Abstract
White adipose tissue (WAT) stores energy in the form of triglycerides, whereas brown tissue (BAT) expends energy, primarily by oxidizing lipids. WAT also secretes many cytokines and acute-phase proteins that contribute to insulin resistance in obese subjects. In this study, we have investigated the mechanisms by which activation of peroxisome proliferator-activated receptor γ (PPARγ) with synthetic agonists induces a brown phenotype in white adipocytes in vivo and in vitro. We demonstrate that this phenotypic conversion is characterized by repression of a set of white fat genes (“visceral white”), including the resistin, angiotensinogen, and chemerin genes, in addition to induction of brown-specific genes, such as Ucp-1. Importantly, the level of expression of the “visceral white” genes is high in mesenteric and gonadal WAT depots but low in the subcutaneous WAT depot and in BAT. Mutation of critical amino acids within helix 7 of the ligand-binding domain of PPARγ prevents inhibition of visceral white gene expression by the synthetic agonists and therefore shows a direct role for PPARγ in the repression process. Inhibition of the white adipocyte genes also depends on the expression of C/EBPα and the corepressors, carboxy-terminal binding proteins 1 and 2 (CtBP1/2). The data further show that repression of resistin and angiotensinogen expression involves recruitment of CtBP1/2, directed by C/EBPα, to the minimal promoter of the corresponding genes in response to the PPARγ ligand. Developing strategies to enhance the brown phenotype in white adipocytes while reducing secretion of stress-related cytokines from visceral WAT is a means to combat obesity-associated disorders.
Adipose tissue plays a principal role in overall metabolic homeostasis, and consequently, perturbations in its functions can lead to metabolic disorders, most notably type 2 diabetes and cardiovascular disease. There are essentially two major types of adipose tissue, white (WAT) and brown (BAT). The former is by far the most predominant, particularly in humans, located within distinct visceral depots surrounding the internal organs and distributed throughout the subcutaneous compartment (10, 11). BAT depots exist in adults within small, defined locations throughout the body and distributed as small pockets within WAT depots (3, 10, 34). Adult human BAT activity is more prominent in women than in men, correlates inversely with age and body fat (especially visceral fat), and can be induced by cold exposure (13, 52, 62, 63). The functions of WAT include storage of energy in the form of triglyceride-containing intracellular droplets and secretion of a plethora of hormones and cytokines (adipokines), such as adiponectin and leptin, that regulate the overall energy balance by signaling to other tissues, most notably the brain, liver, and skeletal muscle (51, 53). BAT functions to burn (oxidize) fat to generate heat. Heat generation is facilitated within brown adipocytes by expression of a brown-specific mitochondrial protein, UCP-1, that uncouples electron transport from ATP production, allowing energy to dissipate as heat (12, 29, 36, 49). Brown fat cells also contain abundant mitochondria that facilitate lipid oxidation and the uncoupling process. Obesity results from an increase in adipose tissue mass due primarily to a sedentary lifestyle in concert with a high-fat/calorie diet, leading to an increased storage of triglycerides within lipid droplets of white adipocytes. An extensive increase in fat mass, particularly in visceral WAT depots, is correlated with an increased likelihood of succumbing to the various metabolic disorders, most notably insulin resistance and type 2 diabetes. This is due in part to infiltration of obese white visceral tissue with macrophages that initiate an inflammatory response leading to secretion of various cytokines and acute-phase proteins by adipocytes that exacerbate the stress response (60).
Formation of white adipocytes is regulated by a cascade of transcription factors, most notably peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα, which together orchestrate expression of hundreds of proteins involved in various functions of the mature fat cell, including lipid metabolism and storage as well as secretion of adipokines (18, 50). Development of the brown adipocyte appears to involve this same cascade of factors, but superimposed on it is an additional pathway of nuclear factors primarily responsible for formation of mitochondria and the enzymes involved in lipid oxidation (17, 20). Most notable among these brown-selective factors is PPARγ coactivator 1α (PGC-1α) and PRDM16. PGC-1α functions in several tissues to coactivate transcription factors that regulate mitochondrial biogenesis and induce thermogenesis in BAT (32). Its absence in mice, however, does not prevent formation of BAT, and thus, PGC-1α alone is not the master regulator of brown adipocyte formation (61). That function appears to belong to PRDM16, which is a recently identified 140-kDa zinc-finger PR domain containing platform protein that coordinates the entire brown gene program (54). Specifically, PRDM16 is induced early during brown adipogenesis and acts to direct both PGC-1α and -β to transcription factors docked on brown genes. Additionally, it also functions to facilitate repression of a select set of genes, including the resistin and angiotensinogen genes, that are much more highly expressed in white adipocytes than in brown cells (26).
PPARγ regulates the underlying adipogenic programs common to both white and brown fat cells, but it also likely participates in expression of tissue-specific genes. In brown adipocytes, PPARγ can regulate expression of UCP-1 through its interaction with PGC-1α (4, 35). A component of the brown program might therefore involve mechanisms that direct PPARγ to associate with brown gene coactivators rather than white gene coactivators (42). One possible means by which such a switch could occur is through association of PPARγ with specific ligands. In fact, several recent studies have reported that exposure of white adipocytes in culture or in mice to potent PPARγ agonists induces a “browning” of the white cells, as indicated by induction of mitochondrial formation and function (19, 67, 68). Other studies have also shown that synthetic PPARγ agonists can repress expression of select genes in white cells, including the resistin, α1-acid glycoprotein, and haptoglobin genes (6, 14, 55).
The specific ligands responsible for regulating the adipogenic activity of PPARγ are not known, but potential natural agonists include derivatives of polyunsaturated fatty acids, such as prostanoids (PGJ2) (27, 28). Much of our understanding of PPARγ function has been derived from the use of synthetic agonists, including the thiazolidinedione (TZD) class of insulin sensitizers (38, 66, 70). Recent studies suggest, however, that synthetic PPARγ agonists are capable of inducing different programs of gene expression from those activated by PPARγ during normal adipogenesis and in mature adipocytes (31). For instance, some target genes are expressed at low levels in adipocytes but are responsive to activation of PPARγ by synthetic agonists. Specifically, genes coding for glycerol kinase (GyK) and the oxidized LDL receptor (OLR-1) are PPARγ target genes that are normally expressed at low abundance in WAT and mature adipocytes in culture (9, 24). Studies by Lazar and coworkers have shown that PPARγ is bound to PPARγ response elements in the promoter of the transcriptionally inactive GyK gene in mature adipocytes, in addition to being bound to the enhancer of the transcriptionally active FABP4/aP2 gene. Treatment of cells with TZDs induces production of PGC-1α, which participates in the dislodgement of corepressors from PPARγ on the GyK gene and recruitment of the p160 family of coactivators (23). These data suggest that endogenous ligands are unable to dislodge corepressors from the GyK gene but do facilitate this process, along with recruitment of p160 coactivators to PPARγ bound to the aP2 gene. The fact that synthetic ligands can facilitate this switch and endogenous ligands cannot suggests that the different agonists are capable of inducing unique conformations within PPARγ to facilitate interaction with specific proteins. It is generally accepted that activation of PPARγ involves a repositioning (reconfiguration) of helix 12 of its ligand-binding domain (LBD) to accommodate dislodgement of corepressors and subsequent interaction with coactivators (41, 66). This exchange process is thought to occur while PPARγ is heterodimerized with RXRα and bound to PPARγ response elements within the promoters/enhancers of target genes. Recent investigations have identified mechanisms by which PPARγ represses the transcriptional activation of inflammatory response genes in macrophages (48). The process involves a synthetic agonist-dependent SUMOylation of the LBD of PPARγ, which targets it, along with NCoR-histone deacetylase 3 (HDAC3) complexes, to transcription factors, most notably NF-κB bound to inflammatory gene promoters. The ability of PPARγ to facilitate transrepression of these genes requires a reconfiguration of helix 7 of the LBD in order to expose the appropriate lysine for SUMOylation (21, 40).
In this study, we identified mechanisms by which synthetic PPARγ agonists repress a select set of white adipocyte genes, including the resistin, angiotensinogen, chemerin, Wdnm1-like protein, and Pank3 genes. In mice, this set of genes (“visceral white”) is more abundantly expressed in visceral WAT depots than in subcutaneous or BAT. Treatment of the mice with synthetic PPARγ agonists represses these genes in visceral WAT and induces a brown adipocyte phenotype, as previously shown by others (67, 68). Repression of the “visceral white” genes by the synthetic agonists requires functional PPARγ and depends on C/EBPα as well as the corepressors CtBP1 and CtBP2. Inhibition of resistin and angiotensinogen gene transcription coincides with a PPARγ-agonist-dependent recruitment of CtBP1 and CtBP2 to complexes containing C/EBPα that are associated with the corresponding promoters.
MATERIALS AND METHODS
Specific reagents were purchased from various vendors, as listed: dexamethasone (DEX), 3-isobutyl-1-methylxanthine (MIX), insulin from Sigma (St. Louis, MO); leupeptin, aprotin, and puromycin from American Bioanalytical (Natick, MA); Dulbecco's modified Eagle's medium (DMEM) from Mediatech, Inc. (Herndon, VA); fetal bovine serum (FBS) from Gemini Bio-Products; calf serum from Invitrogen; and troglitazone from Biomol International.
Cell culture.
