Abstract
Uncontrolled fibroblast growth factor (FGF) signaling can lead to human diseases, necessitating multiple layers of self-regulatory control mechanisms to keep its activity in check. Herein, we demonstrate that FGF9 and FGF20 ligands undergo a reversible homodimerization, occluding their key receptor binding sites. To test the role of dimerization in ligand autoinhibition, we introduced structure-based mutations into the dimer interfaces of FGF9 and FGF20. The mutations weakened the ability of the ligands to dimerize, effectively increasing the concentrations of monomeric ligands capable of binding and activating their cognate FGF receptor in vitro and in living cells. Interestingly, the monomeric ligands exhibit reduced heparin binding, resulting in their increased radii of heparan sulfate-dependent diffusion and biologic action, as evidenced by the wider dilation area of ex vivo lung cultures in response to implanted mutant FGF9-loaded beads. Hence, our data demonstrate that homodimerization autoregulates FGF9 and FGF20's receptor binding and concentration gradients in the extracellular matrix. Our study is the first to implicate ligand dimerization as an autoregulatory mechanism for growth factor bioactivity and sets the stage for engineering modified FGF9 subfamily ligands, with desired activity for use in both basic and translational research.
Fibroblast growth factor (FGF) signaling plays pleiotropic roles throughout the life spans of mammalian organisms, ranging from germ cell maturation, mesoderm induction, body plan formation, and organogenesis during embryonic development to serum phosphate homeostasis and glucose, bile acid, lipid, and cholesterol metabolism in the adult (3, 23, 27, 28, 57, 60, 62). The diversity of FGF signaling is underscored by virtue of the fact that aberrant FGF signaling leads to a wide array of human diseases, including skeletal and olfactory/reproductive syndromes, phosphate wasting disorders, and cancer (16, 60, 67). Recent data also implicate dysregulated FGF signaling in the etiology of neurodegenerative disorders, such as major depressive disorder and Parkinson's disease (10, 63, 64).
Based on pairwise sequence homology and phylogeny, the 18 bona fide mammalian FGFs (FGF1 to FGF10 and FGF16 to FGF23) are divided into six subfamilies (45). Five FGF subfamilies have high-to-moderate affinity for pericellular heparan sulfate (HS) glycosaminoglycans and thus diffuse locally within tissues to act in a paracrine fashion, whereas the poor affinity of the FGF19 subfamily for HS enables this subfamily to act in an endocrine manner (28, 38). All FGFs share a core homology region of about 120 amino acids, which fold into 12 antiparallel β strands (β1 to β12) that are arranged into three sets of four-stranded β sheets (β-trefoil fold) (39). The globular FGF core domain is flanked by highly divergent N- and C-terminal extensions, which are the principal regions responsible for the different biology of FGFs.
FGFs exert their diverse actions by binding and activating FGF receptors (FGFRs) in an HS-dependent fashion (51, 53, 69). There are four distinct mammalian FGFR genes (FGFR1 to FGFR4), each coding for a single-pass transmembrane tyrosine kinase receptor whose ectodomain consists of three immunoglobulin-like domains (D1 to D3) connected by flexible linkers and whose intracellular domain contains the conserved tyrosine kinase domain flanked by the juxtamembrane (JM) and C-terminal regions (38). The 210-amino-acid-long D2-D3 segment of the ectodomain is both necessary and sufficient for ligand binding (20, 51, 52, 58, 70).
FGF signaling is tightly regulated by spatial and temporal expression of ligands, receptors, HS cofactors, and most critically by means of FGF-FGFR binding specificity. The tissue-specific alternative splicing in the D3 domain of FGFR1 to FGFR3 is the main mechanism by which FGF-FGFR binding specificity is regulated. This splicing event gives rise to epithelial “b” isoforms (FGFR1b to FGFR3b) and mesenchymal “c” isoforms (FGFR1c to FGFR3c) (24, 25, 47, 68), which differ from one another at the primary sequences of their key ligand binding regions and thus in their FGF binding specificity/promiscuity profiles. Most FGFs are also expressed in either epithelial or mesenchymal tissues and exhibit specificity for FGFR isoforms expressed in the opposite tissues. This results in the establishment of a bidirectional signaling loop between the epithelium and mesenchyme that is essential for organogenesis and tissue homeostasis. It is well established that FGF7 and FGF10, which are expressed exclusively in the mesenchyme, activate specifically FGFR2b to mediate mesenchymal-to-epithelial signaling in the lung, prostate, and lacrimal, mammary, and salivary glands (19, 29, 35, 36, 59). Several lines of genetic and biochemical evidence suggest that the members of the FGF9 subfamily, which includes FGF9, FGF16, and FGF20, convey the reciprocal signaling from the epithelium to the mesenchyme. In the prostate, the epithelial-specific FGF9 has been shown to activate mesenchymal FGFR3c isoforms (25). In the heart, FGF9, FGF16, and FGF20 in the epicardium and endocardium stimulate myocardial proliferation and differentiation in vivo, acting redundantly through FGFR1c and FGFR2c (32). Analysis of FGF9-deficient mice has identified FGF9 as a reciprocal epithelial-to-mesenchymal signal required for morphogenesis of the lung, cecum, small intestine, and inner ear (14, 49, 65, 71). In addition, studies in zebra fish show that FGF16 and FGF20 are apical ectodermal ridge factors that are required for pectoral fin bud outgrowth and, in general, for cell proliferation and differentiation of the mesenchyme (41, 66).
In light of the key role of the FGF9 subfamily in tissue homeostasis, it is essential to investigate the molecular mechanisms by which the activity of this subfamily is regulated. Our previous structural and in vitro studies of FGF9 showed that homodimerization masks FGF9's key receptor binding sites, suggesting that ligand dimerization may autoinhibit FGF9's biologic activity (50). In this report, we show that, like FGF9, FGF20 also homodimerizes in the crystal and in solution. Characterization of the dimer interface mutations in vitro and in living cells demonstrates that ligand homodimerization autoinhibits FGF9 and FGF20 signaling by suppressing both receptor binding and HS-dependent diffusion in the extracellular matrix (ECM). Our study is the first to implicate ligand dimerization as an autoregulatory mechanism in growth factor bioactivity.
MATERIALS AND METHODS
Protein expression and purification.
FGF20 crystals were grown using bacterially expressed full-length human FGF20 (Met-1 to Thr-211; velafermin, CG-53135 [30]) provided by CuraGen (1). Validation of the structural data was performed using N-terminally six-His-tagged full-length human FGF20 (Met-1 to Thr-211), untagged full-length human FGF9 (Met-1 to Ser-208), and N-terminally six-His-tagged full-length human FGF16 (Met-1 to Arg-207) expressed in Escherichia coli in the lab (50). Single, double, and triple mutations (Table 1) were introduced into both FGF9 and FGF20 using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Competent E. coli BL21(DE3) cells were transformed with wild-type or mutated FGF9, FGF20, or FGF16 expression plasmids. Transformants were grown at 37°C to an optical density of ∼0.5 before inducing protein expression with 1 mM isopropyl-1-thio-beta-d-galactopyronoside. Four hours after induction, the cells were harvested and stored frozen until further use. Spun bacterial cell pellets were lysed using a French press in 25 mM HEPES buffer (pH 7.5) containing 150 mM NaCl, 5 mM EDTA, 10% glycerol, and 0.5 mM phenylmethylsulfonyl fluoride. The soluble fraction containing the wild-type or mutated FGF9 proteins was subsequently precipitated using saturated ammonium sulfate overnight at 4°C. Following centrifugation, the protein pellets containing the FGF9 ligand were redissolved in 25 mM HEPES buffer (pH 7.5) and subjected to heparin affinity chromatography using a HiTrap heparin affinity column (GE Healthcare, Piscataway, NJ), followed by size exclusion chromatography on a HiLoad 16/60 Superdex 75 column (GE Healthcare, Piscataway, NJ) equilibrated with 25 mM HEPES buffer (pH 7.5) and 1 M NaCl. Wild-type and mutated FGF20 proteins, as well as FGF16 and the ligand binding region of FGFR1c (Asp-141 to Arg-365), were refolded from bacterial inclusion bodies, as previously described (22, 51). Correctly folded FGF20, FGF16 ligands, and FGFR1c ectodomain were purified using sequential heparin affinity and size exclusion chromatography. Protein concentrations were determined spectrophotometrically under denaturing conditions using extinction coefficients at 280 nm and verified by running 2 μg of each protein on a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gel (see Fig. S3 in the supplemental material).
TABLE 1.
Description of the FGF9 and FGF20 mutants
| Mutation | Description | Name | Elution vol (ml) | Estimated mass (kDa) |
|---|---|---|---|---|
| FGF20 | ||||
| None | Wild type | FGF20wt | 54.5 | 65.6 |
| E198A | Single mutation | FGF20E198A | 55 | 63.7 |
| L207A | Single mutation | FGF20L207A | 56.5 | 58.1 |
| L208A | Single mutation | FGF20L208A | 55.5 | 61.7 |
| L207A, L208A | Double mutation | FGF20L207A/L208A | 59.5 | 48.2 |
| FGF9 | ||||
| None | Wild type | FGF9wt | 59.5 | 48.2 |
| D195A | Single mutation | FGF9D195A | 62 | 41.4 |
| L200A | Single mutation | FGF9L200A | 66 | 32.4 |
| I204A | Single mutation | FGF9I204A | 65 | 34.4 |
| L205A | Single mutation | FGF9L205A | 65.5 | 33.3 |
| I204A, L205A | Double mutation | FGF9I204A/L205A | 68 | 28.6 |
| L200A, I204A, L205A | Triple mutation | FGF9L200A/I204A/L205A | 68.5 | 27.7 |
| D195A, I204A, L205A | Triple mutation | FGF9D195A/I204A/L205A | 70 | 25.3 |
Crystallization, data collection, structure determination, and refinement.
Crystals of FGF20 were grown by vapor diffusion at 20°C using the hanging drop method. One microliter of FGF20 protein solution (10 mg/ml in 25 mM Na+ phosphate buffer, pH 7.4, and 50 mM Arg-HCl) was mixed with 1 μl of reservoir buffer (0.1 M Tris-HCl [pH 8.5], 0.2 M Li2SO4, 15% glycerol, and 13% polyethylene glycol 2000). FGF20 crystals belong to the primitive rhombohedral R3 space group, with unit cell dimensions of a = b = 102.14 Å and c = 119.80 Å and a solvent fraction of 50% (assuming molecular density of 1.30 g/ml). A single FGF20 crystal was directly flash frozen in the cryostream of liquid nitrogen, and the diffraction data were recorded on a charge-coupled-device detector at beamline X4A at the National Synchrotron Light Source, Brookhaven National Laboratory, and processed using HKL2000 (48). A molecular replacement solution for the two copies of FGF20 in the asymmetric unit was found using the program AMoRe (40) and the FGF9 crystal structure (Protein Data Bank identification [PDB ID] 1IHK) (50) as a search model. The program O (26) was used for model building into the 2Fo-Fc and Fo-Fc maps, and CNS was used for rigid body, positional, and B-factor refinements (4). Tight, noncrystallographic symmetry restraints were imposed throughout the refinement for the backbone atoms of the two FGF20 molecules in the asymmetric unit. Data collection, refinement, and atomic model statistics are summarized in Table 2.
