Abstract
Skeletal muscle atrophy caused by unloading is characterized by both decreased responsiveness to myogenic growth factors (e.g., insulin-like growth factor 1 [IGF-1] and insulin) and increased proteolysis. Here, we show that unloading stress resulted in skeletal muscle atrophy through the induction and activation of the ubiquitin ligase Cbl-b. Upon induction, Cbl-b interacted with and degraded the IGF-1 signaling intermediate IRS-1. In turn, the loss of IRS-1 activated the FOXO3-dependent induction of atrogin-1/MAFbx, a dominant mediator of proteolysis in atrophic muscle. Cbl-b-deficient mice were resistant to unloading-induced atrophy and the loss of muscle function. Furthermore, a pentapeptide mimetic of tyrosine608-phosphorylated IRS-1 inhibited Cbl-b-mediated IRS-1 ubiquitination and strongly decreased the Cbl-b-mediated induction of atrogin-1/MAFbx. Our results indicate that the Cbl-b-dependent destruction of IRS-1 is a critical dual mediator of both increased protein degradation and reduced protein synthesis observed in unloading-induced muscle atrophy. The inhibition of Cbl-b-mediated ubiquitination may be a new therapeutic strategy for unloading-mediated muscle atrophy.
The impairment of growth factor signaling is a near-universal feature of skeletal myopathies induced by unloading (6, 13). Clinical trials have established that during unloading, muscle tissue fails to respond to IGF-1, a dominant myotrophic hormone (7, 19, 34). Under normal conditions and in response to hypertrophic stimuli, IGF-1 promotes muscle growth and suppresses muscle loss largely through the Akt-dependent phosphorylation and cytosolic sequestration of FOXO transcription factors in skeletal myocytes, which leads to the inhibition of FOXO-dependent gene expression (38, 41). In contrast, IGF-1-dependent Akt signaling is impaired during muscle atrophy, which decreases the phosphorylation and increases the transactivation of FOXO target genes. In particular, FOXO regulates the expression of atrophy-related genes (atrogenes) that encode atrogin-1/MAFbx and MuRF-1, which are RING-type ubiquitin ligases that are critical mediators of atrophic myopathies in vivo (3, 14). Atrogin-1 and MuRF-1 regulate the degradation of key proteins involved in striated muscle growth and differentiation, including MyoD, calcineurin, and troponin-I (24, 27, 47). Although diminished growth factor responsiveness and enhanced proteolysis both are major atrophy-related processes, the mechanisms by which skeletal muscle becomes refractory to the trophic actions of muscle growth factors during unloading are not well defined.
In a previous study designed to evaluate changes in skeletal muscle gene expression in rats exposed to a 16-day spaceflight (30), we identified novel potential atrogenes (37) using microarray analysis. The response of skeletal muscle to mechanical stress is accompanied by marked alterations in atrogene expression, and we showed that microgravity induces Siah-1A, MuRF-1 (30), and atrogin-1 (see Table S1 in the supplemental material). Microgravity also resulted in the increased expression of Cbl-b (greater than eightfold). Cbl-b is another RING-type ubiquitin ligase previously established as a negative regulator of receptor tyrosine kinase signaling in a variety of cells (23, 45). These results complement our recent finding that Cbl-b downregulates bone formation through the suppression of IGF-1 signaling in osteoblasts during denervation (43). Given that atrophic muscle is characterized by defective tyrosine kinase IGF-1 receptor pathway signaling, we reasoned that Cbl-b induced by unloading is a critical negative regulator of IGF-1 signal transduction throughout the musculoskeletal locomotor system. Here, we show that the induction of Cbl-b in vivo is required for the loss of muscle mass in response to unloading. Moreover, our results suggest that therapeutic strategies targeting Cbl-b are useful for a variety of muscle atrophy-related conditions.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: atrogenes, muscle atrophy-related genes; Cbl-b, casitus b-lineage lymphoma-b; Cbl-b−/− mice, Cbl-b-deficient mice; CHO-IR cells, Chinese hamster ovary cells permanently expressing insulin receptor; DMEM, Dulbecco's modified Eagle's medium; EGFR, epidermal growth factor receptor; FOXO, Forkhead box O; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; H&E, hematoxylin and eosin; HA, hemagglutinin; IGF-1, insulin-like growth factor 1; IGFR-1, IGF receptor type I; IRS-1, insulin receptor substrate 1; MAFbx (atrogin-1), muscle atrophy F-box protein; MAPK, mitogen-activated protein kinase; MuRF-1, muscle-specific RING finger protein 1; MyHC, myosin heavy chain; P-Akt, phosphorylated Akt; PI3K, phosphatidylinositol 3-kinase; RING, really interesting gene; Siah-1A, seven in absentia homolog 1A; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Syk, spleen tyrosine kinase; TKB, tyrosine kinase binding; and Vav, guanine nucleotide exchange factor.