The cell lines, including Swiss fibroblasts, 3T3-L1 preadipocytes, and mouse embryo fibroblasts (MEFs), were grown in DMEM containing 10% FBS (MEFs and Swiss fibroblasts) or 10% calf serum (preadipocytes) until confluent and were then maintained in the same medium for an additional 2 days. Differentiation was induced at 2 days postconfluence (day 0) by adding fresh DMEM containing 10% FBS, 0.5 mM MIX, 1 μM DEX, and 1.67 μM insulin with or without 5 μM troglitazone, as needed. Troglitazone was added for the last 2 days (day 6 to day 8) of 3T3-L1 adipocyte differentiation, and samples (either RNA or protein) were processed on day 8. In the case of MEFs, troglitazone was added for the first 4 days in order to initiate differentiation and was then removed during the terminal phases of differentiation. In experiments investigating the effect of troglitazone on adipocytes gene expression, the adipocytes were treated with TZD for 2 days between day 8 and day 10. Swiss fibroblasts were exposed to troglitazone for the entire period of differentiation. RNA was then harvested from each of the cell lines at day 10 for quantitative PCR (Q-PCR) analysis. The immortalized primary brown preadipocytes (a gift of C. R. Kahn, Joslin Diabetes Center, Boston, MA) were grown to confluence in differentiation medium composed of DMEM containing 10% FBS supplemented with 20 nM insulin and 1 nM 3,3′,5-triiodo-l-thyronine (T3). Two days postconfluence, the cells were induced to differentiate by exposure to 1 μM DEX, 0.5 mM MIX, 1.67 μM insulin, 0.125 mM indomethacin, and 10% FBS. The cells were refed every 2 days.
Plasmids and viruses.
Expression vectors corresponding to PPARγ and its mutant forms were generated as described previously (65). WT-PPARγ pBabe-Puro (wild-type [WT]-PPARγ), EF-PPARγ pBabe-Puro (EF-PPARγ), or pBabe empty vector-Puro (control) were stably introduced into 3T3-L1 preadipocytes as follows. Plates (10 cm) of human embryonic kidney 293T cells were transiently transfected with 10 μg of the pBabe vectors and the viral packaging vectors pVPack-VSVG vector and pVPack-GAG-POL vector using the TransIT-Express transfection reagent (Mirus Bio Corp.). At 48 h after transfection, virus-containing medium was collected and passed through a 0.45-μm-pore-size syringe filter. Filter-sterilized Polybrene (hexadimethrine bromide; 12 μg/ml) was added to the virus-loaded medium. This medium was then applied to proliferating (40% confluent) 3T3-L1 preadipocytes. At 48 h after infection, cells were treated with trypsin and replated in a medium supplemented with puromycin as the selection antibiotic. Using the same technique, CEBPα−/− MEFs (a gift of Gretchen Darlington, Baylor University) were infected with WT-PPARγ pBabe-Puro vector to generate a cell line referred to as C/EBPα−/− + PPARγ MEFs. These cells were then infected with pRevTRE-CEBPα-Myc-His Hygro (44) or the empty vector, pRevTRE Hygro retrovirus, to generate MEFs that are deficient for C/EBPα (−C/EBPα + PPARγ) or express an ectopic C/EBPα (+C/EBPα + PPARγ) and are adipogenic due to the ectopic expression of PPARγ in both cell lines. Additionally, C/EBPα−/− MEFs were infected with pRevTRE-CEBPα-Myc-His Hygro alone to give rise to C/EBPα−/− MEFs + C/EBPα. Five cell lines were then analyzed: wild-type MEFs (WT-MEFs), C/EBPα−/− MEFs, C/EBPα−/− MEFs (+PPARγ), C/EBPα−/− MEFs (+C/EBPα), and C/EBPα−/− MEFs (+PPARγ + C/EBPα). Lentiviral vectors carrying short hairpin RNAs (shRNAs) specific for mouse CtBP1 and CtBP2 and enhanced green fluorescent protein (EGFP) (used as a negative control) were purchased from Open Biosystems, Inc. Virus stocks were prepared as recommended by the supplier. All shRNAs were expressed using pLKO1-puro, a lentiviral expression vector in which shRNAs are generated under the control of the human U6 promoter. This vector also carries a puromycin resistance marker gene under the control of the human phosphoglycerate kinase gene promoter, which allows the selection of 3T3-L1 transfected cells. To produce infectious lentiviral particles, each shRNA expression plasmid (3 μg) was mixed with the vectors pCMV-dR8.2dvpr (2.7 μg) and pCMV-VSVg (0.3 μg) and transfected into HEK-293T cells using the transfection reagent TransIT-Express (Mirus Bio Corp.) according to the manufacturer's instructions. Medium from these cultures was collected after 2 days, passed through a 0.45-μm filter, and used as a source for lentiviral shRNAs. Knockdown of expression of the target gene was assessed by performing Western blot analyses and Q-PCR analysis.
Oil Red O staining.
Cells were seeded in 35-mm dishes, and at the specified stage of differentiation they were rinsed with phosphate-buffered saline (PBS) and fixed with formalin in PBS for 15 min. After two washes in PBS, the cells were stained for at least 1 h in a freshly diluted Oil Red O solution (six parts Oil Red O stock solution and four parts H2O; Oil Red O Stock solution is 0.5% Oil Red O in isopropyl alcohol). The stain was the removed, and cells were washed twice with water and then photographed.
Western blot analysis.
Monoclonal anti-PPARγ antibody and polyclonal antibodies against CtBP, cyclophilin, β-actin-horseradish peroxidase, and C/EBPα were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CtBP2 antibody was purchased from BD Transduction Laboratories. Polyclonal anti-aP2 serum was kindly provided by David Bernlohr (University of Minnesota), while antiperilipin antibody was kindly provided by Andrew Greenberg (Tufts University, Boston, MA). Equal amounts of protein extracted from the total cell layer were fractionated on 8% or 12% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (PerkinElmer Life Sciences). Following transfer, the membranes were blocked with 10% nonfat dry milk in PBS-0.1% Tween 20 and probed with the antibodies corresponding to the various target proteins. Horseradish peroxidase-conjugated secondary antibodies (Sigma) and an ECL substrate kit (PerkinElmer Life Sciences) were used for detection of specific proteins.
Gene expression analysis.
RNA was isolated from cultured cells using TRIzol reagent (Invitrogen) and from frozen fat tissue using a TRIzol/Qiagen RNeasy tissue kit, both according to the manufacturer's instructions. cDNAs from cultured cells were made from equivalents amounts of total RNA by using a high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. cDNA from frozen fat tissue was synthesized from 3 μg isolated RNA using SuperScript II and oligo(dT) (Invitrogen). Analysis of gene expression was done using Maxima Sybr green qPCR master mix (Fermentas Life Sciences) in the ABI Prism 7300 sequence detector for an initial denaturation at 95°C for 10 min followed by 40 PCR cycles, each cycle consisting of 95°C for 15 s, 60°C for 20 s, and 72°C for 30 s, and SYBR green fluorescence emissions were monitored after each cycle. For each gene, mRNA expression was calculated relative to TATA binding protein (TBP) for murine samples. Amplification of specific transcripts was confirmed by melting-curve profiles (cooling the sample to 68°C and heating slowly to 95°C with measurement of fluorescence) at the end of each PCR. The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis. Primer sequences used for the Q-PCR are provided elsewhere (see Table S1 in the supplemental material).
Mouse studies.
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of University of Texas Southwestern Medical Center at Dallas. All in vivo experiments were performed with littermate-controlled male FVB mice. Mice were housed in groups of two to five in filter-top cages. The colony was maintained in a pathogen-free Assessment and Accreditation of Laboratory Animal Care-accredited facility at the University of Texas Southwestern Medical Center at Dallas under controlled environmental settings (22 to 25°C, 40% to 50% humidity). Mice were maintained on 12-h light and dark cycles with ad libitum access to water and a standard chow diet (5058; LabDiet) or a high-fat diet (D12492; Research Diets Inc.) as appropriate. The high-fat-diet-fed cohort was maintained on this diet for 12 weeks. The PPARγ agonist 2-(2-(4-phenoxy-2-propylphenoxy)ethyl)indole-5-acetic acid (COOH) was a kind gift from Merck (5). The COOH and vehicle were administered by gavage daily at 12 noon for 14 days at 10 mg/kg body weight at week 10 of age. Mice were sacrificed within 6 h after the last gavage, and adipose tissues were collected.
Cellular respiration assay.
A Seahorse Bioscience instrument (model XF24) was used to measure the rate of change of dissolved O2 in medium immediately surrounding adipocytes derived from 3T3-L1 and Swiss fibroblast cell lines (control, WT-PPARγ, and EF-PPARγ) cultured in custom 24-well plates. Measurements were performed using a sensor cartridge, where 24 optical fluorescent O2 sensors are embedded in a sterile disposable cartridge that is configured as individual well “plungers.” For measurements of rates, the plungers gently descend into the wells, forming a chamber that entraps the cells in a ∼7-μl volume. Measurements of the O2 concentration are made periodically over a selected period of time, typically 1 to 2 min, and the rates of oxygen consumption are obtained from the slopes of concentration changes versus time. After the rate measurements are made, the plungers ascend, and the plate is gently agitated to reequilibrate the medium. To prepare the cell plate for an XF24 assay, 1 ml of warmed DMEM buffer (pH 7.4) lacking bicarbonate (8.3 g/liter; Sigma no. D5030), 5.5 mM glucose (Sigma no. G8270), 2 mM GlutaMax-1 (Gibco no. 35050-061), NaCl (1.85 g/liter), and phenol red (15 mg/liter) to 37°C was added to each well. Cells were equilibrated in the medium at 37°C for 30 min, and then baseline metabolic rates were measured over the next ∼30 min and were reported in nmol/min for the oxygen consumption rate. Genomic DNA was extracted from each well using Pico Pure DNA extraction (Arcturus) and was quantified and used to normalize the O2 consumption level/well. Mitochondrial DNA content was assessed as described by Facucho-Oliveira and collaborators (16). Primer sequences used are listed elsewhere (see Table S1 in the supplemental material).
Oligonucleotide pulldown assays.