TABLE 2.
Summary of crystallographic analysis
| Statistics | Value(s) |
|---|---|
| Data collection statistics | |
| Resolution range (Å) | 50.0-2.5 |
| No. of reflections (total/unique) | 168,029/16,050 |
| Completeness (%) | 99.7 (99.9)b |
| Rsym (%)a | 5.6 (28.4)b |
| Signal (l/σl) | 63 (11.8)b |
| Redundancy | 5.2 (4.9)b |
| Refinement statistics | |
| Resolution range (Å) | 25.0-2.5 |
| No. of reflections | 14,309 |
| Rcryst/Rfree (%)c | 25.6/27.3 |
| Root-mean-square deviations | |
| Bond length (Å) | 0.008 |
| Angle (°) | 1.43 |
| B factors (Å2)d | 1.3 |
| Model statistics | |
| No. of protein atoms | 2,510 |
| No. of SO4 ions | 6 |
| No. of H2O molecules | 9 |
| B factors | |
| FGF20 (Å2) | 73 |
| SO4 (Å2) | 120 |
| H2O (Å2) | 64 |
Rsym = 100 × ΣhklΣi{Ii(hkl) − [I(hkl)]}/ΣhklΣiIi(hkl).
Value in parentheses is for the highest-resolution shell: 2.59 to 2.5 Å.
Rcryst/Rfree = 100 × Σhkl {[Fo(hkl)] − [Fc(hkl)]}/Σhkl [Fo(hkl)], where Fo (>0σ) and Fc are the observed and calculated structure factors, respectively. A total of 5% of the reflections were used for calculation of Rfree.
For bonded protein atoms.
Size exclusion chromatography.
Due to the poor solubility of the FGF9 subfamily members in physiological salt buffers, column chromatography was carried out in 25 mM HEPES buffer (pH 7.5) containing 1 M NaCl. To calibrate the column (HiLoad 16/60 Superdex 75; GE Healthcare, Piscataway, NJ), 300 μl of a mixture of protein standards were injected, and the proteins were eluted using an isocratic flow (1 ml/min). The following protein standards from Sigma-Aldrich were used: aprotinin (∼6.5 kDa, 1.4 mg/ml), RNase A (∼13.7 kDa, 2.0 mg/ml), carbonic anhydrase (∼30 kDa, 0.5 mg/ml), ovalbumin (∼44.3 kDa, 2.0 mg/ml), and bovine serum albumin (∼66 kDa, 1.8 mg/ml). The equation y = −1.1632x + 6.2746 was obtained from a linear fit, generated by plotting the logarithm of molecular weights of the standards (y) against their respective elution volumes per the elution volume of blue dextran (∼2 MDa, 0.6 mg/ml) (x). A total of 300 μl of purified wild-type and mutated FGF9 (400 μg/ml = 17 μM), FGF20 (460 μg/ml = 16.2 μM, 230 μg/ml = 8.1 μM, or 115 μg/ml = 4.1 μM), or FGF16 (210 μg/ml = 7.3 μM) proteins were injected, and the molecular sizes for FGF9, -20, and -16 proteins were estimated by solving the above-described equation for “y” and taking its inverse logarithm.
The molecular size estimation of 17 μM wild-type FGF9 or 17 μM FGF9D195A/I204A/L205A alone or with various concentrations of heparin dodecasaccharide (DS) (at 0, 2.1, 4.3, 8.5, 17.0, 34.0, 68.0, and 136 μM) was performed by injecting 350 μl of each mixture onto a HiLoad 16/60 Superdex 200 (GE Healthcare, Piscataway, NJ). The sizes were calculated by taking the inverse logarithm of “y” using an equation, y = −1.5565x + 7.545, which was obtained by calibrating the column, as described above.
SPR experiments and data processing.
Surface plasmon resonance (SPR) measurements were performed on the Biacore 2000 instrument (GE Healthcare, Piscataway, NJ). For ligand-receptor interactions, purified wild-type or mutated FGF9 or FGF20 was immobilized using either amine coupling on a carboxymethyl (CM5) biosensor chip (GE Healthcare, Piscataway, NJ) or oriented coupling using the Ni-nitrilotriacetic acid biosensor chip (GE Healthcare, Piscataway, NJ), according to manufacturer's instructions, at a density of ∼1,800 resonance units and a flow rate of 5 μl/min in acetate buffer (pH 5.5). Eight different concentrations (from 50 nM to 2 μM) of purified FGFR1c were prepared in HBS-EP buffer (10 mM HEPES-NaOH [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] polysorbate 20; GE Healthcare, Piscataway, NJ) and injected over the chip at a flow rate of 50 μl/min. The association and dissociation phases were 250 and 180 s, respectively. The chip was regenerated in acetate buffer (pH 4.5), supplemented with 2 M NaCl. The equilibrium dissociation constants (KD) were calculated using BIAevaluation software 4.1 (GE Healthcare, Piscataway, NJ) by plotting the average maximal response at the end of the association phase (Req) for each concentration against the concentration of FGFR1c. The plots were then examined for goodness (χ2 test) of the model fit. Good fits were obtained, as the χ2 tests were calculated to be less than 10% of Rmax for the reported plots. For each concentration, the signal was corrected against the control surface (fibroblast homologous factor 1b immobilized to 1,800 response units [RUs]) to eliminate any refractive index changes due to nonspecific binding. Similar KD values were obtained using a global fitting of the association and dissociation phases to a Langmuir 1:1 binding model, as previously described (22).
For heparin affinity measurements, biotinylated heparin (Sigma, St. Louis, MO) was noncovalently immobilized on a streptavidin-coated chip (GE Healthcare, Piscataway, NJ) at a density of approximately 140 RUs. Wild-type and mutated FGF9 or FGF20 proteins were then passed over the chip at 80 nM for FGF9 and 60 nM for FGF20. In addition, wild-type FGF9, FGF16, and FGF20 were injected over the chip at 40 nM each. Association and dissociation were allowed to occur for 250 and 350 s, respectively. For each concentration, the signal was corrected against the control surface (immobilized biotin) to eliminate any refractive index changes due to nonspecific binding.
BaF3 receptor stimulation assay.
The interleukin-3 (IL-3)-dependent murine pro-B BaF3 cell line was transduced with lentiviral-based expression vectors for FGFR1c, as previously described (13). Cells stably expressing FGFR1c were selected using 1.2 mg/ml Geneticin in the presence of 5 ng/ml IL-3 for 14 days and then maintained under selection in RPMI medium supplemented with 10% fetal bovine serum, 50 nM beta-mercaptoethanol, 100 U/ml penicillin, 100 mg/ml streptomycin sulfate, 1.2 mg/ml Geneticin, and 5 ng/ml murine IL-3 (R&D Systems, Minneapolis, MN). Cells were washed in phosphate-buffered saline (PBS) to remove IL-3 and plated at 1 × 104 cells per well in triplicate in a 96-well plate in IL-3-free media containing 100 ng/ml wild-type or mutated FGF9 or FGF20 ligands and 5 μg/ml heparin. Bioluminescent measurement of ATP was assessed as an indicator of cell number using the ViaLight Plus cell proliferation/cytotoxicity kit (Lonza Rockland, Inc., Rockland, ME), according to the manufacturer's instructions. Measurements were taken on the day cells were plated (day 0) and every 24 h thereafter up to 72 h following ligand stimulation. Experiments were repeated two independent times.
To study the effect of heparin on receptor activation, BaF3 cells (treated as described above) were stimulated with IL-3-free media containing 100 ng/ml wild-type FGF9 or FGF9D195A/I204A/L205A plus various concentrations of heparin (0.3125, 0.625, 1.25, 2.5, 5, 10, 20, 40, and 80 μg/ml). Bioluminescence measurements were performed, as described above, with measurements taken in triplicate every 24, 48, and 72 h following ligand stimulation. Experiments were repeated two independent times.
pERK stimulation assay.
BaF3/FGFR1c cells were starved of IL-3 and stimulated with 100 ng/ml of wild-type or mutated FGF9 and FGF20 ligands in the presence of 5 μg/ml heparin for 10 min, and lysates were collected. Lysates were then analyzed using the Millipore 8-plex phosphoprotein detection kit. All experiments were performed in duplicate, and each sample was evaluated in triplicate wells.
In-vitro proliferation assay of isolated lung mesenchyme.
Embryonic day 12.5 (E12.5) mouse lung was isolated and treated with trypsin/pancreatin (Sigma, St. Louis, MO) for 5 to 10 min at 4°C. The distal mesenchyme was separated from the distal epithelium using fine tungsten needles, resuspended in the culture medium DMEM (F-12 supplemented with 5% fetal bovine serum and Glutamax), and plated on two-well chamber slides. Mesenchymal cells were cultured up to 70 to 80% confluence (24 to 36 h), then transferred to a defined (serum-free) medium, and cultured for an additional 18 h. The cells were then supplemented with 20 ng/ml of wild-type and mutated FGF9 proteins and cultured for an additional 12 h. Stimulation with 300 nM wild-type FGF9 or FGF9D195A/I204A/L205A was also performed to test whether the differences in FGFR activation among the proteins are still present at a concentration near the dissociation constant (Kd) of ligand dimerization. At the end of this period, bromodeoxyuridine (BrdU) was added to the culture medium, and the cells were incubated for an additional hour. Cells were then washed with PBS and fixed in 2% paraformaldehyde for 20 min. Before BrdU detection, cells were postfixed with −20°C acetone for 5 min. BrdU incorporation was detected using a BrdU detection kit, as described by the manufacturer (GE Healthcare, Piscataway, NJ). In addition, nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Images of BrdU and DAPI staining (10 frames from each slide) were taken using a Leica fluorescent microscope, and BrdU- and DAPI-positive cells were counted using the ImageJ program. The data were analyzed by Student's t test, and the results were determined to be significant if P was <0.01. BrdU labeling of lung mesenchymal cultures was repeated three times.
FGF bead implantation in ex vivo lung cultures and statistical analysis.