Unloading.
In unloading experiments, rodents were subjected to spaceflight for 16 days on the space shuttle Columbia, tail suspension, and denervation, as described previously (22, 30).
Cbl-b-deficient mice.
Mice deficient in the cbl-b gene (Cbl-b−/−), which were generated by inserting a neomycin resistance gene into the third exon of the cbl-b gene, were kindly provided by the National Institutes of Health (Rockville, MD) (5). Cbl-b−/− mice had a mixed 129 and C57BL/6 genetic background, and they were backcrossed more than eight times into the C57BL/6 strain. Therefore, we used C57BL/6 mice (Japan SLC, Shizuoka, Japan) as wild-type mice. The experimental protocols described in this study were approved by the Ethics Review Committee for Animal Experimentation of Tokushima University.
Cell culture.
Cells used in this study, including mouse myoblastic C2C12 cells and COS7, CHO-IR, HEK293, and UMR106 cells, were maintained with DMEM containing 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2 and 95% air. Cells untransfected or transfected with plasmids containing genes of interest were treated with 2 or 10 ng/ml of recombinant human long-R3 IGF-I (Sigma, St Louis, MO) 48 h after the transfection.
Transfection.
C2C12, COS7, CHO-IR, HEK293, and UMR106 cells (40 to 60% of confluence) were transfected using FuGene6 (Roche Diagnostics, Tokyo, Japan) as described previously (21). The plasmids used in this study were pLX2-rat IRS-1, pLX2-rat Y18F IRS-1 (in which tyrosine18 was replaced with phenylalanine), pLX2-rat Y608F IRS-1 (in which tyrosine608 was replaced with pheylalanine), pCEFL-human Cbl-b-HA, pCEFL-human ΔRING-Cbl-b-HA (in which the RING domain of cysteine480 to arginine519 of human Cbl-b was deleted), pCAG-human Cbl-b-HA, and pcDNA3-FLAG-Ubiquitin. To overexpress Cbl-b protein in C2C12 myotubes, we transfected C2C12 myoblasts with pCEFL-Cbl-b-HA using a jetPEI (polyethyleneimine) transfection reagent (Qbiogene, Montreal, Canada), which then were allowed to differentiate into myotubes by incubation in DMEM containing 2% horse serum and 0.1 mg/ml G418 (Gibco, Grand Island, NY) (33).
Gene transduction in vivo.
Striated muscle uniquely can take up and express naked plasmid DNA (48). To avoid an inflammatory reaction, the in vivo overexpression of Cbl-b in rat skeletal muscle was performed as described previously (48) without the use of myotoxic agents. Briefly, 50 μg of plasmid DNA was injected into the middle of the tibialis anterior muscle of 5-week-old anesthetized male Sprague-Dawley rats.
Immunoblotting and immunoprecipitation.
Immunoblot and immunoprecipitation analyses were performed as described previously (20). The following antibodies were used: anti-β-actin (Oncogene Research Products, San Diego, CA), anti-Akt-1 (PharMingen International, Tokyo), anti-phospho-S473-Akt-1, anti-IRS-2, anti-ERK1/2, anti-phosphorylated ERK1/2, anti-p38, anti-phosphorylated p38, anti-JNK, anti-phosphorylated JNK (Cell Signaling Technology, Beverly, MA), anti-HA.11 (BabCo, Richmond, CA), anti-IRS-1 (Calbiochem, La Jolla, CA), anti-FOXO3, anti-phosphorylated FOXO3, anti-phosphorylated tyrosine, anti-PI3K, anti-IGF-IR α-chain (Upstate Biotechnology, Lake Placid, NY), anti-V5 (Invitrogen, Carlsbad, CA), anti-mouse/rat Cbl-b, anti-IGF-IR β-chain, anti-insulin receptor, anti-C3G, anti-EGFR, anti-Vav (Santa Cruz Biotechnology, Santa Cruz, CA), anti-fast/slow-type MyHC, antilaminin, anti-α-tubulin, anti-GAPDH, antiubiquitin, anti-FLAG M2 (Sigma), and anti-c-Cbl (Transduction Lab, Lexington, KY).