Nuclear extracts were prepared with an NE-PER nuclear and cytoplasmic extraction reagent kit (Pierce Biotechnologies). Protein concentrations were determined by the Bradford method (Bio-Rad). Equal amounts of protein (250 μg) were mixed with oligonucleotides containing C/EBP consensus and mutant sequences that had been conjugated to agarose (Santa Cruz Biotechnologies). Purifications were performed as recommended by the manufacturer, and protein complexes were eluted in Laemmli buffer, boiled for 3 min, and analyzed by Western blotting.
Protein interaction analysis.
Adipocytes derived from 3T3-L1 preadipocytes differentiated for 8 days and treated for the last 2 days with either dimethylsulfoxide (DMSO) or troglitazone were fixed by addition of 37% formaldehyde to a final concentration of 1% formaldehyde, followed by incubation at room temperature for 10 min. Cross-linking was stopped by addition of glycine to a final concentration of 0.125 M. Cells were then scraped, and nuclear extracts were prepared and sonicated with components of the EZ-Magna ChIP G chromatin immunoprecipitation (ChIP) kit (Millipore) according to the manufacturer's protocol. The protein/chromatin fractions were incubated in each case with 5 μg of one of the following antibodies: anti-PPARγ (Cell Signaling), anti-C/EBPα (Santa Cruz), anti-CtBP (Santa Cruz), and immunoglobulin G (IgG) rabbit (Millipore) at 4°C overnight with magnetic protein G beads. After extensive washing, magnetic beads/protein complexes were resuspended in Laemmli buffer (1×) and boiled for 10 min. Samples were then analyzed by immunoblot.
ChIP assay.
Adipocytes derived from the C/EBPα-deficient/PPARγ MEFs (−CEBPα and +CEBPα) or from 3T3-L1 preadipocytes (−shEGFP, −shCtBP1, and −shCtBP2) treated for the previous 2 days with either DMSO or troglitazone were fixed by addition of 37% formaldehyde to a final concentration of 1% formaldehyde and incubation at room temperature for 10 min. Cross-linking was stopped by addition of glycine to a final concentration of 0.125 M. Cells were then scraped, and samples were prepared using the EZ-Magna ChIP G ChIP kit (Millipore) according to the manufacturer's protocol. The chromatin fractions were incubated in each case with 2 μg of one of the following antibodies: anti-PPARγ (Santa Cruz), anti-C/EBPγ (Santa Cruz), anti-CtBP (Santa Cruz), anti-acetylated histone H3 (provided by the Millipore kit), and mouse and rabbit IgG (Millipore) at 4°C overnight with magnetic protein G beads. After extensive washing and final elution, the product was treated for 4 h at 65°C to reverse cross-linking. Input DNA and immunoprecipitated DNA were purified using the kit column and analyzed by Q-PCR using Maxima Sybr green qPCR master mix (Fermentas Life Sciences) with different primers (both proximal and distal promoter regions) as described previously (26) (listed in Table S1 in the supplemental material. Data were normalized by input values (for each promoter and with or without Trog), and binding is expressed relative to the nonspecific binding of IgG-immunoprecipitated DNA content. Data are expressed as means ± standard errors (n = 3), with P values of <0.01 considered significant.
RESULTS
Studies by Wilson-Fritch and collaborators showed that TZD treatment of 3T3-L1 adipocytes induces a set of genes coding for proteins involved in mitochondrial function and lipid oxidation, including the Cidea, Cox7a, and Cox8b genes (67). In fact, additional studies by these investigators demonstrated that treatment of mice with PPARγ agonists remodels WAT to adopt a browner phenotype that is capable of greater oxygen consumption at the expense of metabolizing lipids (68). To define the mechanisms by which PPARγ can selectively induce a brown phenotype in white adipocytes, we analyzed the pattern of gene expression produced during the conversion of Swiss fibroblasts expressing ectopic PPARγ (Swiss-PPARγ cells) into adipocytes in response to normal adipogenic inducers versus additional exposure to synthetic ligands. During these investigations, we identified several genes whose expression was selectively repressed by exposure of cells to synthetic ligands including the TZDs, troglitazone, rosiglitazone, and pioglitazone, as well as the non-TZD agonist GM1929. Analysis of this set of genes in 3T3-L1 preadipocytes demonstrated that they are all induced to various extents during 10 days of normal adipogenesis (Table 1, 3T3-L1 D10/D0), but the extent of adipogenic expression is significantly repressed by troglitazone (Table 1, 3T3 L1, +Trog/−Trog). For instance, angiotensinogen mRNA is induced three- to fourfold during normal adipogenesis but repressed at least 60% (ratio of expression with troglitazone to that without troglitazone = 0.4) by exposure of differentiating cells to troglitazone. It was reassuring that this analysis identified the resistin gene within this group, since it had previously been shown to be an adipogenic gene in the mouse and significantly repressed by TZDs (55). In comparison, the levels of two other adipogenic genes, encoding C/EBPα and FABP4/aP2, are unresponsive to the synthetic agonist. We chose to further investigate five genes that were representative of the group shown in Table 1, including the resistin, angiotensinogen, chemerin, Wdnm1-like protein, and Pank3 genes (2, 15, 22, 55, 69, 71).
TABLE 1.
Genes induced during white adipocyte (3T3-L1 cell) differentiation and inhibited by troglitazone activation of PPARγa
| Protein (gene) | 3T3-L1
|
WAT/BAT | |
|---|---|---|---|
| +Trog/−Trog ratio | D10/D0 ratio | ||
| Pank3 (Pank3*) | 0.1 | 4.7 | 3.9 |
| Adcyap1r1 (Adcyap1r1) | 0.1 | 12.2 | 2.6 |
| Haptoglobin (Hp) | 0.2 | 6.5 | 2.4 |
| Vnn3 (Vnn3) | 0.2 | 8.4 | 15.4 |
| Sult1a1 (Sult1a1) | 0.2 | 44.3 | 4.3 |
| Resistin (Retn*) | 0.3 | 2,063 | 21.6 |
| Adrb3 (Adrb3) | 0.3 | 988.2 | 10 |
| Lumican (Lum) | 0.3 | 6.4 | 4 |
| Apcdd1 (Apcdd1) | 0.3 | 20.6 | 41.4 |
| Aoc3 (Aoc3) | 0.3 | 18 | 3.1 |
| Chemerin (Rarres2*) | 0.3 | 33.5 | 9.5 |
| Cyp2f2 (Cyp2f2) | 0.4 | 14.6 | 25.6 |
| Angiotensinogen (Agt*) | 0.4 | 3.7 | 215.4 |
| Wdnm1-like protein* | 0.4 | 643.8 | 34.5 |
| Mc2r (Mc2r) | 0.5 | 165.3 | 7.3 |
| Gpr43-Ffar2 (Ffar2) | 0.5 | 10.8 | 4.6 |
| Sortilin 1 (Sort1) | 0.6 | 9.2 | 3.7 |
| PPARγ (Pparγ) | 0.6 | 9.7 | 0.6 |
| C/EBPα (Cebpα) | 1.7 | 35.4 | 1 |
| Fabp4 (Fabp4) | 1.8 | 522.9 | 0.8 |
Genes marked with an asterisk are those chosen for further study. The ratio of relative level of expression (Q-PCR relative number) of selected genes in 3T3-L1 adipocytes cultured in the presence (+) or absence (−) of 5 μM troglitazone (Trog) or in 3T3-L1 adipocytes (day 10 [D10]) versus preadipocytes (day 0 [D0]) is shown. Total RNA was extracted from WAT (mesenteric depot) and BAT of five mice. Expression of the genes was analyzed by Q-PCR. Data are presented as the WAT/BAT ratios of gene expression.
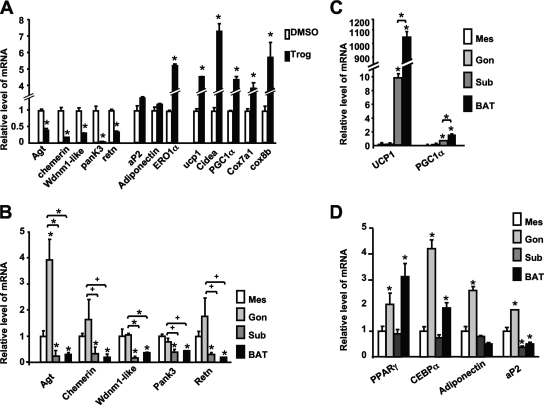
As mentioned above, treatment of white adipocytes, such as 3T3-L1 cells, with TZDs induces production of mitochondrial proteins accompanied by increased oxygen consumption and fatty acid oxidation. To determine whether the TZD-repressed genes are also inhibited during the TZD-associated remodeling of white adipocytes into “brown-like” cells, we exposed mature 3T3-L1 adipocytes to troglitazone for 48 h and quantified various mRNAs by Q-PCR. Figure 1A shows the expected induction of brown adipocyte-enriched genes, including the UCP-1, PGC-1α, Cidea, Cox7a1 and Cox8b genes, by the TZD; but more importantly, there is an accompanying repression of genes encoding angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin without any significant effect on adiponectin or FABP4/aP2 expression. These data suggest that this subgroup of white adipocyte genes is likely expressed at low levels in BAT compared to levels in WAT, as was shown for three of them (resistin, angiotensinogen, and Wdnm1-like protein genes) by Kajimura and collaborators (26). To assess the pattern of expression of these TZD-repressed genes in different adipose depots, RNA was extracted from brown and different white adipose depots, including subcutaneous, mesenteric, and gonadal, of chow-fed mice. Figure 1B presents the Q-PCR analysis of the RNA and demonstrates that the five TZD-repressed genes, encoding angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin, are expressed at much higher levels in mesenteric and gonadal white depots than in the brown depot. It is interesting, however, that the level of expression in the subcutaneous depot is as low as that in the brown depot. We also demonstrate that select brown adipocyte genes, encoding PGC-1α and UCP-1, are more abundantly expressed in the subcutaneous depots than in the visceral depots but not to the levels expressed in BAT (Fig. 1C). The adipogenic genes common to both WAT and BAT, most notably the PPARγ, C/EBPα, adiponectin, and FABP4/aP2 genes, are abundantly produced in all depots (Fig. 1D). Additionally, the five TZD-repressed genes are induced to significantly higher levels during the differentiation of white preadipocytes than the levels attained during brown adipogenesis (see Fig. S1 in the supplemental material). Based on these observations, we have chosen to refer to these genes as “visceral white.”