Timed, pregnant C57BL/6 wild-type mice were sacrificed on E13.0 to E13.5, and the embryos were harvested. Lung primordia (n = 8 for each condition) were isolated from embryos by microdissection under sterile conditions. Embryonic lungs were placed on a 0.8-μm Millipore membrane (Millipore, Billerica, MA), supported by a metal grid (36), and cultured in defined medium. Defined medium was prepared, as follows: CMRL was supplemented with 0.1% lipid-rich albumin, insulin-transferrin-selenium, human transferrin, nonessential amino acids, Glutamax, and antibiotic-antimycotic (Invitrogen, Carlsbad, CA). Lungs were cultured in an air-fluid interface at the level of the explants. The cultures were maintained in 100% humidity, with an atmosphere of 95% air and 5% CO2 for 2 to 4 days. The medium was changed daily.
To monitor the effects of the “monomerizing” mutations on the HS-dependent diffusion of FGF9 ligands, we compared the areas of dilation within whole-lung explants after the implantation of heparin acrylic beads containing mutated and wild-type FGF9 proteins. Beads were incubated with 200 ng/μl of wild-type or mutated FGF9 proteins overnight, washed in PBS, and implanted into the center areas of lung explants. After 24 to 48 h of incubation, explants were photographed using a SPOT digital camera and a Leica microscope, and images were imported into Canvas X (ACD Systems, British Columbia, Canada). The dilated area within each explant was outlined and then measured using ImageJ software (Image Processing and Analysis in Java). All experiments were repeated five times. The measurements of the dilated areas induced by mutated and wild-type FGF9 proteins were averaged, and data were processed for statistic analysis using Student's t test. Results were determined to be significant if P was <0.01.
Limited proteolysis of FGF9 and FGF20 proteins.
To assess ligands’ sensitivity to proteases, 2 μg of FGF9 or 2.3 μg of FGF20 protein (in 25 mM HEPES buffer, pH 7.5, containing 1 M NaCl) were incubated for 2 h at ambient temperature with increasing amounts of thermolysin, elastase (0 μg, 0.0016 μg, 0.016 μg, 0.16 μg, 1.6 μg, and 16 μg) or trypsin (0 μg, 0.00005 μg, 0.0005 μg, 0.005 μg, 0.05 μg, and 0.5 μg). The reactions were stopped by adding SDS loading buffer and boiling for 5 min at 95°C. The digests were electrophoresed through a 12% SDS-polyacrylamide gel electrophoresis gel. The gel was then stained with Coomassie brilliant blue R-250 staining solution, and images were captured with a Canon scanner.
Mass spectrometry analysis.
To delineate trypsin cleavage sites within FGF9, 2 μg of wild-type FGF9 was partially digested with 0.5 μg of trypsin for 2 h at ambient temperature. The reaction was stopped with 0.5 μl of 10% formic acid and was subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis. Specifically, the sample was desalted and concentrated with ZipTipC4 (Millipore, Billerica, MA), according to standard protocols. A total of 1 μl of eluted protein was mixed with 1 μl of alpha-cyano-4-hydroxycinnamic acid matrix (Agilent Technologies, Santa Clara, CA) and spotted onto a 384-well MALDI plate (Bruker Daltonics, Billerica, MA). All spectra were acquired with an Autoflex MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, MA) in linear, positive-ion mode.
Protein structure accession number.
The atomic coordinates and structure factors for FGF20 have been deposited into the RCSB Protein Data Bank (http://www.rcsb.org/pdb/) with accession number PDB ID 3F1R.
RESULTS
FGF20 dimerizes in the crystal and in solution.
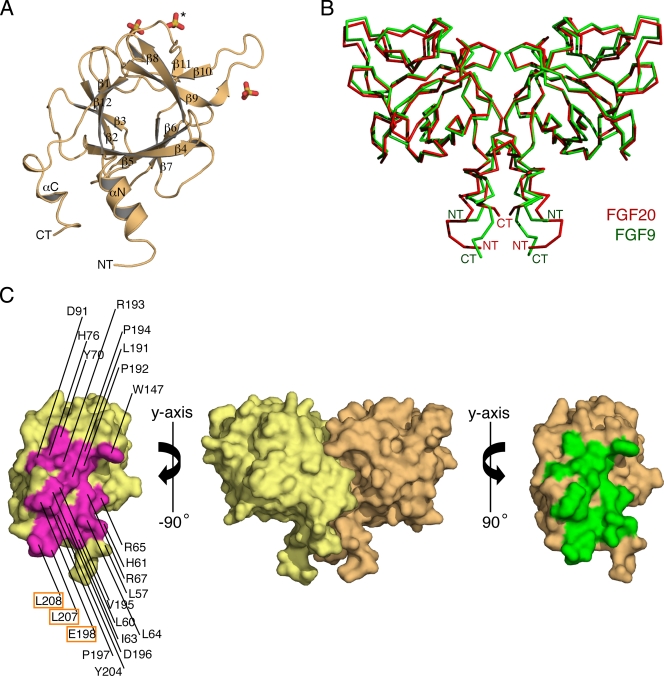
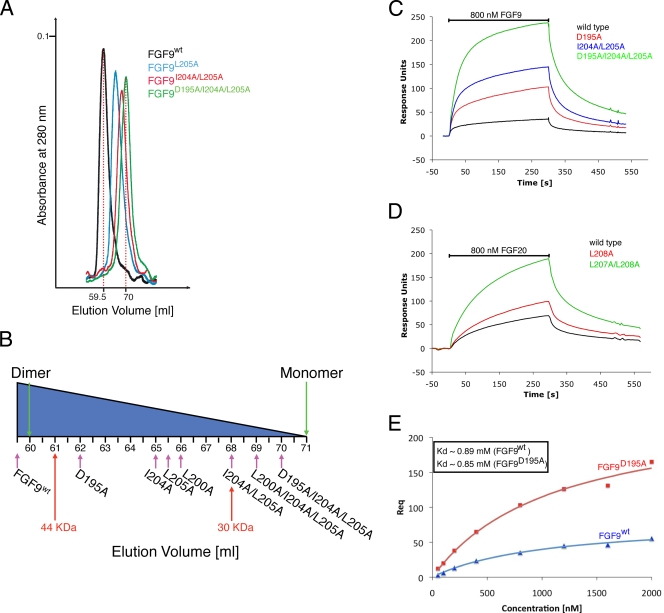
Recombinant full-length human FGF20 (Met-1 to Thr-211) is being produced by CuraGen Corporation under the generic name velafermin (CG-53135) for treatment of chemotherapy and radiation-induced oral mucositis (30). FGF20 crystallized in the primitive rhombohedral space group R3 with two FGF20 molecules per asymmetric unit. The crystal structure of FGF20 was solved by molecular replacement, using the FGF9 crystal structure as a search model (PDB ID 1IHK) (50) and has been refined to 2.5 Å. The final model consists of two FGF20 molecules (Pro-52 to Leu-208), six sulfate ions, and nine water molecules. Data collection and refinement statistics are given in Table 2. As anticipated, based on its high sequence identity to FGF9 (69%), FGF20 adopts a β-trefoil fold that is flanked by N- and C-terminal helices (Fig. 1A). The helices are oriented in antiparallel fashion and interact with each other through several hydrophobic and hydrogen-bonding contacts (Fig. 2C). Interestingly, FGF20, analogous to FGF9 (50), also crystallized as a twofold symmetric dimer (Fig. 1B and C). The dimer interface buries 2,198 Å2 of surface area and has a shape complementarity value of 0.67 and a hydrophobic/polar fraction of 1.4:1 (9, 33). These dimer interface parameters are consistent with a physiological interface.
FIG. 1.
FGF20 crystal structure. (A) Cartoon diagram of an FGF20 monomer. The β strands of FGF20 are labeled according to the conventional strand nomenclature for FGFs. The αN and αC helices are unique to FGF9 and FGF20 (11). NT and CT denote the N and C termini, respectively. Three sulfate ions are shown in stick representation and colored as follows: sulfur is in yellow and oxygen is in red. The sulfate ion that is bound into the predicted HBS of FGF20 is indicated by an asterisk. (B) Superimposition of the ribbon structures of FGF9 and FGF20 dimers. FGF9 is in green, and FGF20 is in red. NT and CT denote the N and C termini, respectively. (C) Surface representation of the noncrystallographic FGF20 dimer. The two FGF20 protomers are colored yellow and orange. To expose the dimer interface, the protomers are pulled apart and rotated 90 degrees about the vertical axis, as indicated. Residues participating in the dimerization in each protomer are colored magenta and green, respectively, and are labeled on one of the protomers. Dimerization residues that were mutated are boxed. This figure was created using PyMOL.
FIG. 2.
Both the core and the tail regions of FGF20 participate in dimerization. (A) A cartoon representation of the FGF20 dimer interface, including the core and tail regions, is encircled in blue ovals. Residues mediating dimer formation are shown in stick representation and are colored magenta and green. Two sulfate ions that are bound to the predicted HBSs of FGF20 molecules in the crystal structure are shown in stick representation. The Cα traces of FGF20 residues predicted to participate in HS interactions are colored blue. (B) Detailed view of the dimer interface involving the ligand's core region. An aromatic and a pi-cation interaction (in surface representation) between W147 from one monomer and Y70 and R193 of the second monomer, respectively, are indicated. (C) Detailed view of the dimer interface involving the ligand's N- and C-terminal regions flanking the trefoil core. The view is looking down along the twofold axis that passes through the N- and C-terminal extensions from both of the FGF20 protomers. To generate this view, the dimer in panel A is rotated by 90 degrees around the horizontal axis. The hydrophobic interaction between L207 of one protomer and L64 of the other is highlighted by showing the surfaces. Hydrogen bonds between R65 and R67 from the N terminus of one protomer and D196 and E198 in the C terminus of the other protomer are indicated with dashed lines. Atom colorings are as follows: sulfur in yellow, oxygen in red, and nitrogen in blue.
Reminiscent of the FGF9 dimer, the two FGF20 monomers engage in homotypic interactions through their core regions, as well as through their N- and C-terminal regions flanking the core (Fig. 1B and 2A). W147, situated in the β8-β9 turn, plays a pivotal role at the dimer interface portion that is mediated by the core region. W147 from one monomer engages in aromatic and pi-cation interactions with Y70 and R193, respectively, of the second monomer (Fig. 2B). Another prominent contact at this interface is the hydrophobic contact between L191 (in strand β12) of the two protomers (Fig. 2B).
The main driving forces of FGF20 dimerization, however, are the extensive hydrophobic and hydrogen-bonding contacts involving the N- and C-terminal regions flanking the trefoil core (Fig. 2C). At this section of the dimer interface, L57, L60, I63, and L64 from the αN helix, together with P197, Y204, L207, and L208 from the C terminus, form a continuous hydrophobic surface that binds the corresponding surface from the other protomer in the dimer. Hydrogen bonds between R65 and R67 from the N terminus of one protomer and D196 and E198 in the C terminus of the other protomer further promote dimerization. In this dimer, the heparin binding sites (HBSs) of the monomers are located on the same plane, and each contains a bound sulfate ion (Fig. 2A).