Histochemical analysis.
Immunohistochemistry for rat or mouse skeletal muscle was conducted as described previously (21). Sections were counterstained with H&E. The cross-sectional area of myofibers was measured using the imaging software WinRoof (version 5.0; Mitani Corp, Fukui, Japan).
Real-time RT-PCR.
To measure the mRNA amount in small samples, real-time reverse transcription-PCR (RT-PCR) was performed with SYBR green dye using an ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA) as described previously (31). The oligonucleotide primers used for amplification are listed in Table S3 in the supplemental material.
Cell-free ubiquitination assay.
We performed a cell-free ubiquitination assay (39). We prepared immunoprecipitated Cbl-b and IRS-1 bound to anti-HA- and anti-IRS-1-immunoglobulin G-linked protein A beads from extracts (1 mg protein) of HEK293 cells transfected with pCAG-Cbl-b-HA and pLX2-IRS-1 (5 μg). Slurries of immunoprecipitated Cbl-b and IRS-1 were incubated at 37°C for 4 h in reaction buffer (50 μl) containing an ATP-regenerating system, recombinant mouse E1 (500 ng), UbcH7 (5 μg; E2), glutathione S-transferase-tagged ubiquitin (10 μg), and/or ubiquitin-aldehyde (Ubc-CHO; 1 μg; a deubiquitinase inhibitor), which were purchased from Boston Biochem Inc. (Cambridge, MA). After terminating the reaction, 20 μl of boiled supernatant was separated by SDS-PAGE and visualized by immunoblotting with an anti-IRS-1 or anti-Cbl-b antibody.
Inhibition of Cbl-b-mediated IRS-1 ubiquitination by synthetic oligopeptides.
We synthesized pentapeptides corresponding to tyrosine phosphorylation domains of IRS-1: peptide a, DG-phosphorylated YMP (DGpYMP); Y-F-mutated peptide a, DGFMP; dephosphorylated peptide a, DGYMP; peptide b, GEpYVN; peptide c, EEpYMN; peptide d, LNpYID; peptide e, STpYAS; and peptide f, VGpYLR (42). These synthetic peptides were kindly provided by Otsuka Pharmaceutical Co. (Tokushima, Japan) and Ajinomoto Co. (Kawasaki, Japan). We then screened the inhibitory activity using a cell-free ubiquitination assay, as described above. Pentapeptides, which had inhibitory activity on ubiquitination in the cell-free system, were subjected to further analysis in cell cultures and intramuscular injection in denervated mice.
Recording and analysis of muscle contraction.
Isolated soleus muscle was stimulated directly by an electrical field generated between two parallel electrodes connected to an SEN-320 stimulator and bath drive amplifier SEG-3101 (Nihon Kohden, Tokyo, Japan) (15). Specific tetanic force was calculated as the maximal force per cross-sectional area (15).
Statistical analysis.
All data are expressed as means ± standard deviations of three to five individual samples per group. Differences between groups were analyzed by one-way analysis of variance using SPSS software (release 6.1; SPSS Japan, Tokyo, Japan). Differences between values for two groups were tested by Scheffé's test. A P value of <0.05 was considered statistically significant.
RESULTS
Mechanical unloading of skeletal muscle results in upregulation of the ubiquitin ligase Cbl-b.