FIG. 1.
Troglitazone inhibits expression of select “visceral white” genes while inducing brown gene expression. (A) Q-PCR analysis of mRNA levels of genes encoding “visceral white” (chemerin, Wdnm1-like protein, resistin [retn]), angiotensinogen [agt], and Pank3), classic adipocyte (Fabp4/aP2 and adiponectin), brown fat-selective (UCP-1, cidea, PGC-1α, Cox7a1, and Cox8b), or Ero1α proteins expressed in 3T3-L1 adipocytes exposed to either DMSO or 5 μM troglitazone (Trog) for 2 days. Data are represented as n-fold changes in mRNA expression compared to mRNA expressed in DMSO-treated cells. Each bar represents the mean value ± standard errors (SE) (n = 3; *, P < 0.01 relative to control results [DMSO]). Q-PCR analysis of expression of mRNAs corresponding to the “visceral white” genes (B), “brown” genes (C), or adipocyte markers (D) in different mouse adipose tissues, including BAT, mesenteric (Mes), gonadal (Gon), and subcutaneous (Sub), is shown. Data are represented as n-fold changes in mRNA expression compared to mRNA expressed in mesenteric adipose tissue. Each bar represents the mean ± SE (n = 5; *, P < 0.01; +, P < 0.05 relative to mesenteric values for each gene).
As discussed earlier, Wilson-Fritch and colleagues showed that PPARγ agonists remodel white adipocytes in vivo by enhancing mitochondrial biogenesis, which is accompanied by enhanced oxygen consumption and lipid oxidation (68). To assess whether the select “visceral white” genes are also suppressed during this remodeling process, we administered a potent non-TZD PPARγ agonist, COOH (5), by gavage to chow-fed mice for 2 weeks, at which stage animals were sacrificed and adipose tissues isolated for analysis of mRNA expression using Q-PCR assays. Figure 2A and B shows an extensive decrease in expression of angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin mRNAs in the mesenteric and gonadal white fat depots in response to the synthetic agonist, along with a corresponding increase in UCP1 mRNA expression, indicating a shift to a brown phenotype in these white depots. A similar effect of the agonist on the “visceral white” genes is not as apparent in the subcutaneous depots, likely because the expression of these genes is already at the low levels normally expressed in the brown depot (Fig. 2C and D).
FIG. 2.
A synthetic PPARγ agonist inhibits “visceral white” gene expression in vivo. Mice were given a PPARγ agonist (COOH) and vehicle at 10 weeks of age for 14 days at 10 mg/kg. Total RNA was extracted from WAT of different depots (mesenteric [A], gonadal [B], subcutaneous [C], and brown [D]). Expression of the “visceral white” genes (see Fig. 1) was analyzed by Q-PCR. Data are represented as n-fold changes in mRNA expression compared to mRNA expressed in non-COOH (vehicle)-treated mice (five mice for each group). Each bar represents mean ± standard errors (n = 5; *, P < 0.01; +, P < 0.05 relative to results for non-COOH-treated mice).
The data in Fig. 2 showing suppression of the “visceral white” genes in mesenteric and gonadal depots in response to treatment of mice with the COOH compound are consistent with a direct role for PPARγ in the repression process. Previously, we demonstrated that the response of PPARγ to synthetic agonists involves select amino acids (E365 and F372) within helix 7 of its LBD (65). Consequently, we questioned whether a PPARγ molecule in which E365 and F372 are modified to alanines (EF-PPARγ) could repress “visceral white” gene expression. To address this question and to demonstrate a role for PPARγ in the repression process, we analyzed mRNA expression in Swiss fibroblasts ectopically expressing either WT-PPARγ or EF-PPARγ. As observed in earlier studies (45, 65), we demonstrated that ectopic expression of WT-PPARγ in the fibroblasts was capable of inducing their conversion to adipocytes in response to treatment with DMI alone (i.e., activation by endogenous ligands and/or coactivators) whereas expression of EF-PPARγ was unresponsive to endogenous activity and required activation with a synthetic agonist to induce conversion of the fibroblasts into lipid-laden cells (data not shown). The data in Fig. 3A show that expression of angiotensinogen, resistin, chemerin, Pank3, and Wdnm1-like protein was induced manyfold in response to activation of WT-PPARγ by endogenous effectors (DMI alone) along with the classic adipogenic gene, encoding FABP4/aP2. Exposure of differentiating WT-PPARγ cells to troglitazone significantly inhibited expression of the five “visceral white” genes, and it also resulted in a dramatic induction of brown genes, including those encoding UCP-1, PGC-1α, Cidea, and Cox7a1, as well as FGF21 (Fig. 3A). Additionally, induction of the brown genes coincided with a significant increase in O2 consumption in response to the troglitazone activation of WT-PPARγ (Fig. 3B). As expected, exposure of EF-PPARγ cells to DMI alone (without troglitazone) failed to induce adipogenesis and the “visceral white” genes since this PPARγ mutant did not respond to endogenous ligand and/or coactivator activity. Activation of EF-PPARγ with troglitazone induced adipogenesis based on expression of FABP4/aP2 (Fig. 3A); interestingly, though, it also induced expression of the “visceral white” genes to levels detected in WT-PPARγ fibroblasts undergoing optimum differentiation (without troglitazone) (Fig. 3A). These data suggest that mutation of E365 and F372 within PPARγ appears to eliminate the repressive response to synthetic ligands (troglitazone) while retaining the activation response. Additionally, it appears that mutation of E and F prevents induction of UCP-1, Cidea, Cox7a1, PGC-1α, and FGF21 mRNAs, as well as preventing enhancement of mitochondrial biogenesis and O2 consumption in response to troglitazone (Fig. 3B).
FIG. 3.
PPARγ mediates the troglitazone-associated repression of the “white visceral” genes and induction of the brown phenotype in Swiss 3T3 adipocytes. (A) Q-PCR analysis of “visceral white” and “brown” gene expression in Swiss fibroblasts expressing WT-PPARγ (WT), EF-PPARγ (EF), or a control vector differentiated by the presence or absence of troglitazone (Trog). Data are represented as relative amounts of mRNA, in which each bar represents the mean (n = 3; *, P < 0.01 relative to results for DMSO-treated cells for each cell line). (B) Cellular respiration (left) and mitochondrial DNA content as assessed by Q-PCR (right) in WT-PPARγ (WT), EF-PPARγ (EF), or control (C) Swiss 3T3 adipocytes. Data are represented as n-fold induction compared to data obtained for Swiss control cells treated with DMSO. Each bar represents the mean ± standard error (n = 3; *, P < 0.01).
The fact that EF-PPARγ is incapable of inducing “visceral white” gene expression in Swiss fibroblasts in response to endogenous effectors (ligands and/or coactivators) prevented us from assessing the effect of the mutations within helix 7 on repression versus activation activity of PPARγ. As an additional approach, we chose to constitutively overexpress EF-PPARγ and WT-PPARγ, as well as a control vector, in 3T3-L1 preadipocytes. In fact, ectopic WT-PPARγ induced some spontaneous differentiation in proliferating preadipocytes, as indicated by an enhanced production of aP2/FABP4 (see Fig. S2A in the supplemental material), presumably in response to a low level of endogenous ligands in these cells. In comparison, EF-PPARγ is incapable of activating adipogenesis in proliferating preadipocytes since it is unresponsive to endogenous ligands (65). The levels of expression of WT-PPARγ and EF-PPARγ are similar in both proliferating and differentiated adipocytes (see Fig. S2A and B in the supplemental material). Exposure of the three 3T3-L1 cell populations (control vector, WT-PPARγ, and EF-PPARγ) to DMI results in conversion of almost the entire population of cells to adipocytes (Fig. 4A). Additionally, cells expressing ectopic WT-PPARγ differentiate to a greater extent based on their higher expression levels for perilipin, FABP/aP2, and adiponectin than those expressing either the control vector or EF-PPARγ (see Fig. S2B in the supplemental material). To assess the repressive activity of PPARγ in mature adipocytes, each of the 3T3-L1 cell populations was induced to differentiate into adipocytes and then treated with or without troglitazone for 48 h, and mRNA expression was analyzed using Q-PCR assays. Figure 4B demonstrates that troglitazone inhibits expression of angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin mRNAs in control 3T3-L1 adipocytes and those expressing ectopic WT-PPARγ but has no effect on their expression in cells containing EF-PPARγ. These data are consistent with those in Fig. 3 showing that mutation of critical amino acids within helix 7 of the LBD eliminates the repressive activity of PPARγ in response to troglitazone while retaining the activation activity. Additionally, the data suggest that EF-PPARγ might also act as a dominant-negative regulator of the repressive activity of endogenous PPARγ.
FIG. 4.
PPARγ mediates the troglitazone (Trog)-associated inhibition of “visceral white” and induction of “brown” gene expression by troglitazone in 3T3-L1 adipocytes. Oil Red O staining (A) and Q-PCR analysis (B) of “visceral white” and “brown” gene expression (as in Fig. 1) in adipocytes derived from the 3T3-L1 cell lines L1-C (control), L1-WT (WT-PPARγ), and L1-EF (EF-PPARγ) treated with or without troglitazone for 2 days are shown. Each bar represents the mean ± standard error (SE) (n = 3), and 1 represents baseline expression (DMSO-treated cells) for each cell line (*, P < 0.01 relative to control [DMSO]). (C) Cellular respiration from differentiated 3T3-L1 cells expressing WT-PPARγ (L1-WT), EF-PPARγ (L1-EF), or control vector (L1-Cont) was analyzed (left panel), and the mitochondrial DNA level were assessed by Q-PCR (right panel). Data are represented as changes compared to data obtained from DMSO-treated cells. Each bar represents the mean ± SE (n = 3; *, P < 0.01 relative to control [DMSO]).