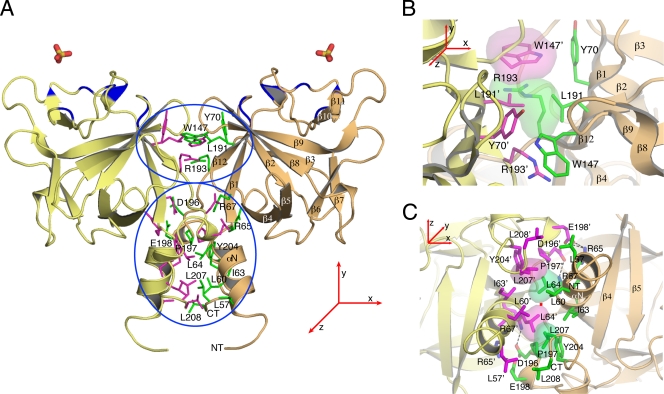
We used size exclusion chromatography to test whether FGF20 dimerizes in solution. When injected at 460 μg/ml, FGF20 eluted as a single peak of 65.6 kDa (Fig. 3B and C), consistent with the theoretical size of 56.8 kDa for the FGF20 dimer. This finding suggests that FGF20 is mostly dimeric at this concentration. Next, we injected incrementally diluted samples of FGF20 onto the sizing column (see Materials and Methods) and observed that the elution peak of FGF20 shifted to its predicted monomer position, indicative of concentration-dependent homodimerization in solution (see Table S1 in the supplemental material). Reminiscent of FGF9 (50), FGF20 does not resolve into distinct monomer and dimer peaks on the sizing column, suggesting that the dynamics of monomer-dimer exchange are faster than the time scale of the size exclusion chromatography.
FIG. 3.
FGF20 dimerizes in solution. (A) Structure-based sequence alignment of human FGF9 subfamily members. Periods indicate identical residues to the corresponding FGF9 sequence. The locations of the 12 β strands defining the FGF trefoil fold and N- and C-terminal α-helices flanking the core are indicated atop of the sequence alignment. Dimerization interface residues of FGF9 and FGF20 are in orange, and residues selected for mutagenesis are in boldface type. The predicted receptor binding residues are in cyan. Residues that mediate dimerization and are predicted to bind receptors are colored half orange and half cyan. The predicted heparin/HS-binding residues are in yellow. The dashes represent a gap introduced to optimize the alignment. (B) Size exclusion chromatograms of wild-type FGF20 (FGF20wt) and selected FGF20 mutants (FGF20L207A, FGF20L207A/L208A) are shown in the indicated colors (see Table 1 for an explanation of FGF9/20 nomenclature in the text). (C) Schematic plot showing the retention times of all FGF20 proteins, indicated with magenta arrows. Red arrows indicate the position of the sizing standards. Blue arrows show the predicted positions of FGF20 monomer (28.4 kDa) and dimer (56.8 kDa). FGF16 elutes at fraction 71, corresponding to a molecular size of 24 kDa. The predicted sizes of the monomeric and dimeric wild-type FGF16 (FGF16wt) are 28.6 kDa and 57.2 kDa, respectively.
To confirm further that FGF20 dimerizes, we carried out preliminary sedimentation velocity experiments at ligand concentrations of 2, 6.7, and 11.4 μM in 25 mM HEPES buffer (pH 7.5) and 1 M NaCl at 20°C. A Beckman Coulter analytical ultracentrifuge, model XLI, was used. The sedimenting boundary traces from the UV absorbance detection system were analyzed with the SEDFIT/SEDPHAT suite of programs (56). The sedimentation behavior of FGF20 was found to be that of a rapid monomer-dimer equilibrium, with a Kd value between 30 and 100 nM (data not shown). Thus, FGF20 dimerizes more strongly than FGF9, whose Kd is reported to be 680 nM, according to equilibrium sedimentation (50).
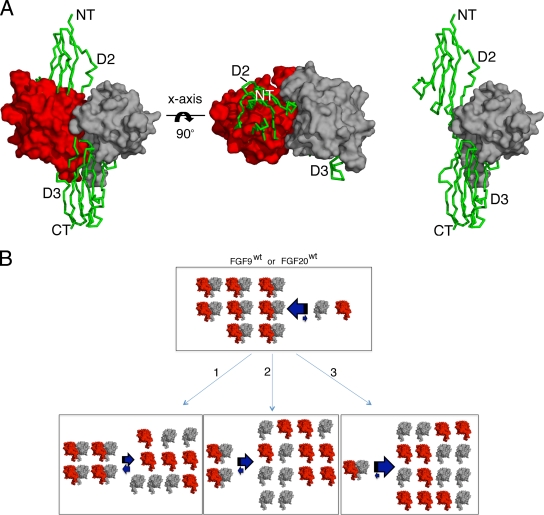
Presently, there is no crystal structure of FGF20 or FGF9 bound to their cognate FGFRs. Nevertheless, when one of the two monomers in the FGF20 dimer (Fig. 4A) or FGF9 dimer (data not shown) is superimposed onto FGF2 in the FGF2-FGFR1c-heparin complex (PDB ID 1FQ9) (55), it is observed that as a dimer, FGF20 is incompatible with receptor binding. This is due to a major overlap between the dimer interface and FGF20's predicted receptor binding sites in the core and N terminus of the ligand (Fig. 3A and 4A). Thus, FGF20 must interconvert between monomer (active) and dimer (inactive) forms to be capable of engaging the receptor. These data led us to propose that homodimerization autoinhibits the ability of FGF9 and FGF20 to bind FGFR by downregulating the effective concentrations of monomeric ligands, the form capable of binding and activating their cognate receptors (Fig. 4B).
FIG. 4.
The FGF20 homodimer is incompatible with FGF receptor binding. (A) One of the FGF20 protomers (in red) from the FGF20 dimer (in red and gray) is superimposed onto FGF2 in the FGF2-FGFR1c-heparin structure (PDB ID 1FQ9). Only a single FGFR1c molecule from the FGF2-FGFR1c-heparin structure is shown. In the left and middle panels, one of the FGF20 monomers (in red) is observed to physically block FGFR binding. For comparison, a 1:1 ratio model of the FGF20-FGFR1c complex is shown in the right panel. The Cα trace of FGFR1c is shown as a green ribbon, with domain 2 and domain 3 labeled as D2 and D3, respectively. NT and CT denote the N and C termini, respectively. The view in the middle panel is related to left panel by a 90-degree rotation along the horizontal axes. (B) Schematic model showing that dimerization downregulates the concentrations of FGF9 and FGF20 monomers capable of receptor binding. A representation of dimeric FGF9 and FGF20 proteins incapable of receptor binding is adapted from panel A, left, while the FGF9 and FGF20 monomers competent for receptor binding are adapted from panel A, right. The three bottom panels illustrate how single (1), double (2), and triple (3) dimer interface mutations would shift the equilibrium toward ligand monomers. The thick arrows indicate the direction of the shift in the equilibrium. FGF9wt, wild-type FGF9; FGF20wt, wild-type FGF20.
To test this hypothesis, we decided to weaken FGF9 and FGF20 homodimerization by introducing mutations into the residues at their dimer interfaces. To this end, the FGF9 and FGF20 dimer interfaces were surveyed for residues that exclusively participate in each ligand's dimerization but are not predicted to mediate ligand binding to FGFR. Four residues in the C-terminal region of FGF9 (D195, L200, I204, L205) and six residues in the C-terminal region of FGF20 (D196, E198, P197, Y204, L207, L208) were found to best satisfy these criteria. All four residues in FGF9 and three of the six residues in FGF20 (E198, L207, and L208) were substituted with alanine either individually or in combination (Table 1). According to our hypothesis, at any given protein concentration, the mutated ligands are predicted to be less dimeric than the wild-type ligands, and as a result, the variants are expected to bind and activate more FGFRs than wild-type ligands (Fig. 4B). Consequently, the mutated ligands should have increased biological activity relative to that of the wild type.
Mutations in the dimer interface of FGF9 and FGF20 increase the effective concentrations of monomeric ligands capable of receptor binding.
We first analyzed the impact of the mutations on ligand dimerization in solution using size exclusion chromatography (Fig. 3B and C and 5A and B). Relative to the wild-type FGF9 and FGF20 ligands, the retention times of all mutated FGF9 and FGF20 proteins increased in the following order: wild-type FGF20 > FGF20E198A > FGF20L208A > FGF20L207A > FGF20L207A/L208A and wild-type FGF9 > FGF9D195A > FGF9I204A > FGF9L205A > FGF9L200A > FGF9I204A/L205A > FGF9L200A/I204A/L205A > FGF9D195A/I204A/L205A.
FIG. 5.
Dimer interface mutations increase the populations of receptor binding-competent monomers. (A) Size exclusion chromatograms of wild-type FGF9 (FGF9wt) and selected FGF9 mutants (FGF9L205A, FGF9I204A/L205A, FGF9D195A/I204A/L205A) are shown in the indicated colors. (B) Schematic plot showing the retention times of all FGF9 proteins, indicated with magenta arrows. Red arrows indicate the positions of the sizing standards. Green arrows show the predicted positions of the monomer and dimer for FGF9 (monomer = 23.4 kDa; dimer = 46.8 kDa). (C) Representative SPR sensorgrams of 800 nM FGFR1c injections over CM5 sensor chips containing covalently immobilized wild-type FGF9 (in black), FGF9D195A (in red), FGF9I204A/L205A (in blue), and FGF9D195A/I204A/L205A (in green). (D) Representative SPR sensorgrams of 800 nM FGFR1c injection over CM5 sensor chips containing covalently immobilized wild-type FGF20 (in black), FGF20L208A (in red), and FGF20L207A/L208A (in green). The biosensor chip response is indicated on the y axis (response units) as a function of time (x axis). (E) Steady-state binding curves of two representative FGF9 proteins (wild-type FGF9 in blue and FGF9D195A in red). The maximum response at equilibrium (Req) is plotted on the y axis as a function of concentration (nM), and the KD values determined from the fitted curves are given. All data were analyzed utilizing BIAevaluation software.
These data indicate that all variants are less dimeric than their wild-type counterparts and that the mutations have an additive effect on ligand monomerization. For example, while wild-type FGF9 migrates at an estimated size of 48.2 kDa, corresponding to a dimer (the predicted size is 47 kDa), the FGF9L205A single mutant and the FGF9D195A/I204A/L205A triple mutant migrate at estimated sizes of 33.3 kDa and 25.3 kDa, respectively (Fig. 5A and B; Table 1). These data confirm that the dimer interface observed in the crystal structure also mediates ligand dimerization in solution.
