to the Centers for Disease Control and Prevention. Thus, it is important to understand the occurrence and distribution of MAb2-positive legionellae in the environment and to learn how the MAb2 epitope imparts enhanced virulence in these bacteria. This is the first comparison of MAb2 reactivity, lag-1 alleles, and sequence-based types among clinical and environmental L. pneumophila serogroup 1 isolates from the United States. Even though we identified a large number of STs unique to the United States, the trends in distribution of the MAb2 epitope and lag-1 gene were in agreement with results previously observed in Europe. Specifically, our analysis showed that the number of clinical isolates recognized by MAb2 was significantly greater than the number of environmental isolates recognized by MAb2. This finding correlates with previous observations showing that the MAb2 epitope is overrepresented in L. pneumophila serogroup 1 strains causing human disease (3, 4, 6, 14, 15, 17). We also confirmed that all MAb2-positive isolates harbored a lag-1 gene (2, 26). Similar to the MAb2 epitope, the lag-1 gene is significantly overrepresented in L. pneumophila serogroup 1 clinical isolates.
To understand the basis for the prevalence of MAb2-positive isolates among clinical specimens, we analyzed the distributions of the MAb2 epitope and the lag-1 gene within the genetic lineages of L. pneumophila serogroup 1. The data from Fig. 4 indicate that there are no separate lineages for MAb2-positive and MAb2-negative L. pneumophila serogroup 1. The data are supportive of a scenario in which some MAb2-positive L. pneumophila serogroup 1 strains became MAb2 negative upon losing the lag-1 gene, as opposed to MAb2-negative strains converting to MAb2-positive ones after the acquisition of a lag-1 locus.
The analysis of L. pneumophila serogroup 1 environmental isolates should provide insight into the evolution of MAb2 and lag-1. The ability to infect humans does not contribute to the evolutionary selection of legionellae, as the bacteria are opportunistic pathogens of humans and are not transmitted from person to person. Instead, evolutionary selection may result from pressures due to the ecological niche or protozoan host of legionellae. Among 50 L. pneumophila serogroup 1 environmental isolates analyzed in this study, only 3/50 (6%) were MAb2 positive and only 4/50 (8%) carried the lag-1 gene. This suggests a trend of environmental isolates discarding the MAb2 epitope and lag-1 gene. Perhaps expression of lag-1-encoded O-acetyltransferase represents a burden for L. pneumophila serogroup 1 competing in the environment. Analysis of the lag-1 locus composition suggests that there is a difference in the rates with which L. pneumophila serogroup 1 strains discard the lag-1 gene and the MAb2 epitope. Strains with a Philadelphia-like lag-1 locus may lose the lag-1 gene during a relatively frequent event of homologous recombination between ORF2 and ORF2-like genes (2), whereas strains with Lens- and Arizona-like loci probably lose MAb2 reactivity more slowly by accumulating point mutations within the lag-1 gene (Fig. 1b and c). Supportive of this theory is the widespread distribution of ST1 L. pneumophila serogroup 1. These strains may be so prevalent due to their ability to quickly discard the lag-1 gene, as they appear to exclusively harbor the Philadelphia-like locus.
If the MAb2 epitope is not necessary for L. pneumophila serogroup 1 success in the environment, it is still possible that the MAb2 epitope, the lag-1 gene, or a neighboring gene is important for L. pneumophila serogroup 1 virulence. Therefore, it is essential to develop means to better identify and track the distribution of MAb2- and lag-1-positive strains. The lag-1 gene PCR will allow rapid detection of these strains in both clinical and environmental samples. The lag-F and lag-R primers we designed targeting an internal 584-bp region of the lag-1 gene successfully amplified fragments from all 78 lag-1-positive L. pneumophila serogroup 1 isolates. Similarly, Thürmer et al. in a recently published paper reported successful amplification of the full-length lag-1 gene from 30 MAb3/1-positive L. pneumophila serogroup 1 isolates (35). Some MAb2-negative isolates have a nonfunctional full-length lag-1 gene, which would falsely identify these isolates as MAb2 positive. However, our data suggest that most isolates have lost the lag-1 gene through recombination events and that point mutations are rare. We found only one out of 73 MAb2-negative isolates that carried a full-length lag-1 gene. In contrast, Thürmer et al. demonstrated that 4/13 MAb3/1-negative isolates were lag-1 positive (35), while another recent study showed that all 18 MAb3/1-negative L. pneumophila serogroup 1 isolates analyzed were lag-1 negative (24). Further studies are needed to determine the proportion of MAb2-negative lag-1-positive isolates among L. pneumophila serogroup 1.