The fact that C/EBPα cooperates with PPARγ in regulating adipogenic gene expression prompted us to question whether it plays a role in repressing this set of “visceral white” genes. To address this question, we analyzed gene expression in white adipocytes derived from MEFs that contain or lack expression of C/EBPα. This first required the constitutive expression of PPARγ in C/EBPα-deficient MEFs (C/EBPα−/− MEFs) to enhance their adipogenic potential. The resulting cell line was then infected with either a control retrovirus or one expressing C/EBPα in order to generate adipogenic MEFs that either lack (PPARγ/−C/EBPα) or contain (PPARγ/+C/EBPα) C/EBPα. Additionally, we also analyzed WT-MEFs and the parental C/EBPα-deficient MEFs (C/EBPα−/− MEFs). Fig S3 in the supplemental material demonstrates that exposure of each of the MEFs to DMI and troglitazone induces expression of perilipin (a marker of adipocyte formation) in cells that produce PPARγ (ectopic or endogenous) in the absence and presence of C/EBPα. The parental C/EBPα−/− MEFs express virtually undetectable amounts of PPARγ and consequently are incapable of any significant level of adipogenesis. In contrast, WT-MEFs respond to the adipogenic inducers by producing PPARγ and C/EBPα and thus are capable of differentiating into adipocytes. In order to assess the inhibitory effect of troglitazone on expression of the “visceral white” genes, the MEF cell lines were induced to differentiate by exposure to troglitazone for 4 days, at which stage the cells were allowed to develop into mature adipocytes in the absence of TZD for an additional 4 days. Each population of cells was then treated with troglitazone or DMSO for an additional 2 days and total RNA analyzed using Q-PCR assays. Troglitazone does not repress the five “visceral white” genes (encoding angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin) in MEF-derived adipocytes lacking CEBPα (C/EBPα−/− MEFs + PPARγ), whereas TZD is an effective repressor of these genes in adipocytes that express C/EBPα (C/EBPα−/− MEFs + PPARγ + C/EBPα, C/EBPα−/− MEFs + C/EBPα, and WT-MEFs). It is important to point out that the absence of C/EBPα does not prevent the induction of the visceral white genes during adipogenesis, since there is a similar level of basal expression of these genes in adipocytes derived from MEFs lacking (C/EBPα−/− MEFs + PPARγ) or expressing (C/EBPα−/− MEFs + C/EBPα) C/EBPα (see Fig. S3 in the supplemental material). Interestingly, it appears that there is no observable role for C/EBPα in regulating the troglitazone-associated induction of the UCP-1 or Cidea genes in adipocytes since the extent of induction of these genes is approximately the same in adipocytes that lack (C/EBP−/− MEFs + PPARγ) or express C/EBPα (C/EBPα−/− MEFs + C/EBPα and WT-MEFs) (Fig. 5). The data in Fig. S3 in the supplemental material do, however, show a clear role of C/EBPα in regulating adiponectin expression, as was previously reported (7, 39, 46).
FIG. 5.
CEBPα is required for troglitazone (Trog)-associated inhibition of “visceral white” gene expression. Adipocyte differentiation was induced in WT-MEFs or C/EBPα-deficient MEFs expressing ectopic CEBPα (CEBPα−/− MEF + CEBPα) or ectopic PPARγ (CEBPα−/− MEF + PPARγ) or both (CEBPα−/− MEF + CEBPα+ PPARγ). At day 8, cultured adipocytes derived from all MEFs were treated with DMSO or troglitazone for 2 days. RNA was harvested and analyzed by Q-PCR for expression of “visceral white” and “brown” genes, as indicated. Data are represented as n-fold changes in mRNA expression due to troglitazone treatment compared with mRNA expressed in DMSO-treated cells for each gene. Each bar represents the mean ± standard error (n = 3; *, P < 0.01 relative to results for DMSO control).
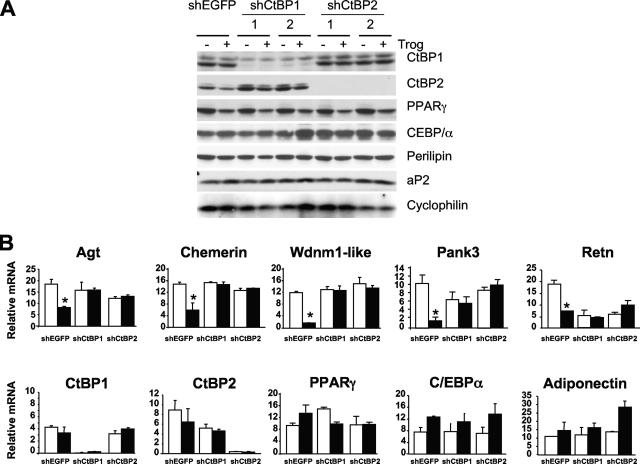
A recent study by Kajimura and colleagues identified the carboxy-terminal binding proteins CtBP1 and CtBP2 as corepressors facilitating the PRDM16-associated repression of resistin during formation of brown adipocytes (26). These repressors are also expressed in white adipocytes; consequently, we questioned whether they have a role to play in the PPARγ-associated repression of the “visceral white” genes. Consequently, we knocked down expression of either CtBP1 or CtBP2 in 3T3-L1 preadipocytes using lentivirus-directed stable expression of corresponding shRNAs. The resulting cell lines were differentiated into adipocytes and exposed to troglitazone or DMSO for 2 days, at which stage RNA or protein was harvested for subsequent analysis. Figure 6A shows the extensive repression of CtBP1 or CtBP2 in knockdown cell lines corresponding to two independent shRNA targets of each gene (1 and 2). An absence of either CtBP appears to have no observable effect on the normal adipogenic process, as indicated by expression of the PPARγ, C/EBPα, perilipin, and aP2 proteins (Fig. 6A) or PPARγ, C/EBPα, and adiponectin mRNAs (Fig. 6B) in the knockdown cells at levels comparable to those in control 3T3-L1 adipocytes (shEGFP). In contrast, troglitazone is incapable of suppressing expression of angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin mRNAs in the absence of either CtBP1 or CtBP2 compared to the extensive repression of these genes by the synthetic ligand in control shEGFP adipocytes (Fig. 6B).
FIG. 6.
Inhibition of “visceral white” and induction of “brown” gene expression by troglitazone (Trog) in 3T3-L1 cells requires CtBP1 and CtBP2. 3T3-L1 control preadipocytes (shEGFP), CtBP1-deficient (shCtBP1 targets 1 and 2) preadipocytes, and CtBP2-deficient (shCtBP2 targets 1 and 2) preadipocytes were induced to differentiate as described in Materials and Methods for 6 days and were then treated with or without (DMSO) 5 μM troglitazone (Trog) for an additional 2 days. Total cellular proteins were collected at day 8 for Western blot analysis with the indicated antibodies (A), and total RNA was subjected to Q-PCR analysis (B). Each bar represents the mean ± standard error (n = 3), and 1 represents baseline expression (DMSO-treated cells) for each cell lines (*, P < 0.01 relative to results for the control [DMSO]).
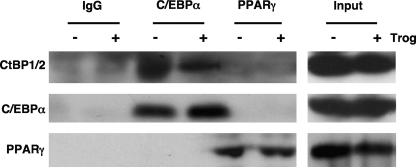
The data in Fig. 5 and 6 suggest that the PPARγ-associated repression of the “visceral white” genes requires involvement of CtBPs as well as C/EBPα. A possible mechanism includes interaction of CtBP1/2 with C/EBPα or PPARγ. To address this possibility, we immunoprecipitated complexes associating with C/EBPα or PPARγ in 3T3-L1 adipocytes treated or not with troglitazone and analyzed the associated proteins on Western blots. The data in Fig. 7 show that CtBPs can interact with C/EBPα in 3T3-L1 adipocytes independently of troglitazone but apparently not with PPARγ (Fig. 7). We also questioned whether the CtBPs associate with C/EBPα complexes that are bound to corresponding response elements in DNA. To address this question, we performed an oligonucleotide pulldown assay using oligonucleotides corresponding to consensus and mutant DNA binding sites for C/EBPs. Nuclear extracts from 3T3-L1 adipocytes exposed to troglitazone or vehicle for 2 days were prepared and incubated with the consensus or mutant oligonucleotides conjugated to agarose beads. The protein complexes bound to the oligonucleotides were then analyzed on Western blots as described previously (1). Fig. S4 in the supplemental material shows the selective binding of both the 42-kDa and 30-kDa isoforms of C/EBPα to the consensus C/EBP binding site in nuclear extracts obtained from cells exposed to troglitazone or vehicle (DMSO). Interestingly, it appears that CtBP1/2 associates at low levels with complexes that also selectively bind to the C/EBP response element only in nuclear extracts exposed to troglitazone.
FIG. 7.
C/EBPα interacts with CtBP1/2 in 3T3-L1 adipocytes. Nuclear extracts isolated from 3T3-L1 adipocytes treated or not with troglitazone (Trog) were subjected to immunoprecipitation for purification of specific complexes using antibody against C/EBPα, PPARγ, or IgG as outlined in Materials and Methods. The immunoprecipitated proteins in each complex were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and CtBP1/2 was detected by Western blotting using a pan-CtBP antibody.