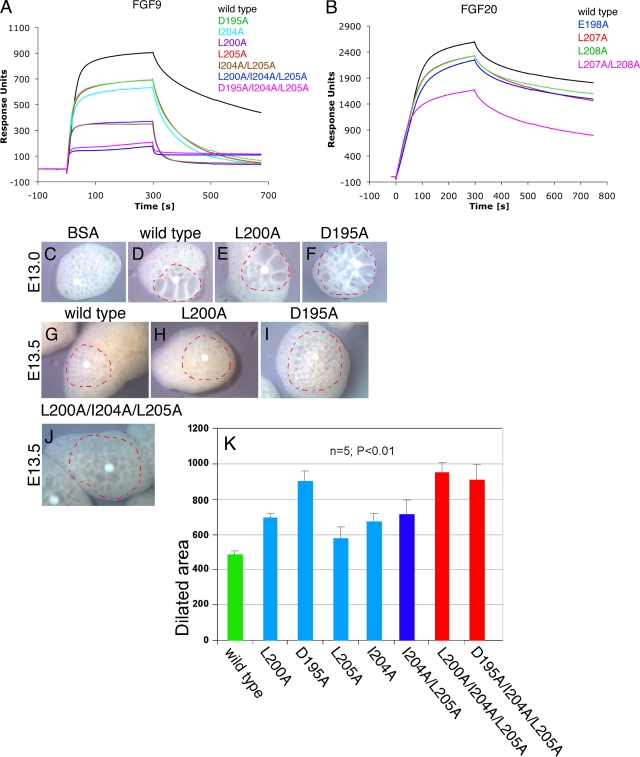
To confirm that the dimer interface mutations have shifted the equilibrium toward monomers, the ligand form capable of receptor binding, we employed SPR spectroscopy to compare the binding of wild-type and mutated ligands to FGFR1c. Wild-type and mutated ligands were immobilized either covalently (using amine coupling) or noncovalently (employing Ni-nitrilotriacetic acid-oriented coupling) onto biosensor chips at a density of ∼1,800 RUs, and serial dilutions of FGFR1c were passed over the chip. Both methods of immobilization gave similar results, and only the results using the amine coupling are shown. At all concentrations of FGFR1c tested (from 50 nM to 2 μM), all mutated FGF9 and FGF20 ligands produced greater signals than the wild-type FGF9 and FGF20 molecules. For example, with 800-nM ligand injection, the responses of FGF9D195A, FGF9I204A, FGF9L205A, and FGF9L200A single mutants were 2.9-, 4.1-, 4.2-, and 6.4-fold greater, respectively, than the response of the wild type (Fig. 5C and data not shown). FGF9I204A/L205A and FGF9L200A/I204A/L205A variants exhibited about 6.9-fold-higher plasmon resonance than the wild type, while FGF9D195A/I204A/L205A showed an enhancement of 9-fold (an average from two experiments) (Fig. 5C and data not shown). Similarly, in the case of FGF20, the single mutants FGF20E198A, FGF20L207A, and FGF20L208A, as well as the double mutant FGF20L207A/L208A, caused increases of 1.5-, 1.4-, 1.8-, and 2.7-fold, respectively, in plasmon resonance upon receptor binding compared to that of wild-type FGF20 (Fig. 5D and data not shown). Importantly, the increases in the plasmon resonance signal conferred by the mutations were proportional to the monomerizing potentials of the mutated ligands, as indicated in Fig. 3C and 5B. Moreover, the KD values of mutated ligands toward FGFR1c were similar to those of the wild-type ligands. For example, the following KD values were determined from the fitted equilibrium binding plots: 0.89 μM and 0.85 μM for wild-type FGF9 and FGF9D195A proteins, respectively (Fig. 5E). This was predictable because the mutated ligands were designed to conserve the FGF-FGFR contacts, without introducing any additional contacts with the receptor. Thus, the mutations were expected merely to increase the available pool of monomeric ligands that are capable of receptor binding (Fig. 4B, bottom). Taken together, these data support our hypothesis that dimerization serves as a mechanism to downregulate the effective concentrations of monomeric receptor binding-competent forms of FGF9 and FGF20.
Mutated FGF9 and FGF20 ligands exhibit increased bioactivity.
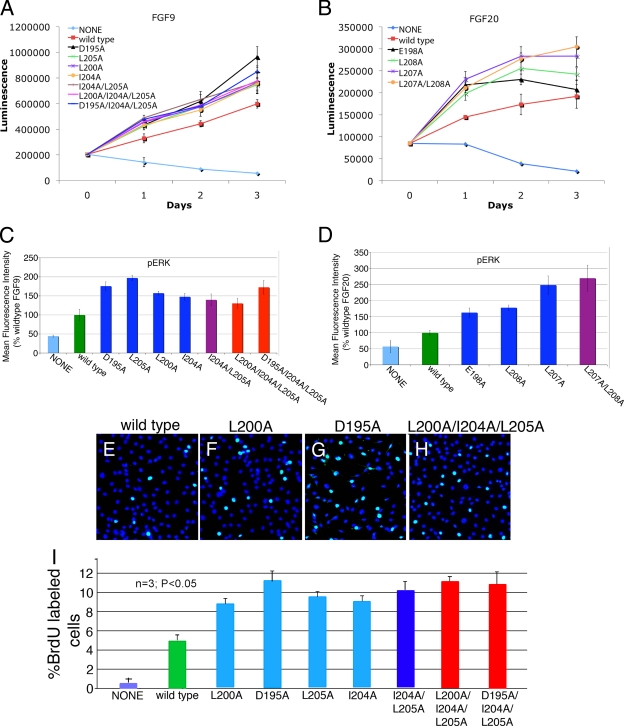
The IL-3-dependent murine pro-B BaF3 cell line does not express FGFRs and HS and therefore cannot be propagated by FGFs. Forced expression of FGFRs, however, makes this cell line responsive to FGF in the presence of exogenous heparin. Consequently, BaF3 cell-based proliferation has served as a standard assay system for studying FGFR activation by FGF (46). To further verify that the mutations shift the monomer-dimer equilibrium toward monomers, the ligand form capable of receptor binding, we compared the ability of wild-type and mutated ligands to activate FGFR1c expressed ectopically in BaF3 cells in either the short term by assessing the extracellular signal-regulated kinase (ERK) activation after 10 min of stimulation or the long term by monitoring cell proliferation over the course of 3 days. BaF3 cells transduced with FGFR1c were starved of IL-3 and stimulated with wild-type or mutated FGF9/20 ligands in the presence of 5 μg/ml heparin. After 10 min of stimulation, the amount of phosphorylated ERK (pERK) was determined, while proliferation was assessed every day for 3 days. Throughout the experiment, all seven FGF9 variants exhibited a modest but reproducible increase in receptor activation (ranging from enhancement of 1.3- to 2.0-fold in pERK after 10 min of stimulation [Fig. 6C] and from enhancement of 1.2- to 1.6-fold on the third day of proliferation assessment [Fig. 6A]) relative to that of the wild-type ligand. Each FGF20 variant also showed modestly increased activity relative to that of the wild type, reaching an enhancement of ∼2.7-fold in pERK (Fig. 6D) and ∼1.6-fold in cell proliferation for the FGF20L207A/L208A double mutant at day 3 of the experiment (Fig. 6B).
FIG. 6.
FGF9 and FGF20 mutants exhibit enhanced proliferative activities. (A) Growth curve of BaF3 cells overexpressing FGFR1c stimulated with 100 ng/ml of wild-type or mutated FGF9 and 5 μg/ml heparin in the absence of IL-3. (B) Growth curve for BaF3/FGFR1c cells stimulated with 100 ng/ml of wild-type or mutated FGF20 and 5 μg/ml heparin in the absence of IL-3. Experiments were performed in triplicate and repeated twice, with similar results. (C, D) Analysis of phospho-p44/42 mitogen-activated protein kinase (pERK) upon stimulation with 100 ng/ml of exogenous wild-type and mutated FGF9 (C) or wild-type and mutated FGF20 proteins (D) supplemented with 5 μg/ml of heparin. pERK activation is represented as a measure of mean fluorescence intensity. (E to H) Fluorescence images showing BrdU incorporation in lung mesenchymal cells treated with wild-type FGF9 (E), FGF9L200A (F), FGF9D195A (G), and FGF9L200A/I204A/L205A (H). BrdU staining is in green, and DAPI staining is in blue. (I) Quantification of BrdU incorporation in wild-type FGF9 and FGF9 variants. Percent BrdU labeled in lung mesenchymal cells after lack of treatment (None) or after 12 h of treatment with wild-type FGF9 and FGF9 mutants. The “n” indicates the number of experiments used for averaging.
Gene knockout studies in mice have shown that FGF9 is required for the proliferation of lung mesenchyme (7). To compare the activity of FGF9 variants in a more biologically relevant setting, we examined BrdU incorporation by lung mesenchymal cells in response to mutated and wild-type FGF9 proteins. Lung mesenchyme was isolated enzymatically on E12.5 and plated on chamber slides. Cells were grown to 70 to 80% confluence, transferred to defined medium, and then stimulated with 20 ng/ml of FGF9 proteins for 12 h, after which BrdU incorporation was performed. As shown in Fig. 6E to I, all FGF9 variants induced DNA synthesis, with an average of twice as many cells as wild-type FGF9. These data suggest that the weakening of ligand dimerization results in an increase in the biological activities of mutated ligands in cultured cells.
Dimerization promotes the interaction of FGF9 and FGF20 with HS and restricts HS-dependent diffusion of these ligands in ECM.
The differential increases in the population of monomers detected in size exclusion and SPR experiments for FGF9 and FGF20 mutants do not manifest in corresponding differential increases in the biological activities of mutated ligands in cultured cells. For example, according to SPR data, the FGF9D195A single and FGF9D195A/I204A/L205A triple mutants exhibit increases of 2.9-fold and 9-fold in their monomeric forms, respectively, relative to that of the wild-type ligand. However, both mutants show comparable enhancements over the wild type (1.6- and 1.4-fold, respectively) in the BaF3 receptor activation assay.
One key difference that could account for the lack of correlation between the SPR assay and the receptor activation assay is that SPR measures only a 1:1 FGF/FGFR binding event, whereas the receptor activation assay is a readout for the net effect of multiple binding events involved in 2:2:2 FGF/FGFR/HS dimerization, including FGF-HS interactions. Therefore, any negative effects of the monomerizing mutations on the FGF-HS interactions could offset the positive contributions of mutations on FGF9 and FGF20's biological activity.