The primary purpose for the SBT of 150 L. pneumophila serogroup 1 isolates in this study was to establish a phylogenetic relationship between these bacteria to analyze the distributions of the MAb2 epitope and the lag-1 gene among L. pneumophila serogroup 1 lineages. Even within this relatively small sampling pool, we identified a large number of STs unique to the United States. Specifically, 34 out of 59 (58%) STs identified in this study did not match any reported isolates in the EWGLI database. A recent study of Canadian L. pneumophila serogroup 1 isolates also identified a large number of unique STs (36). Moreover, eBURST analysis helped us to identify a clonal complex of L. pneumophila serogroup 1 (clonal complex A; Fig. 3b) specific to the northeastern region of the North American continent (both the United States and Canada). Future studies will allow us to determine whether these strains remain endemic to North America or spread to other continents.
ST1 appears to be the most prevalent ST in the world. It was the most common ST identified in our study and is the most common reported to the EWGLI database (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php). The Canadian study mentioned above reported that the prevalence of clinical isolates that belong to ST1 has decreased in the last 30 years (36). This observation is also in keeping with our hypothesis that loss of MAb2 increases environmental fitness but makes the strain less virulent. The diversity of STs among clinical isolates being greater than the diversity of environmental isolates in our study is mostly due to the proportion of ST1 isolates among environmental samples (40%) being higher than that among clinical specimens (19%) (Tables 1 and 2).
Several STs described in this study are commonly associated with outbreaks of Legionnaires' disease. ST213 and ST222, from clonal complex A (Fig. 3b), were associated with outbreaks in both the United States and Canada (36). Two of the four ST36 clinical isolates were also implicated in U.S. outbreaks. These observations support the hypothesis of Harrison et al. that a select group of STs have an enhanced ability to cause legionellosis in humans (16). In addition, we observed that STs tend to associate with a single lag-1 allele type. This suggests the existence of clonal groups of L. pneumophila serogroup 1 that rarely recombine (7). Further studies should indicate if combining SBT and lag-1 data will define a hierarchy of virulence genotypes that require greater scrutiny to prevent human disease. Currently, all guidelines for the prevention of legionellosis address legionellae as if all species and serogroups were of equivalent virulence. This is clearly not the case. Prevention measures should target the genotypes which cause the majority of human disease.
Acknowledgments
We thank Norman Fry for help and insightful discussions of sequence-based typing and eBURST analysis. We also thank Claressa Lucas and Genyan Yang for proofreading the manuscript.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Bartram, J., Y. Chartier, J. V. Lee, K. Pond, and S. Surman-Lee (ed.). 2007. Legionella and the prevention of legionellosis. World Health Organization, Geneva, Switzerland.