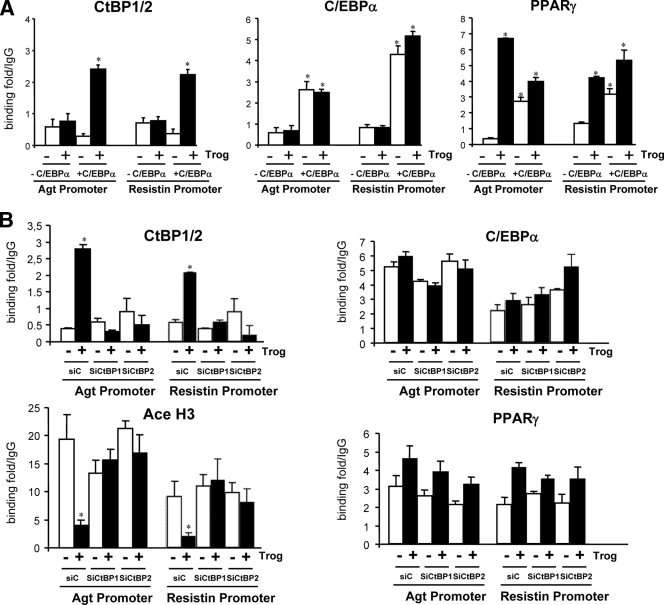
Our next goal was to define the mechanisms by which troglitazone activation of PPARγ represses transcription of the “visceral white” genes, with a particular focus on the role of C/EBPα and the CtBPs. Previous studies by Hartman and coworkers identified C/EBPα as a regulator of resistin mRNA production in adipocytes and in so doing also characterized a C/EBP regulatory element in the proximal promoter of the corresponding gene (25). Other studies have also suggested a role for C/EBPs in regulating angiotensinogen expression (33). Consequently, it seemed appropriate to determine whether C/EBPα and/or CtBPs associate with the promoters of each of these “visceral white” genes under conditions that repress their expression. In the case of C/EBPα, we performed ChIP assays on two of the MEF cell lines described in Fig. 5 that ectopically express PPARγ in the presence or absence of C/EBPα (+C/EBPα and −C/EBPα). The MEFs were differentiated into adipocytes, treated or not with troglitazone for the last 2 days of differentiation, and then subjected to ChIP assays. Figure 8A demonstrates that C/EBPα associates with the proximal regions of resistin and angiotensinogen genes to approximately the same extent in adipocytes treated or not with troglitazone. Interestingly, PPARγ can also associate with each of the promoters most effectively in adipocytes that also express C/EBPα, since there is very little association in C/EBPα-deficient adipocytes that are not exposed to troglitazone. More importantly, binding of CtBP1/2 to each promoter occurs only in response to troglitazone and in cells that also express C/EBPα (Fig. 8A). Additionally, binding of CtBP1/2 to the chromatin was observed only in the proximal promoter region of each gene and not in a more distal region (data not shown).
FIG. 8.
The binding of CtBP1 and CtBP2 to resistin and angiotensinogen promoters requires CEBPα and is induced by troglitazone. (A) C/EBPα−/− MEFs expressing either PPARγ alone (CEBPα−/− MEFs + PPARγ) or PPARγ and CEBPα (CEBPα−/− MEFs + CEBPα + PPARγ) were differentiated into adipocytes as for Fig. 4 and then treated with or without troglitazone (Trog) for 2 days. ChIP assays were performed by immunoprecipitating protein-associated DNA with antibody against CEBPα, PPARγ, CtBP1/2, or IgG (negative control). DNA was amplified by primer sets designed for resistin and angiotensinogen proximal promoters and analyzed by Q-PCR. Each bar represents the mean ± standard error (n = 3; *, P < 0.01). Binding is expressed relative to the nonspecific binding of IgG-immunoprecipitated DNA content. (B) 3T3-L1 preadipocytes in which CtBP1 (siCtBP1) or CtBP2 (siCtBP2) has been knocked down using lentiviral shRNAs, as well as control (siC) cells, were differentiated into adipocytes following standard procedures and were then treated with or without troglitazone for 2 days. ChIP assays were performed using antibody against CEBPα, PPARγ, CtBP1/2, or IgG (negative control) as described for part A.
To determine whether binding of CtBP complexes to the resistin and/or angiotensinogen promoters required the participation of both CtBP1 and CtBP2, we performed additional ChIP assays with 3T3-L1 adipocytes in which CtBP1 or CtBP2 was knocked down using shRNAs, as illustrated in Fig. 6. Interestingly, the data in Fig. 8B show a significant increase in association of both CtBP1 and CtBP2 with angiotensinogen and resistin promoters only in response to troglitazone. In fact, knockdown of CtBP1 or CtBP2 in 3T3-adipocytes prevented association of either CtBP with each promoter. Additionally, there was a significant decrease in acetylation of histone H3 of both promoters in response to troglitazone that depended on the presence of either CtBP1 or CtBP2 (Fig. 8). As observed for MEFs induced to differentiate into adipocytes (Fig. 8A), both C/EBPα and PPARγ associated with angiotensinogen and resistin promoters in 3T3-L1 adipocytes with or without troglitazone treatment (Fig. 8B).
DISCUSSION
The goal of these studies was to identify mechanisms by which synthetic PPARγ agonists can induce a brown adipose phenotype in white adipocytes. To this end, the study has identified a subset of white adipocyte genes, including the angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin genes, that are selectively repressed in vitro and in vivo by synthetic PPARγ agonists. Expression of these “visceral white” genes is significantly lower in BAT than in visceral WAT. It is noteworthy that their level of expression in subcutaneous WAT is as low as that in BAT and consequently a potent PPARγ agonist is incapable of repressing their expression any further in either of these tissues. The studies further show that repression of the “visceral white” genes involves participation of C/EBPα and two members of the carboxy-terminal binding protein family, CtBP1 and CtBP2. Additionally, use of a PPARγ mutant (EF-PPARγ) demonstrates a role for PPARγ in the expression of a brown phenotype in white adipocytes, including repression of the “visceral white” genes and induction of select brown genes and stimulation of mitochondrial biogenesis and oxygen consumption. ChIP analysis of two of the “visceral white” genes, encoding resistin and angiotensinogen, suggest that this troglitazone-induced repression involves association of C/EBPα with the proximal promoter of each gene. Additionally, repression correlates with recruitment of CtBP1/2, well-recognized repressors (8), to each promoter in response to troglitazone but only in cells that also express C/EBPα. Such recruitment might occur as a result of direct interaction between C/EBPα and CtBP1/2 (Fig. 7; see also Fig. S4 in the supplemental material).
It is well accepted that PPARγ regulates expression of multiple gene programs involved in establishing the white adipose phenotype in 3T3-L1 cells, presumably in response to as yet unidentified endogenous ligands or association of specific coactivators with unliganded PPARγ (64). These programs encode proteins required for lipid droplet formation and storage, lipogenesis, and secretion of a diverse group of adipokines. It appears that activation of PPARγ with synthetic ligands, including non-TZD and TZD agonists, can reprogram gene expression so that white adipocytes adopt a browner phenotype. This process appears to involve induction of genes such as UCP-1 that are normally silent, as well as repression of genes that are very active, such as that encoding resistin. To facilitate such a dramatic change in PPARγ target gene expression, the synthetic agonists must reconfigure the PPARγ molecule so that it can both bind to previously masked regulatory elements in gene promoters and/or facilitate association with different coactivators or corepressors. In the case of the resistin and angiotensinogen genes, troglitazone appears to repress their transcription by recruiting both CtBP1 and CtBP2 to the minimal promoter in a region which also associates with C/EBPα. In fact, recruitment of the CtBPs to this region not only depends on TZD activation of PPARγ but requires C/EBPα. The precise nature of the complexes engaged at these promoters in the presence or absence of the TZD needs to be further investigated, but possible associations include a direct interaction between C/EBPα and CtBP1/2 (Fig. 7). Additionally, there is a significant decrease in acetylation of histone H3 of both promoters in response to troglitazone that depends on expression of both CtBP1 and CtBP2, which implies a role for HDAC in repressing the white genes in a CtBP-dependent manner (Fig. 8).
Selective repression of the white genes likely involves a PPARγ-dependent association of specific corepressor complexes with only the C/EBPα molecules that are bound to chromatin within the upstream regions of these genes. Such a mechanism might include PPARγ that is bound to response elements several base pairs distal to the proximal promoter and interacts with C/EBPα complexes as a result of extensive looping of the chromatin (30, 37). Interaction of PPARγ with the specific C/EBPα complexes could be directed by other factors associated with regulatory domains within each target gene. It is possible that PPARγ associates with repressor complexes through direct protein-protein interactions that do not depend on binding of PPARγ to DNA, similar to the mechanism involved in PPARγ-dependent repression of cytokine genes in macrophages (48). These studies demonstrated that PPARγ inhibits the activities of other transcription factors, such as NF-κB and AP-1, which positively regulate the cytokine genes through a mechanism referred to as ligand-dependent transrepression. This process does not require binding of PPARγ to corresponding response elements in the promoters of the cytokine genes; instead, it involves docking of PPARγ to complexes that are associated with NF-κB or AP-1. Furthermore, recruitment of corepressors to these sites requires a reconfiguration of amino acids within helix 7 of PPARγ in response to binding of ligands (21, 40). It is conceivable, therefore, that a comparable ligand-dependent transrepression operates to inhibit expression of the “visceral white” genes during the PPARγ-associated remodeling of white adipose tissue to a brown phenotype. In this case, binding of the agonist to PPARγ facilitates the association of CtBP1/2 corepressor complexes to C/EBPα docked on the promoters of the target genes, including resistin and angiotensinogen. As is the case for the cytokine genes in macrophages, the ligand-dependent repression of the “visceral white” genes appears to involve helix 7 of the LBD of PPARγ (Fig. 3 and 4).