To consider this possibility, we used SPR to compare the impact of monomerizing mutations on the interaction of FGF9 and FGF20 ligands with immobilized heparin, a commonly used surrogate for HS. As shown in Fig. 7A and B, relative to the ability of wild-type ligands, all of the mutants had a reduced ability to interact with immobilized heparin. Importantly, the least-dimeric variants exhibited the greatest loss in HS binding (compare Fig. 7A with 5B and Fig. 7B with 3C), indicating that dimerization enhances the binding of FGF9 and FGF20 to HS. Since SPR signaling is directly proportional to the mass of bound analyte, the reduction in the plasmon signal observed for the mutants may not be necessarily due to the actual loss in the affinity for heparin but rather could simply reflect the fact that the wild type binds predominantly as a dimer, while the mutated variant binds as a monomer. To confirm that the monomerizing mutations truly impact the affinity of the FGF-HS interactions, we compared the ionic strength required to elute wild-type FGF9 and the FGF9D195A/I204A/L205A mutant from a heparin affinity column. This analysis showed that FGF9D195A/I204A/L205A elutes at 780 mM NaCl, which is about 220 mM less salt than that required to elute wild-type FGF9 (data not shown), suggesting that homodimerization augments the affinity of the FGF9 subfamily members for HS.
FIG. 7.
The FGF9 and FGF20 mutants bind heparin more weakly and induce wider dilation of lung epithelium in ex vivo lung explants than the wild type. (A) SPR analysis of wild-type and mutated FGF9 interactions with heparin. Presented are the sensorgrams of the representative 80 nM injections of FGF9 proteins onto a heparin-immobilized chip. (B) Sensorgrams of 60 nM injections of wild-type and mutated FGF20 proteins passed over a heparin-coated chip. The biosensor chip response is indicated on the y axis (response units) as a function of time (x axis). (C to J) Mouse embryonic lungs were explanted on E13.0 and E13.5, and heparin acrylic beads containing bovine serum albumin (BSA) (C), wild-type FGF9 (D, G), or the indicated FGF9 mutants (E, F, H, I, and J) were inserted into the central area of the explant. Outlined in red are dilated areas around the beads, measured after explants were cultured for an additional 24 to 48 h. (K) A statistical quantification of the dilated area around the bead after application of wild-type FGF9 or FGF9 dimerization mutants.
Emerging data show that FGF-HS interactions can regulate the biology of FGFs by controlling the diffusion rate and thus FGF gradients in the HS-abundant ECM. This is illustrated by the FGF19 subfamily, whose members interact poorly with HS and consequently are not trapped in the ECM of the secreting cells, thus enabling them to act in an endocrine fashion (15, 28). Moreover, we have recently demonstrated that a difference in the shapes of HS-dependent gradients underlies the divergent roles of FGF7 and FGF10 in branching morphogenesis (H. P. Makarenkova and M. Mohammadi, unpublished data). Since our data show that dimerization enhances FGF9's interactions with HS, we decided to address the role of dimerization in regulating ligand diffusion in ECM. It has been recently reported that implantation of heparin acrylic beads loaded with FGF9 into ex vivo lung organ explants causes significant dilation of the lung epithelial structures around the bead (8). We adopted this assay to compare the diffusion rates of wild-type and mutated FGF9 ligands. We reasoned that due to their decreased HS-binding affinity, the mutated FGF9 ligands will diffuse and induce lung dilation further away from the FGF source. Wild-type or mutated FGF9 ligands were bound onto heparin acrylic beads, which were then implanted into whole-lung explants. Compared to wild-type FGF9, all mutated ligands induced dilation of a wider area around the bead (Fig. 7C to K). The dilated areas around the FGF9D195A, FGF9L200A/I204A/L205A, and FGF9D195A/I204A/L205A beads were approximately 30 to 35% larger than that around the bead containing wild-type protein (compare Fig. 7D with F and Fig. 7G with I or J; see also Fig. 7K). Together, these experiments suggest that dimerization confines the radius of FGF9 biologic action by promoting ligand's HS binding and thus restricting its HS-dependent diffusion.
Heparin shifts the equilibrium toward the dimer.
As HSs constitute an integral part of FGF signaling in vivo (reviewed in references 18 and 37), they are a likely component to influence the dimerization state of the ligand in a cell. The way HSs are thought to accomplish this is by serving as a matrix for concentrating the ligand on the cell surface and ECM.
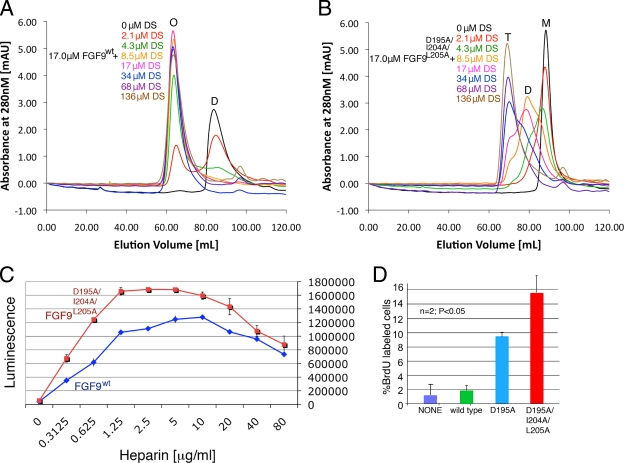
We reasoned that if dimerization increases heparin binding, then the presence of heparin should increase dimerization. To test whether HS can shift the monomer-dimer equilibrium toward the dimer, we added increasing concentrations of heparin DS to wild-type FGF9 and analyzed the mixtures by size exclusion chromatography. Consistent with our prediction, DS shifts the wild-type FGF9 toward the oligomeric state (Fig. 8A). We next tested the ability of DS to dimerize FGF9D195A/I204A/L205A and found that DS is less effective in dimerizing the mutated ligand (Fig. 8B). Specifically, while a 1:1 ratio of wild-type FGF9 to DS was sufficient to entirely shift the FGF9 toward the highest multimer (∼8 molecules of FGF9) (Fig. 8A), the same amount of DS shifts the FGF9D195A/I204A/L205A peak only to approximately a 50:50 mixture of monomer/dimer (Fig. 8B). These data are consistent with our size exclusion (Fig. 3B and C) and SPR data (Fig. 7A and B), showing that the mutated ligands are more monomeric and have reduced affinity for HS. Therefore, we can conclude that the observed differences in bioactivity of the wild-type and mutated ligands are due to the differences in their ability to homodimerize.
FIG. 8.
Heparin shifts the monomer-dimer equilibrium toward the dimer. (A, B) Size exclusion chromatograms of 17 μM of wild-type FGF9 (A) or FGF9D195A/I204A/L205A (B), with various amounts (0, 2.1, 4.3, 8.5, 17, 34, 68, and 136 μM) of heparin DS. M, D, T, and O represent monomer, dimer, tetramer, and octamer peaks, respectively. (C) Growth curve of BaF3 cells overexpressing FGFR1c stimulated with 100 ng/ml of wild-type FGF9 (in blue) or FGF9D195A/I204A/L205A (in red) and various heparin concentrations (0, 0.3125, 0.625, 1.25, 2.5, 5, 10, 20, 40, and 80 μg/ml) in the absence of IL-3. (D) Quantification of BrdU incorporation in variants stimulated with 300 nM wild-type FGF9, FGF9D195A, or FGF9D195A/I204A/L205A. Percent BrdU labeled in lung mesenchymal cells after lack of treatment (None) or after 12 h of treatment with wild-type FGF9 and FGF9 mutants. The “n” indicates the number of experiments used for averaging.
To study the effect of HS on the FGFR activation in BaF3 system, we compared the extents of activation for wild-type FGF9 and FGF9D195A/I204A/L205A by varying the amount of added heparin while keeping the ligand concentration constant. We report that the amount of heparin required to produce maximal bioluminescence responses for both proteins is in the range of 0.625 to 5 μg/ml, while at over 30 μg/ml of added heparin, this difference becomes negligible (Fig. 8C). These findings are consistent with our hypothesis that at a low concentration of heparin, there is a large proportion of the monomeric fraction of FGF9D195A/I204A/L205A compared to that of wild-type FGF9. When the concentration of heparin is raised so that both wild-type FGF9 and FGF9D195A/I204A/L205A exist mostly as dimers, the difference in activation is no longer existent.
Lastly, we chose to perform the cell-based assay using a 300 nM ligand concentration, which is close to the Kd for FGF9 dimerization (680 nM). At this concentration, ∼25% of the wild-type ligand should exist as dimer, whereas the mutants should still be mostly monomeric. Our data show that compared to wild-type FGF9, FGF9D195A and FGF9D195A/I204A/L205A still produced an enhancement of 4.5- and 7.5-fold in the ratio of proliferating cells, respectively (Fig. 8D).
The positive effect conferred by the monomerizing mutations on receptor binding alongside their negative effects on HS binding would then explain why the increases in the population of the monomers observed in solution (size exclusion and SPR experiments) do not manifest in corresponding increases in the biological activities of mutated ligands in cultured cells.
DISCUSSION
Structural and biochemical studies show that the components of growth factor signaling pathways are subject to autoinhibition to combat uncontrolled activation. Indeed, loss of these autoinhibitory mechanisms is often the culprit of human pathologies. For example, in FGFR kinases, pathogenic mutations lead to constitutive kinase action by disengaging the autoinhibitory molecular brake at the kinase hinge region (5). Numerous naturally occurring mutations in the JM regions of KIT, PDGFRα, and FLT3 relieve the autoinhibition provided by JM region interaction with the ATP binding cleft and active site, resulting in constitutive kinase activity that gives rise to cancer (reviewed in reference 21). Recent studies also begin to show the existence of autoinhibition in extracellular regions of receptor tyrosine kinases (RTKs). In the absence of epidermal growth factor, the extracellular domain of the epidermal growth factor receptor is maintained in an autoinhibited conformation by specific intramolecular contacts between the “dimerization arm” in domain II and a homologous region in domain IV to prevent high-affinity binding of the ligand (6, 12). In FGFR, the D1 and D1-D2 linker region, although not necessary for ligand binding, can indirectly modify the ligand and heparin binding affinities of the D2-D3 region through intramolecular interactions (reviewed in reference 38). D1 is proposed to operate in concert with the acid box region in the D1-D2 linker to occlude the ligand binding site and HBS of FGFRs, thereby preventing accidental receptor activation in the basal state (44).
In this study, we provide evidence of autoinhibition in FGF signaling at the level of the ligand by demonstrating that homodimerization autoinhibits signaling activity of FGF9 and FGF20 ligands. We show that dimerization represses receptor binding and limits HS-dependent diffusion of FGF9 and FGF20. We also report here that FGF20 dimerizes as much as 10-fold more strongly than FGF9, according to analytical ultracentrifugation (AUC), although different AUC methodologies were used under slightly different conditions. Interestingly, we found that FGF16, the third member of the subfamily, migrates as a monomer on the gel filtration column (Fig. 3B). Preliminary SPR studies of FGF16 binding affinity to heparin show that FGF16 binds to heparin the weakest compared to FGF9 and FGF20 (see Fig. S1 in the supplemental material). The primary sequences of the predicted HBS of the FGF9 subfamily members are highly conserved (Fig. 3A), and therefore, the differences in HS binding among subfamily members cannot be explained by sequence variation at the HBS. Importantly, these data further corroborate our model that dimerization positively regulates the FGF9 subfamily's binding affinity for HS. Sequence alignment of the FGF9 subfamily shows that FGF16 diverges from FGF9 and FGF20 at its C terminus, the key region mediating ligand dimerization (Fig. 3A). Based on these findings, we propose that the members of the FGF9 subfamily have different propensities to homodimerize, suggesting that gradations of autoinhibitory control by dimerization exist for FGF9/16/20 ligands.