- 2.Bernander, S., K. Jacobson, J. H. Helbig, P. C. Luck, and M. Lundholm. 2003. A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns. J. Clin. Microbiol. 412503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchardt, J., J. H. Helbig, and P. C. Luck. 2008. Occurrence and distribution of sequence types among Legionella pneumophila strains isolated from patients in Germany: common features and differences to other regions of the world. Eur. J. Clin. Microbiol. Infect. Dis. 2729-36. [DOI] [PubMed] [Google Scholar]
- 4.Castellani Pastoris, M., M. McIntyre, and P. Goldoni. 1990. Legionella pneumophila serogroup 1 population in Italy by monoclonal subtyping. Epidemiol. Infect. 105169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cianciotto, N. P., J. K. Stamos, and D. W. Kamp. 1995. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr. Microbiol. 30247-250. [DOI] [PubMed] [Google Scholar]
- 6.Dournon, E., W. F. Bibb, P. Rajagopalan, N. Desplaces, and R. M. McKinney. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157496-501. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, M. T., N. K. Fry, and T. G. Harrison. 2008. Clonal population structure of Legionella pneumophila inferred from allelic profiling. Microbiology 154852-864. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 10.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields, B. S., and M. R. Moore. 2006. Control of legionellae in the environment: a guide to the US guidelines. ASHRAE Trans. 112691-699. [Google Scholar]
- 12.Gaia, V., N. K. Fry, B. Afshar, P. C. Luck, H. Meugnier, J. Etienne, R. Peduzzi, and T. G. Harrison. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 432047-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaia, V., N. K. Fry, T. G. Harrison, and R. Peduzzi. 2003. Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J. Clin. Microbiol. 412932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison, T. G., B. Afshar, N. Doshi, N. K. Fry, and J. V. Lee. 2009. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000-2008). Eur. J. Clin. Microbiol. Infect. Dis. 28781-791. [DOI] [PubMed] [Google Scholar]
- 15.Harrison, T. G., N. Doshi, N. K. Fry, and C. A. Joseph. 2007. Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin. Microbiol. Infect. 1378-85. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, T. G., N. K. Fry, B. Afshar, W. Bellamy, N. Doshi, and A. P. Underwood. 2005. Typing of Legionella pneumophila and its role in elucidating the epidemiology of Legionnaire's disease, p. 94-99. In N. P. Cianciotto, Y. A. Kwaik, P. H. Edelstein, B. S. Fields, D. F. Geary, T. G. Harrison, C. A. Joseph, R. M. Ratcliff, J. E. Stout, and M. S. Swanson (ed.), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC.
- 17.Helbig, J. H., S. Bernander, M. Castellani Pastoris, J. Etienne, V. Gaia, S. Lauwers, D. Lindsay, P. C. Luck, T. Marques, S. Mentula, M. F. Peeters, C. Pelaz, M. Struelens, S. A. Uldum, G. Wewalka, and T. G. Harrison. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21710-716. [DOI] [PubMed] [Google Scholar]
- 18.Helbig, J. H., P. C. Luck, Y. A. Knirel, W. Witzleb, and U. Zahringer. 1995. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol. Infect. 11571-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joly, J. R., R. M. McKinney, J. O. Tobin, W. F. Bibb, I. D. Watkins, and D. Ramsay. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23768-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-32. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 22.Kozak, N. A., R. F. Benson, E. Brown, B. G. Shelton, and B. S. Fields. 2008. Distribution of sequence-based types (SBT) and lag-1 alleles among Legionella pneumophila serogroup 1 clinical and environmental isolates collected in the United States, poster D-139. Abstr. 108th Gen. Meet. Am. Soc. Microbiol. ASM, Washington, DC. [DOI] [PMC free article] [PubMed]
- 23.Kozak, N. A., E. Brown, R. F. Benson, B. G. Shelton, and B. S. Fields. 2007. Presence of a lag-1 gene as a virulence marker for Legionella pneumophila serogroup 1 strains, poster D-144. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. ASM, Washington, DC.
- 24.Kuchana, R., and S. A. Uldum. 2007. PCR method for detection of monoclonal subgroups of Legionella pneumophila serogroup 1, abstr. Or19, p. 39. 22nd Annu. Meet. Eur. Working Group Legionella Infect., Stockholm, Sweden.