Induction of the brown genes during the remodeling of the white adipocytes is quite likely initiated by the PPARγ agonist-dependent induction of PGC-1α, a coactivator of PPARγ, which coordinates expression of genes encoding mitochondrial biogenesis and lipid oxidation when overexpressed in white adipocytes (43). The data presented here suggest that activation of the PGC-1α gene, which is normally silent in white cells, requires a unique reconfiguration of PPARγ to allow it to associate with coactivators other than those already being employed for white adipocyte gene expression such as FABP4/aP2. Such a reconfiguration also depends on amino acids in helix 7 of the LBD of PPARγ (Fig. 3 and Fig. 4). Following the induction of PGC-1α, it seems likely that it then cooperates with PPARγ to induce expression of the brown genes, which may involve a selective coactivation or derepression of the previously silent genes in the white adipocyte. In fact, studies by Lazar and coworkers suggest that the TZD-dependent induction of OLR-1 and GyK, two genes that are normally silent in white adipocytes, involves a PGC-1α-associated dislodgement of NCoR/HDAC3 repressor complexes from the corresponding promoters (23).
A recent study by Spiegelman and collaborators suggests that an alternative mechanism might contribute to remodeling process presented so far. As discussed previously, these investigators have identified a 140-kDa zinc-finger protein, PRDM16, which induces a program of gene expression resulting in increased mitochondrial biogenesis/oxygen consumption, consistent with a brown phenotype, when ectopically expressed in white preadipocytes in culture or white adipose tissue in mice (54). PRDM16 appears to function as a master regulator of brown formation and function through its ability to simultaneously induce UCP-1 and Cidea as well as other brown genes while suppressing select white fat genes including resistin. The mechanism regulating this process appears to involve mutually exclusive interactions of PRDM16 with either PGC-1α/β to activate the brown genes or with CtBPs to suppress the white genes (26). It is conceivable, therefore, that the mechanism by which PPARγ agonists induce the brown phenotype in white adipocytes might be due to a corresponding PPARγ-dependent induction of PRDM16. Analysis of RNA from TZD treated 3T3-L1 adipocytes as well as adipose tissues from the PPARγ ligand-treated mice, however, failed to detect any significant quantities of PRDM16 mRNA compared to the levels expressed in brown adipocytes or BAT (data not shown). It appears, therefore, that synthetic ligands are capable of inducing the association of PPARγ in white adipocytes with similar complexes to those that associate with PRDM16 in brown adipocytes including PGC1α/β as well as CtBPs. It is also possible that PPARγ induces another large multivalent protein equivalent to PRDM16 that, as yet, has not been identified possibly another member of the PRDM family. The studies presented here do raise the question of whether the mechanism by which PRDM16 represses the “visceral white” genes during brown adipocyte formation is similar to that employed by PPARγ, and specifically whether it recruits CtBPs to C/EBPα, which is docked on its target genes.
It is interesting that the subgroup of white adipocyte genes represented by angiotensinogen, chemerin, Wdnm1-like protein, Pank3, and resistin are expressed at significantly lower levels in BAT compared to visceral WAT. Repression of these genes appears also to be equally as representative of the brown phenotype as induction of UCP-1 and mitochondrial biogenesis. These five ‘′visceral white'’ genes are members of a larger group listed in Table 1 that were initially identified based on repression of their expression in white adipocytes by TZDs. It is important to highlight that several of these genes code for acute phase reactants and adipokines (i.e., resistin, angiotensinogen, haptoglobin and chemerin), whose expression in many cases is enhanced during obesity and correlates with insulin resistance (2, 22, 47, 56, 58, 59). It is also of note that expression of the ‘′visceral white'’ genes is significantly enhanced in the mesenteric depot of mice fed a chronic high fat diet, which caused a dramatic increase in weight gain (data not shown). Moreover, their expression is much higher in visceral depots than in subcutaneous white adipose depots (hence their name) and thereby correlates with the possible involvement of the former tissues in metabolic syndrome (57). Consequently, the PPARγ ligand-associated inhibition of these ‘′visceral white'’ genes in visceral depots most likely contributes to the insulin sensitizing activity of the ligands. The question arises, however: why do brown adipocytes express such low levels of these proteins under normal physiological circumstances. It is conceivable that they do not fulfill any of the functions of the brown cell or possibly counteract some unique brown-specific function.
An important strategy in the fight to combat obesity-related disorders including insulin resistance, type 2 diabetes and dislipidemias is to develop therapeutics that divert dietary fat from storage depots to tissues that expend energy. The data presented here and discussed in a recent perspective (17) support the notion that browning of white adipocytes might fulfill this goal. It is already apparent that existing insulin sensitizers such as the TZDs act in part by inducing this conversion, but unfortunately these drugs have significant side effects including increased fat mass, water retention and life-threatening cardiac problems. Knowledge of the molecular mechanisms regulating the conversion of white to brown adipocytes and the identification of regulators such as CtBPs will no doubt provide additional targets for the development of novel therapeutics.
Supplementary Material
Acknowledgments
We acknowledge the support of the Genome Center at Boston University School of Medicine and the valuable help of Marc Lenburg and Karen Schlauch in analyzing the microarray data. We are grateful to Olivier Bezy for stimulating discussions and critical reading of the manuscript. We thank Yaguang Si for advice and assistance with O2 measurement. We also thank the UT Southwestern Metabolic Core for phenotyping efforts.
This work was supported by USPHS grants no. DK51586 and DK58825 to S.R.F. and by grant no. DK55758 and the UTSW TORS Consortium Grant (no. 1PL1DK081182) to P.E.S. K.E.D. was supported by a UTSW TORS/Interdisciplinary Research Training Program grant, no. T90-DK081181, and by a National Research Service Award (NRSA) fellowship, no. F32 DK081279-01.
Footnotes
Published ahead of print on 29 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bezy, O., C. Vernochet, S. Gesta, S. R. Farmer, and C. R. Kahn. 2007. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol. Cell. Biol. 276818-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozaoglu, K., K. Bolton, J. McMillan, P. Zimmet, J. Jowett, G. Collier, K. Walder, and D. Segal. 2007. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology 1484687-4694. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, B., and J. Nedergaard. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84277-359. [DOI] [PubMed] [Google Scholar]
- 4.Cao, W., K. W. Daniel, J. Robidoux, P. Puigserver, A. V. Medvedev, X. Bai, L. M. Floering, B. M. Spiegelman, and S. Collins. 2004. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 243057-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carley, A. N., L. M. Semeniuk, Y. Shimoni, E. Aasum, T. S. Larsen, J. P. Berger, and D. L. Severson. 2004. Treatment of type 2 diabetic db/db mice with a novel PPARγ agonist improves cardiac metabolism but not contractile function. Am. J. Physiol. Endocrinol. Metab. 286E449-E455. [DOI] [PubMed] [Google Scholar]
- 6.Castriota, G., G. M. Thompson, Y. Lin, P. E. Scherer, D. E. Moller, and J. P. Berger. 2007. Peroxisome proliferator-activated receptor gamma agonists inhibit adipocyte expression of alpha1-acid glycoprotein. Cell Biol. Int. 31586-591. [DOI] [PubMed] [Google Scholar]
- 7.Cha, H. C., N. R. Oak, S. Kang, T. A. Tran, S. Kobayashi, S. H. Chiang, D. G. Tenen, and O. A. Macdougald. 2008. Phosphorylation of CCAAT/enhancer-binding protein α regulates GLUT4 expression and glucose transport in adipocytes. J. Biol. Chem. 28318002-18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 2007. Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 391593-1607. [DOI] [PubMed] [Google Scholar]
- 9.Chui, P. C., H. P. Guan, M. Lehrke, and M. A. Lazar. 2005. PPARγ regulates adipocyte cholesterol metabolism via oxidized LDL receptor 1. J. Clin. Investig. 1152244-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinti, S. 2005. The adipose organ. Prostaglandins Leukot. Essent. Fatty Acids 739-15. [DOI] [PubMed] [Google Scholar]
- 11.Cinti, S. 2001. The adipose organ: morphological perspectives of adipose tissues. Proc. Nutr. Soc. 60319-328. [DOI] [PubMed] [Google Scholar]
- 12.Costford, S., A. Gowing, and M. E. Harper. 2007. Mitochondrial uncoupling as a target in the treatment of obesity. Curr. Opin. Clin. Nutr. Metab. Care 10671-678. [DOI] [PubMed] [Google Scholar]
- 13.Cypess, A. M., S. Lehman, G. Williams, I. Tal, D. Rodman, A. B. Goldfine, F. C. Kuo, E. L. Palmer, Y. H. Tseng, A. Doria, G. M. Kolodny, and C. R. Kahn. 2009. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 3601509-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.do Nascimento, C. O., L. Hunter, and P. Trayhurn. 2004. Regulation of haptoglobin gene expression in 3T3-L1 adipocytes by cytokines, catecholamines, and PPARγ. Biochem. Biophys. Res. Commun. 313702-708. [DOI] [PubMed] [Google Scholar]
- 15.Engeli, S., R. Negrel, and A. M. Sharma. 2000. Physiology and pathophysiology of the adipose tissue renin-angiotensin system. Hypertension 351270-1277. [DOI] [PubMed] [Google Scholar]
- 16.Facucho-Oliveira, J. M., J. Alderson, E. C. Spikings, S. Egginton, and J. C. St John. 2007. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J. Cell Sci. 1204025-4034. [DOI] [PubMed] [Google Scholar]