The monomerizing mutations map onto the dimer region that is opposite of the ligands’ predicted HS-binding sites (Fig. 2A), suggesting that the mutations impact HS binding of the ligands indirectly. One likely mechanism would be if ligand monomerization increases the intrinsic thermal motion of the mutant ligands. The increased flexibility of the HBSs of the mutated FGF9 and FGF20 ligands could then impose an entropic loss upon HS binding and could account for the reductions in HS binding of the mutated ligands. Sensitivity to proteases can be used to study protein flexibility. We tested the impact of the mutations on the ligands’ intrinsic flexibility by comparing the sensitivity of wild-type and mutated ligands to specific (trypsin) and nonspecific (thermolysin and elastase) proteases. This assay showed that the more monomeric the mutants are, the more susceptible to proteolysis they become (see Fig. S2 and S4A in the supplemental material). Specifically, cleavage profiles of partially digested wild-type FGF9 and FGF9D195A/I204A/L205A (2 μg each) with 0.5 μg trypsin show that while wild-type FGF9 gives a spread of at least seven identifiable digestion bands, FGF9D195A/I204A/L205A is almost entirely digested (see Fig. S4A, compare lanes 3 and 9, in the supplemental material). In order to identify the cleavage sites/products of limited trypsin digestion of wild-type FGF9 at this concentration, we employed MALDI-TOF analysis. We identified three of the digested fragments (G48-S208, G59-S208, and T70-R190). The cleavage sites for two later products appear to map within the dimer interface (see Fig. S4B to E in the supplemental material). Taken together, these data demonstrate that while an increase in the thermal flexibility of monomers could account for their reduced HS binding relative to that of wild-type ligands, it cannot be ruled out, however, that the appearance of expedited digestion of the mutated derivative is due to the better access of the protease to its substrate within the dimer interface. We predict that a combination of two events is a likely possibility; however, further studies are necessary.
Our data set the stage for the engineering of analogues of FGF9/20 with several potential therapeutic applications. Recombinant human FGF20, velafermin, is currently in phase 1/2 clinical development for the treatment of chemotherapy/radiation-induced oral mucositis due to FGF20 cyto- and radioprotective properties (1, 2, 34). Thus, FGF20 analogues may well be the next generation of velafermin, with increased bioactivities for use in radiation-induced mucositis.
FGF20 analogues may play beneficial role for the treatment of Parkinson's disease. Single-nucleotide polymorphisms in the FGF20 gene have been associated with Parkinson's disease (64). Consistent with this finding, in situ hybridization experiments have shown that FGF20, as well as its cognate receptor (FGFR1c), is expressed in the substantia nigra pars compacta, a region of the brain where dopaminergic (DA) neurons degenerate in Parkinson's disease (42, 43). Studies with cultured DA neurons show that FGF20 is capable of promoting the survival of rat DA neurons in serum-free medium or in the presence of cytotoxic concentrations of glutamate (43). FGF20 has also been shown to be able to differentiate pluripotent stem cells into DA neurons (61) and improve DA neuron survival after transplantation (54). Hence, FGF20 analogues could be used for differentiation, propagation, and maintenance of DA neurons for cell replacement therapy for Parkinson's disease.
Additionally, FGF20 analogues can prove to be valuable tools in cardiovascular biology. Myocardial FGF9 subfamily signaling regulates coronary vascular development by triggering a wave of hedgehog activation that is essential for vascular endothelial growth factor A (VEGF-A), VEGF-B, VEGF-C, and angiopoietin-2 expression (31). FGF9/16/20 agonists can serve as agents to increase coronary vessel density and perfusion. Lastly, engineered FGF9 ligands may also be used to stimulate the growth of lung stem/progenitor cells. Taken together, our data provide a framework for the discovery of novel FGF9 and FGF20 agents with desired biological activity for use in both basic and translational research.
While the manuscript was in revision, a study by Harada et al. was reported (17). This work further strengthens our hypothesis that homodimerization controls the FGF9 subfamily's receptor binding and HS-dependent diffusion in the ECM.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant DE13686, a Postdoctoral Fellowship (PF-07-215-01-TBE) from the American Cancer Society (to S.A.B.) and a research agreement with CuraGen Corporation (to M.M.).
We are grateful to R. Abromowitz and J. Schwanof for synchrotron beamline X4A assistance at the National Synchrotron Light Source, Brookhaven National Laboratory, a DOE facility. The beamline is supported by the New York Structural Biology Consortium. We are also thankful to Fuming Zhang and Robert J. Linhardt for the help with processing of SPR data and to a summer student, Michael B. Houy, for help with lung experiments. Finally, we are obliged to Danny Reinberg for allowing us to use the AUC instrument and Peter Schuck for help with the processing of analytical centrifugation data.
Footnotes
Published ahead of print on 29 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, E., E. G. Fey, P. Valax, Z. Yim, J. D. Peterson, M. Mesri, M. Jeffers, M. Dindinger, N. Twomlow, A. Ghatpande, W. J. LaRochelle, S. T. Sonis, and H. S. Lichenstein. 2003. Preclinical characterization of CG53135 (FGF-20) in radiation and concomitant chemotherapy/radiation-induced oral mucositis. Clin. Cancer Res. 93454-3461. [PubMed] [Google Scholar]
- 2.Ara, G., B. A. Watkins, H. Zhong, T. R. Hawthorne, C. E. Karkaria, S. T. Sonis, and W. J. Larochelle. 2008. Velafermin (rhFGF-20) reduces the severity and duration of hamster cheek pouch mucositis induced by fractionated radiation. Int. J. Radiat. Biol. 84401-412. [DOI] [PubMed] [Google Scholar]
- 3.Bottcher, R. T., and C. Niehrs. 2005. Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 2663-77. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., J. Ma, W. Li, A. V. Eliseenkova, C. Xu, T. A. Neubert, W. T. Miller, and M. Mohammadi. 2007. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell 27717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, H. S., and D. J. Leahy. 2002. Structure of the extracellular region of HER3 reveals an interdomain tether. Science 2971330-1333. [DOI] [PubMed] [Google Scholar]
- 7.Colvin, J. S., A. C. White, S. J. Pratt, and D. M. Ornitz. 2001. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 1282095-2106. [DOI] [PubMed] [Google Scholar]
- 8.del Moral, P. M., S. P. De Langhe, F. G. Sala, J. M. Veltmaat, D. Tefft, K. Wang, D. Warburton, and S. Bellusci. 2006. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev. Biol. 29377-89. [DOI] [PubMed] [Google Scholar]
- 9.Dill, K. A. 1985. Theory for the folding and stability of globular proteins. Biochemistry 241501-1509. [DOI] [PubMed] [Google Scholar]
- 10.Evans, S. J., P. V. Choudary, C. R. Neal, J. Z. Li, M. P. Vawter, H. Tomita, J. F. Lopez, R. C. Thompson, F. Meng, J. D. Stead, D. M. Walsh, R. M. Myers, W. E. Bunney, S. J. Watson, E. G. Jones, and H. Akil. 2004. Dysregulation of the fibroblast growth factor system in major depression. Proc. Natl. Acad. Sci. USA 10115506-15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faham, S., R. J. Linhardt, and D. C. Rees. 1998. Diversity does make a difference: fibroblast growth factor-heparin interactions. Curr. Opin. Struct. Biol. 8578-586. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, K. M., M. B. Berger, J. M. Mendrola, H. S. Cho, D. J. Leahy, and M. A. Lemmon. 2003. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11507-517. [DOI] [PubMed] [Google Scholar]
- 13.Gartside, M. G., H. Chen, O. A. Ibrahimi, S. A. Byron, A. V. Curtis, C. L. Wellens, A. Bengston, L. M. Yudt, A. V. Eliseenkova, J. Ma, J. A. Curtin, P. Hyder, U. L. Harper, E. Riedesel, G. J. Mann, J. M. Trent, B. C. Bastian, P. S. Meltzer, M. Mohammadi, and P. M. Pollock. 2009. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol. Cancer Res. 741-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geske, M. J., X. Zhang, K. K. Patel, D. M. Ornitz, and T. S. Stappenbeck. 2008. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development 1352959-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz, R., A. Beenken, O. A. Ibrahimi, J. Kalinina, S. K. Olsen, A. V. Eliseenkova, C. Xu, T. A. Neubert, F. Zhang, R. J. Linhardt, X. Yu, K. E. White, T. Inagaki, S. A. Kliewer, M. Yamamoto, H. Kurosu, Y. Ogawa, M. Kuro-o, B. Lanske, M. S. Razzaque, and M. Mohammadi. 2007. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 273417-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grose, R., and C. Dickson. 2005. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 16179-186. [DOI] [PubMed] [Google Scholar]
- 17.Harada, M., H. Murakami, A. Okawa, N. Okimoto, S. Hiraoka, T. Nakahara, R. Akasaka, Y. Shiraishi, N. Futatsugi, Y. Mizutani-Koseki, A. Kuroiwa, M. Shirouzu, S. Yokoyama, M. Taiji, S. Iseki, D. M. Ornitz, and H. Koseki. 2009. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat. Genet. 41289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmer, N. J. 2006. Insights into the role of heparan sulphate in fibroblast growth factor signalling. Biochem. Soc. Trans. 34442-445. [DOI] [PubMed] [Google Scholar]
- 19.Hogan, B. L., and J. M. Yingling. 1998. Epithelial/mesenchymal interactions and branching morphogenesis of the lung. Curr. Opin. Genet. Dev. 8481-486. [DOI] [PubMed] [Google Scholar]
- 20.Hou, J., M. Kan, F. Wang, J. M. Xu, M. Nakahara, G. McBride, K. McKeehan, and W. L. McKeehan. 1992. Substitution of putative half-cystine residues in heparin-binding fibroblast growth factor receptors. Loss of binding activity in both two and three loop isoforms. J. Biol. Chem. 26717804-17808. [PubMed] [Google Scholar]
- 21.Hubbard, S. R. 2004. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 5464-471. [DOI] [PubMed] [Google Scholar]