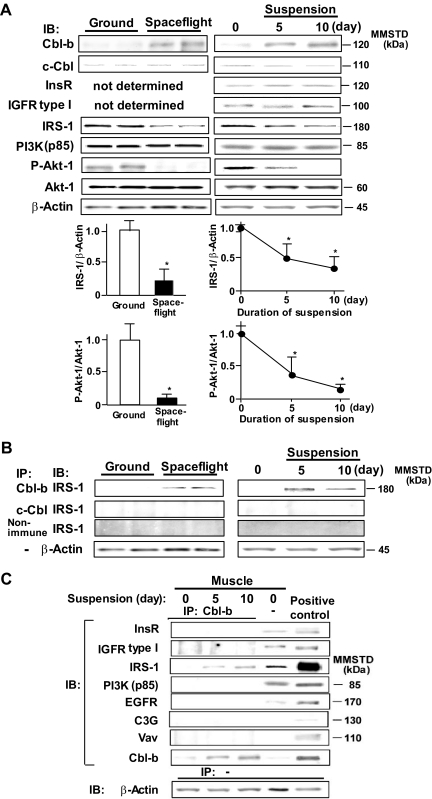
We first confirmed that unloading (spaceflight and tail suspension) increased Cbl-b protein and mRNA expression levels. Cbl-b and the related c-Cbl, another member of the Cbl family, were expressed in various tissues, but their levels in rat gastrocnemius muscle were relatively low under normal conditions (21). The unloading of the muscle by spaceflight (16 days) resulted in an approximately 10-fold induction of gastrocnemius Cbl-b at the mRNA level and only a slight increase for c-Cbl (see Fig. S1A in the supplemental material). Similarly, the expression of Cbl-b, but not c-Cbl, gradually increased during the first 14-day period of tail suspension and was sustained for the following week (see Fig. S1B in the supplemental material). Consistently with changes observed in mRNA expression, levels of Cbl-b protein in gastrocnemius muscle were markedly increased in response to spaceflight. Cbl-b protein continued to increase during the entire 10-day period of tail suspension (Fig. 1A) and was detected at myofibers in gastrocnemius of rats that were subjected to tail suspension for 10 days (see Fig. S1C in the supplemental material). In contrast, neither spaceflight nor tail suspension altered c-Cbl protein levels, suggesting that the expression of Cbl-b, but not c-Cbl, specifically is sensitive to unloading.
FIG. 1.
Unloading decreases IGF-1 signaling and increases IRS-1-Cbl-b interaction. Male Sprague-Dawley rats were subjected to spaceflight or tail suspension for the indicated time periods. (A) Homogenates from gastrocnemius muscle were subjected to SDS-PAGE and immunoblotted (IB) using the indicated antibodies. Densitometric ratios of IRS-1 to β-actin or P-Akt-1 to Akt-1 are shown. Data are means ± standard deviations (n = 3). *, P < 0.05 compared to results for control rats or those tested at time zero. (B) Homogenates from gastrocnemius muscle of spaceflight and suspended rats were immunoprecipitated (IP) with the indicated antisera. Immunoprecipitates were subjected to IB for IRS-1. (C) Cbl-b immunoprecipitates of muscle homogenates and lysates of Cbl-b-transfected CHO-IR cells (positive control) were subjected to IB for various proteins that have been reported to interact with Cbl-b. Results shown in this figure are representative of three experiments. MMSTD, molecular mass standards. InsR, insulin receptor.
Unloading-induced Cbl-b preferentially binds and degrades IRS-1.
We next determined the effect of unloading on the expression of IGF-1 signaling molecules, including IRS-1, the regulatory p85 subunit of PI3K, and Akt-1 in gastrocnemius muscles of animals exposed to spaceflight/tail suspension. Whereas unloading did not influence the levels of p85 and Akt-1 protein, spaceflight and tail suspension each significantly decreased the levels of IRS-1 protein (Fig. 1A). The changes in IRS-1 mRNA expression were smaller than the decrease in IRS-1 protein levels (see Table S2 in the supplemental material), suggesting a posttranslational degradation of IRS-1 protein. To determine the physiological impact of IRS-1 downregulation, we determined the effect of unloading on the phosphorylation state of Akt-1, a major downstream effector of IRS-1 signaling. Compared to results for control animals, and consistently with observations following denervation (32), both spaceflight and tail suspension significantly decreased the ratio of phosphorylated to total Akt-1.