- 17.Farmer, S. R. 2008. Molecular determinants of brown adipocyte formation and function. Genes Dev. 221269-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmer, S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukui, Y., S. Masui, S. Osada, K. Umesono, and K. Motojima. 2000. A new thiazolidinedione, NC-2100, which is a weak PPAR-gamma activator, exhibits potent antidiabetic effects and induces uncoupling protein 1 in white adipose tissue of KKAy obese mice. Diabetes 49759-767. [DOI] [PubMed] [Google Scholar]
- 20.Gesta, S., Y. H. Tseng, and C. R. Kahn. 2007. Developmental origin of fat: tracking obesity to its source. Cell 131242-256. [DOI] [PubMed] [Google Scholar]
- 21.Ghisletti, S., W. Huang, S. Ogawa, G. Pascual, M. E. Lin, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2007. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol. Cell 2557-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goralski, K. B., T. C. McCarthy, E. A. Hanniman, B. A. Zabel, E. C. Butcher, S. D. Parlee, S. Muruganandan, and C. J. Sinal. 2007. Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J. Biol. Chem. 28228175-28188. [DOI] [PubMed] [Google Scholar]
- 23.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes Dev. 1 9453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan, H. P., Y. Li, M. V. Jensen, C. B. Newgard, C. M. Steppan, and M. A. Lazar. 2002. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat. Med. 81122-1128. [DOI] [PubMed] [Google Scholar]
- 25.Hartman, H. B., X. Hu, K. X. Tyler, C. K. Dalal, and M. A. Lazar. 2002. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 27719754-19761. [DOI] [PubMed] [Google Scholar]
- 26.Kajimura, S., P. Seale, T. Tomaru, H. Erdjument-Bromage, M. P. Cooper, J. L. Ruas, S. Chin, P. Tempst, M. A. Lazar, and B. M. Spiegelman. 2008. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 221397-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kliewer, S., S. Sundseth, S. Jones, P. Brown, G. Wisely, C. Koble, P. Devchand, W. Wahli, T. Willson, J. Lenhard, and J. Lehmann. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. USA 944318-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kliewer, S. A., J. M. Lenhard, T. M. Wilson, I. Patel, D. C. Morris, and J. M. Lehmann. 1995. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor γ and promotes adipocyte differentiation. Cell 83813-819. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, L. P., and M. E. Harper. 2000. Mitochondrial uncoupling proteins in energy expenditure. Annu. Rev. Nutr. 20339-363. [DOI] [PubMed] [Google Scholar]
- 30.Lefterova, M. I., Y. Zhang, D. J. Steger, M. Schupp, J. Schug, A. Cristancho, D. Feng, D. Zhuo, C. J. Stoeckert, Jr., X. S. Liu, and M. A. Lazar. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 222941-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrke, M., and M. A. Lazar. 2005. The many faces of PPARγ. Cell 123993-999. [DOI] [PubMed] [Google Scholar]
- 32.Lin, J., C. Handschin, and B. M. Spiegelman. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1361-370. [DOI] [PubMed] [Google Scholar]
- 33.Narayanan, C. S., Y. Cui, S. Kumar, and A. Kumar. 2000. cAMP increases the expression of human angiotensinogen gene through a combination of cyclic AMP responsive element binding protein and a liver specific transcription factor. Mol. Cell Biochem. 21281-90. [PubMed] [Google Scholar]
- 34.Nedergaard, J., T. Bengtsson, and B. Cannon. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293E444-E452. [DOI] [PubMed] [Google Scholar]
- 35.Nedergaard, J., N. Petrovic, E. M. Lindgren, A. Jacobsson, and B. Cannon. 2005. PPARγ in the control of brown adipocyte differentiation. Biochim. Biophys. Acta 1740293-304. [DOI] [PubMed] [Google Scholar]
- 36.Nedergaard, J., D. Ricquier, and L. P. Kozak. 2005. Uncoupling proteins: current status and therapeutic prospects. EMBO Rep. 6917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, R., T. A. Pedersen, D. Hagenbeek, P. Moulos, R. Siersbaek, E. Megens, S. Denissov, M. Borgesen, K. J. Francoijs, S. Mandrup, and H. G. Stunnenberg. 2008. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 222953-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395137-143. [DOI] [PubMed] [Google Scholar]
- 39.Park, B.-H., L. Qiang, and S. R. Farmer. 2004. Phosphorylation of C/EBPβ at a consensus ERK/GSK3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol. Cell. Biol. 248671-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116511-526. [DOI] [PubMed] [Google Scholar]
- 42.Picard, F., M. Gehin, J. Annicotte, S. Rocchi, M. F. Champy, B. W. O'Malley, P. Chambon, and J. Auwerx. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111931-941. [DOI] [PubMed] [Google Scholar]
- 43.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92829-839. [DOI] [PubMed] [Google Scholar]
- 44.Qiang, L., and S. R. Farmer. 2006. C/EBPα-dependent induction of glutathione S-transferase ζ/maleylacetoacetate isomerase (GSTζ/MAAI) expression during the differentiation of mouse fibroblasts into adipocytes. Biochem. Biophys. Res. Commun. 340845-851. [DOI] [PubMed] [Google Scholar]
- 45.Qiang, L., H. Wang, and S. R. Farmer. 2007. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-Lα. Mol. Cell. Biol. 274698-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao, L., P. S. Maclean, J. Schaack, D. J. Orlicky, C. Darimont, M. Pagliassotti, J. E. Friedman, and J. Shao. 2005. C/EBPα regulates human adiponectin gene transcription through an intronic enhancer. Diabetes 541744-1754. [DOI] [PubMed] [Google Scholar]
- 47.Rajala, M. W., and P. E. Scherer. 2003. Minireview: the adipocyte—at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology 1443765-3773. [DOI] [PubMed] [Google Scholar]
- 48.Ricote, M., and C. K. Glass. 2007. PPARs and molecular mechanisms of transrepression. Biochim. Biophys. Acta 1771926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricquier, D. 2005. Respiration uncoupling and metabolism in the control of energy expenditure. Proc. Nutr. Soc. 6447-52. [DOI] [PubMed] [Google Scholar]
- 50.Rosen, E. D., and O. A. MacDougald. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7885-896. [DOI] [PubMed] [Google Scholar]
- 51.Rosen, E. D., and B. M. Spiegelman. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito, M., Y. Okamatsu-Ogura, M. Matsushita, K. Watanabe, T. Yoneshiro, J. Nio-Kobayashi, T. Iwanaga, M. Miyagawa, T. Kameya, K. Nakada, Y. Kawai, and M. Tsujisaki. 2009. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 581526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scherer, P. E. 2006. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 551537-1545. [DOI] [PubMed] [Google Scholar]
- 54.Seale, P., S. Kajimura, W. Yang, S. Chin, L. M. Rohas, M. Uldry, G. Tavernier, D. Langin, and B. M. Spiegelman. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 638-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steppan, C. M., S. T. Bailey, S. Bhat, E. J. Brown, R. R. Banerjee, C. M. Wright, H. R. Patel, R. S. Ahima, and M. A. Lazar. 2001. The hormone resistin links obesity to diabetes. Nature 409307-312. [DOI] [PubMed] [Google Scholar]
- 56.Steppan, C. M., and M. A. Lazar. 2002. Resistin and obesity-associated insulin resistance. Trends Endocrinol. Metab. 1318-23. [DOI] [PubMed] [Google Scholar]
- 57.Tran, T. T., Y. Yamamoto, S. Gesta, and C. R. Kahn. 2008. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 7410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trayhurn, P., and J. H. Beattie. 2001. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. Proc. Nutr. Soc. 60329-339. [DOI] [PubMed] [Google Scholar]
- 59.Trayhurn, P., and I. S. Wood. 2004. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 92347-355. [DOI] [PubMed] [Google Scholar]
- 60.Trujillo, M. E., and P. E. Scherer. 2006. Adipose tissue-derived factors: impact on health and disease. Endocr. Rev. 27762-778. [DOI] [PubMed] [Google Scholar]
- 61.Uldry, M., W. Yang, J. St-Pierre, J. Lin, P. Seale, and B. M. Spiegelman. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3333-341. [DOI] [PubMed] [Google Scholar]
- 62.van Marken Lichtenbelt, W. D., J. W. Vanhommerig, N. M. Smulders, J. M. Drossaerts, G. J. Kemerink, N. D. Bouvy, P. Schrauwen, and G. J. Teule. 2009. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 3601500-1508. [DOI] [PubMed] [Google Scholar]
- 63.Virtanen, K. A., M. E. Lidell, J. Orava, M. Heglind, R. Westergren, T. Niemi, M. Taittonen, J. Laine, N. J. Savisto, S. Enerback, and P. Nuutila. 2009. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 3601518-1525. [DOI] [PubMed] [Google Scholar]
- 64.Walkey, C. J., and B. M. Spiegelman. 2008. A functional peroxisome proliferator-activated receptor-gamma ligand-binding domain is not required for adipogenesis. J. Biol. Chem. 28324290-24294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, H., L. Qiang, and S. R. Farmer. 2008. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Mol. Cell. Biol. 28188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willson, T. M., M. H. Lambert, and S. A. Kliewer. 2001. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem. 70341-367. [DOI] [PubMed] [Google Scholar]
- 67.Wilson-Fritch, L., A. Burkart, G. Bell, K. Mendelson, J. Leszyk, S. Nicoloro, M. Czech, and S. Corvera. 2003. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 231085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson-Fritch, L., S. Nicoloro, M. Chouinard, M. A. Lazar, P. C. Chui, J. Leszyk, J. Straubhaar, M. P. Czech, and S. Corvera. 2004. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 1141281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, Y., and C. M. Smas. 2008. Wdnm1-like, a new adipokine with a role in MMP-2 activation. Am. J. Physiol. Endocrinol. Metab. 295E205-E215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu, H. E., M. H. Lambert, V. G. Montana, D. J. Parks, S. G. Blanchard, P. J. Brown, D. D. Sternbach, J. M. Lehmann, G. B. Wisely, T. M. Willson, S. A. Kliewer, and M. V. Milburn. 1999. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell 3397-403. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, Y. M., C. O. Rock, and S. Jackowski. 2005. Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J. Biol. Chem. 28032594-32601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.