- 22.Ibrahimi, O. A., F. Zhang, A. V. Eliseenkova, N. Itoh, R. J. Linhardt, and M. Mohammadi. 2004. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum. Mol. Genet. 132313-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inagaki, T., M. Choi, A. Moschetta, L. Peng, C. L. Cummins, J. G. McDonald, G. Luo, S. A. Jones, B. Goodwin, J. A. Richardson, R. D. Gerard, J. J. Repa, D. J. Mangelsdorf, and S. A. Kliewer. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2217-225. [DOI] [PubMed] [Google Scholar]
- 24.Iseki, S., A. O. Wilkie, J. K. Heath, T. Ishimaru, K. Eto, and G. M. Morriss-Kay. 1997. Fgfr2 and osteopontin domains in the developing skull vault are mutually exclusive and can be altered by locally applied FGF2. Development 1243375-3384. [DOI] [PubMed] [Google Scholar]
- 25.Jin, C., F. Wang, X. Wu, C. Yu, Y. Luo, and W. L. McKeehan. 2004. Directionally specific paracrine communication mediated by epithelial FGF9 to stromal FGFR3 in two-compartment premalignant prostate tumors. Cancer Res. 644555-4562. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 27.Kharitonenkov, A., T. L. Shiyanova, A. Koester, A. M. Ford, R. Micanovic, E. J. Galbreath, G. E. Sandusky, L. J. Hammond, J. S. Moyers, R. A. Owens, J. Gromada, J. T. Brozinick, E. D. Hawkins, V. J. Wroblewski, D. S. Li, F. Mehrbod, S. R. Jaskunas, and A. B. Shanafelt. 2005. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 1151627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuro-o, M. 2008. Endocrine FGFs and klothos: emerging concepts. Trends Endocrinol. Metab. 19239-245. [DOI] [PubMed] [Google Scholar]
- 29.Kuslak, S. L., J. L. Thielen, and P. C. Marker. 2007. The mouse seminal vesicle shape mutation is allelic with Fgfr2. Development 134557-565. [DOI] [PubMed] [Google Scholar]
- 30.Lalla, R. V. 2005. Velafermin (CuraGen). Curr. Opin. Investig. Drugs 61179-1185. [PubMed] [Google Scholar]
- 31.Lavine, K. J., A. C. White, C. Park, C. S. Smith, K. Choi, F. Long, C. C. Hui, and D. M. Ornitz. 2006. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 201651-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavine, K. J., K. Yu, A. C. White, X. Zhang, C. Smith, J. Partanen, and D. M. Ornitz. 2005. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell 885-95. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence, M. C., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234946-950. [DOI] [PubMed] [Google Scholar]
- 34.Maclachlan, T., B. Narayanan, V. L. Gerlach, G. Smithson, R. W. Gerwien, O. Folkerts, E. G. Fey, B. Watkins, T. Seed, and E. Alvarez. 2005. Human fibroblast growth factor 20 (FGF-20; CG53135-05): a novel cytoprotectant with radioprotective potential. Int. J. Radiat. Biol. 81567-579. [DOI] [PubMed] [Google Scholar]
- 35.Mailleux, A. A., B. Spencer-Dene, C. Dillon, D. Ndiaye, C. Savona-Baron, N. Itoh, S. Kato, C. Dickson, J. P. Thiery, and S. Bellusci. 2002. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development 12953-60. [DOI] [PubMed] [Google Scholar]
- 36.Makarenkova, H. P., M. Ito, V. Govindarajan, S. C. Faber, L. Sun, G. McMahon, P. A. Overbeek, and R. A. Lang. 2000. FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development 1272563-2572. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi, M., S. K. Olsen, and R. Goetz. 2005. A protein canyon in the FGF-FGF receptor dimer selects from an a la carte menu of heparan sulfate motifs. Curr. Opin. Struct. Biol. 15506-516. [DOI] [PubMed] [Google Scholar]
- 38.Mohammadi, M., S. K. Olsen, and O. A. Ibrahimi. 2005. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 16107-137. [DOI] [PubMed] [Google Scholar]
- 39.Murzin, A. G., A. M. Lesk, and C. Chothia. 1992. beta-Trefoil fold. Patterns of structure and sequence in the Kunitz inhibitors interleukins-1 beta and 1 alpha and fibroblast growth factors. J. Mol. Biol. 223531-543. [DOI] [PubMed] [Google Scholar]
- 40.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50157-163. [Google Scholar]
- 41.Nomura, R., E. Kamei, Y. Hotta, M. Konishi, A. Miyake, and N. Itoh. 2006. Fgf16 is essential for pectoral fin bud formation in zebrafish. Biochem. Biophys. Res. Commun. 347340-346. [DOI] [PubMed] [Google Scholar]
- 42.Ohmachi, S., T. Mikami, M. Konishi, A. Miyake, and N. Itoh. 2003. Preferential neurotrophic activity of fibroblast growth factor-20 for dopaminergic neurons through fibroblast growth factor receptor-1c. J. Neurosci. Res. 72436-443. [DOI] [PubMed] [Google Scholar]
- 43.Ohmachi, S., Y. Watanabe, T. Mikami, N. Kusu, T. Ibi, A. Akaike, and N. Itoh. 2000. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem. Biophys. Res. Commun. 277355-360. [DOI] [PubMed] [Google Scholar]
- 44.Olsen, S. K., O. A. Ibrahimi, A. Raucci, F. Zhang, A. V. Eliseenkova, A. Yayon, C. Basilico, R. J. Linhardt, J. Schlessinger, and M. Mohammadi. 2004. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. USA 101935-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ornitz, D. M., and N. Itoh. 2001. Fibroblast growth factors. Genome Biol. 2REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ornitz, D. M., J. Xu, J. S. Colvin, D. G. McEwen, C. A. MacArthur, F. Coulier, G. Gao, and M. Goldfarb. 1996. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 27115292-15297. [DOI] [PubMed] [Google Scholar]
- 47.Orr-Urtreger, A., M. T. Bedford, T. Burakova, E. Arman, Y. Zimmer, A. Yayon, D. Givol, and P. Lonai. 1993. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 158475-486. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 49.Pirvola, U., X. Zhang, J. Mantela, D. M. Ornitz, and J. Ylikoski. 2004. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev. Biol. 273350-360. [DOI] [PubMed] [Google Scholar]
- 50.Plotnikov, A. N., A. V. Eliseenkova, O. A. Ibrahimi, Z. Shriver, R. Sasisekharan, M. A. Lemmon, and M. Mohammadi. 2001. Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J. Biol. Chem. 2764322-4329. [DOI] [PubMed] [Google Scholar]
- 51.Plotnikov, A. N., S. R. Hubbard, J. Schlessinger, and M. Mohammadi. 2000. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101413-424. [DOI] [PubMed] [Google Scholar]
- 52.Plotnikov, A. N., J. Schlessinger, S. R. Hubbard, and M. Mohammadi. 1999. Structural basis for FGF receptor dimerization and activation. Cell 98641-650. [DOI] [PubMed] [Google Scholar]
- 53.Rapraeger, A. C., A. Krufka, and B. B. Olwin. 1991. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 2521705-1708. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Pernaute, R., H. Lee, M. Patterson, C. Reske-Nielsen, T. Yoshizaki, K. C. Sonntag, L. Studer, and O. Isacson. 2008. Parthenogenetic dopamine neurons from primate embryonic stem cells restore function in experimental Parkinson's disease. Brain 1312127-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlessinger, J., A. N. Plotnikov, O. A. Ibrahimi, A. V. Eliseenkova, B. K. Yeh, A. Yayon, R. J. Linhardt, and M. Mohammadi. 2000. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6743-750. [DOI] [PubMed] [Google Scholar]
- 56.Schuck, P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320104-124. [DOI] [PubMed] [Google Scholar]
- 57.Shimada, T., M. Kakitani, Y. Yamazaki, H. Hasegawa, Y. Takeuchi, T. Fujita, S. Fukumoto, K. Tomizuka, and T. Yamashita. 2004. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Investig. 113561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stauber, D. J., A. D. DiGabriele, and W. A. Hendrickson. 2000. Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl. Acad. Sci. USA 9749-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinberg, Z., C. Myers, V. M. Heim, C. A. Lathrop, I. T. Rebustini, J. S. Stewart, M. Larsen, and M. P. Hoffman. 2005. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development 1321223-1234. [DOI] [PubMed] [Google Scholar]
- 60.Su, N., X. Du, and L. Chen. 2008. FGF signaling: its role in bone development and human skeleton diseases. Front. Biosci. 132842-2865. [DOI] [PubMed] [Google Scholar]
- 61.Takagi, Y., J. Takahashi, H. Saiki, A. Morizane, T. Hayashi, Y. Kishi, H. Fukuda, Y. Okamoto, M. Koyanagi, M. Ideguchi, H. Hayashi, T. Imazato, H. Kawasaki, H. Suemori, S. Omachi, H. Iida, N. Itoh, N. Nakatsuji, Y. Sasai, and N. Hashimoto. 2005. Dopaminergic neurons generated from monkey embryonic stem cells function in a Parkinson primate model. J. Clin. Investig. 115102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thisse, B., and C. Thisse. 2005. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 287390-402. [DOI] [PubMed] [Google Scholar]
- 63.Turner, C. A., N. Calvo, D. O. Frost, H. Akil, and S. J. Watson. 2008. The fibroblast growth factor system is downregulated following social defeat. Neurosci. Lett. 430147-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Walt, J. M., M. A. Noureddine, R. Kittappa, M. A. Hauser, W. K. Scott, R. McKay, F. Zhang, J. M. Stajich, K. Fujiwara, B. L. Scott, M. A. Pericak-Vance, J. M. Vance, and E. R. Martin. 2004. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am. J. Hum. Genet. 741121-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White, A. C., J. Xu, Y. Yin, C. Smith, G. Schmid, and D. M. Ornitz. 2006. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 1331507-1517. [DOI] [PubMed] [Google Scholar]
- 66.Whitehead, G. G., S. Makino, C. L. Lien, and M. T. Keating. 2005. fgf20 is essential for initiating zebrafish fin regeneration. Science 3101957-1960. [DOI] [PubMed] [Google Scholar]
- 67.Wilkie, A. O. 2005. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 16187-203. [DOI] [PubMed] [Google Scholar]
- 68.Xu, X., M. Weinstein, C. Li, M. Naski, R. I. Cohen, D. M. Ornitz, P. Leder, and C. Deng. 1998. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125753-765. [DOI] [PubMed] [Google Scholar]
- 69.Yayon, A., M. Klagsbrun, J. D. Esko, P. Leder, and D. M. Ornitz. 1991. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64841-848. [DOI] [PubMed] [Google Scholar]
- 70.Yeh, B. K., M. Igarashi, A. V. Eliseenkova, A. N. Plotnikov, I. Sher, D. Ron, S. A. Aaronson, and M. Mohammadi. 2003. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl. Acad. Sci. USA 1002266-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, X., T. S. Stappenbeck, A. C. White, K. J. Lavine, J. I. Gordon, and D. M. Ornitz. 2006. Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development 133173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.