- 25.Lück, P. C., T. Freier, C. Steudel, Y. A. Knirel, E. Luneberg, U. Zahringer, and J. H. Helbig. 2001. A point mutation in the active site of Legionella pneumophila O-acetyltransferase results in modified lipopolysaccharide but does not influence virulence. Int. J. Med. Microbiol. 291345-352. [DOI] [PubMed] [Google Scholar]
- 26.Lück, P. C., M. Schuppler, and J. H. Helbig. 2002. Changes in the lag-1 locus of Legionella pneumophila serogroup 1 strains result in different lipopolysaccharides recognized by monoclonal antibodies but do not influence virulence, p. 52-55. In R. Marre (ed.), Legionella. ASM Press, Washington, DC.
- 27.Lüneberg, E., N. Zetzmann, D. Alber, Y. A. Knirel, O. Kooistra, U. Zahringer, and M. Frosch. 2000. Cloning and functional characterization of a 30 kb gene locus required for lipopolysaccharide biosynthesis in Legionella pneumophila. Int. J. Med. Microbiol. 29037-49. [DOI] [PubMed] [Google Scholar]
- 28.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60561-588. [DOI] [PubMed] [Google Scholar]
- 29.Para, M. F., and J. F. Plouffe. 1983. Production of monoclonal antibodies to Legionella pneumophila serogroups 1 and 6. J. Clin. Microbiol. 18895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratzow, S., V. Gaia, J. H. Helbig, N. K. Fry, and P. C. Luck. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 451965-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 32.Sanden, G. N., P. K. Cassiday, and J. M. Barbaree. 1993. Rapid immunodot technique for identifying Bordetella pertussis. J. Clin. Microbiol. 31170-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stout, J. E., J. Joly, M. Para, J. Plouffe, C. Ciesielski, M. J. Blaser, and V. L. Yu. 1988. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila. J. Infect. Dis. 157486-495. [DOI] [PubMed] [Google Scholar]
- 34.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 35.Thürmer, A., J. H. Helbig, E. Jacobs, and P. C. Luck. 2009. PCR-based ‘serotyping’ of Legionella pneumophila. J. Med. Microbiol. 58588-595. [DOI] [PubMed] [Google Scholar]
- 36.Tijet, N., P. Tang, M. Romilowych, C. Duncan, F. Jamieson, D. E. Low, and C. Guyard. 2008. Molecular evolution and epidemiology of clinical Legionella pneumophila serotype I (L. pneumophila serogroup 1) isolates from Ontario: a 30 years retrospective analysis using sequence based typing, abstr. C-290. Abstr. 108th Gen. Meet. Am. Soc. Microbiol. ASM, Washington, DC.
- 37.Vassileva, M., K. Torii, M. Oshimoto, A. Okamoto, N. Agata, K. Yamada, T. Hasegawa, and M. Ohta. 2006. Phylogenetic analysis of Bacillus cereus isolates from severe systemic infections using multilocus sequence typing scheme. Microbiol. Immunol. 50743-749. [DOI] [PubMed] [Google Scholar]
- 38.Watkins, I. D., J. O. Tobin, P. J. Dennis, W. Brown, R. Newnham, and J. B. Kurtz. 1985. Legionella pneumophila serogroup 1 subgrouping by monoclonal antibodies—an epidemiological tool. J. Hyg. (London) 95211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186127-128. [DOI] [PubMed] [Google Scholar]
- 40.Zähringer, U., Y. A. Knirel, B. Lindner, J. H. Helbig, A. Sonesson, R. Marre, and E. T. Rietschel. 1995. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res. 392113-139. [PubMed] [Google Scholar]
- 41.Zou, C. H., Y. A. Knirel, J. H. Helbig, U. Zahringer, and C. S. Mintz. 1999. Molecular cloning and characterization of a locus responsible for O acetylation of the O polysaccharide of Legionella pneumophila serogroup 1 lipopolysaccharide. J. Bacteriol. 1814137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]