To assess the potential involvement of Cbl-b in the unloading-induced loss of IRS-1 protein, we performed ex vivo coimmunoprecipitation experiments with anti-IRS-1 and anti-Cbl-b antibodies in animals subjected to control (normal loading) and unloading conditions. Compared to that of the control, a strong Cbl-b-IRS-1 interaction was observed in gastrocnemius muscle lysates from animals exposed to spaceflight and tail suspension (Fig. 1B). In contrast, anti-c-Cbl antibody and nonimmune serum failed to immunoprecipitate IRS-1. Cbl-b is known to interact with various signaling molecules, including EGFR and Vav (4, 12). We also determined whether Cbl-b interacts preferentially with IRS-1 in rat gastrocnemius muscle. As expected, a preferential binding of Cbl-b to IRS-1, but not EGFR, was noted in gastrocnemius muscle (Fig. 1C). Interactions between Cbl-b and either insulin or IGF-1 receptors did not occur in skeletal muscle (Fig. 1C).
Cbl-b regulates expression of atrogin-1 via impaired IGF-1 signaling.
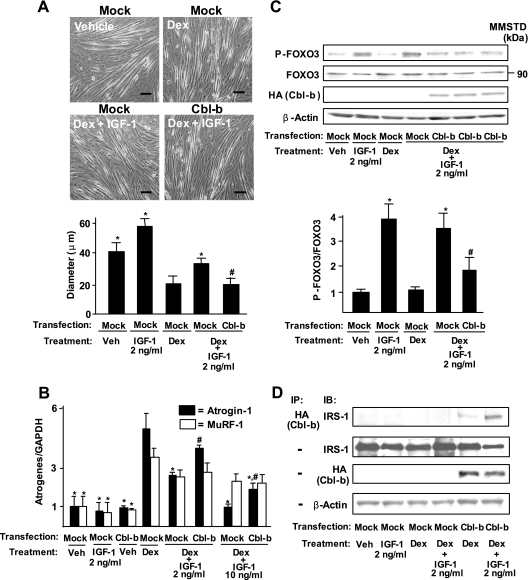
To determine whether muscle atrophy is associated with the upregulation of Cbl-b expression, we used dexamethasone-treated C2C12 myotubes, which is a well-established in vitro atrophy model (36). Myotubes expressed relatively low levels of Cbl-b proteins (see Fig. S2A in the supplemental material). Similarly to unloading-induced atrophy in vivo, dexamethasone treatment stimulated the expression of Cbl-b at mRNA and protein levels (see Fig. S2B in the supplemental material). Moreover, dexamethasone treatment resulted in a decrease in the thickness of myotubes (Fig. 2A) and the upregulation of atrogin-1 and MuRF-1 expression in cultured myotubes (Fig. 2B). IGF-1 at more than 2 ng/ml blocked dexamethasone-induced atrophy (measured by the thickness of myotubes) and the induction of atrogin-1 expression in cultured myotubes (Fig. 2A and B). Interestingly, the overexpression of Cbl-b significantly attenuated the protective effects of IGF-1 on the dexamethasone-mediated atrophy of myotubes and the expression of atrogin-1 (Fig. 2A and B). Cbl-b overexpression in the absence of dexamethasone did not induce the expression of atrogenes (Fig. 2B) and decrease the thickness of C2C12 myotubes (data not shown). The expression of atrogin-1 was reflected by the dephosphorylation of FOXO3 through a decrease in IGF-1 signaling (Fig. 2C). In contrast, IGF-1 treatment did not influence the dexamethasone-induced expression of MuRF-1 (Fig. 2B). In the absence of IGF-1, overexpressed Cbl-b interacted weakly with IRS-1 in dexamethasone-treated myotubes (Fig. 2D). IGF-1 significantly increased the interaction between overexpressed Cbl-b and IRS-1, leading in turn to a reduction in IRS-1 levels.
FIG. 2.
Cbl-b disrupts the myotrophic action of IGF-1 on dexamethasone-induced protein breakdown in C2C12 myotubes. Mock vector- or pCEFL-Cbl-b-HA-transfected C2C12 myotubes were treated with vehicle (phosphate-buffered saline and ethanol) or 0.1 mM dexamethasone (Dex) for 24 h. In some cases, they were treated simultaneously with recombinant human long-R3 IGF-I at 2 or 10 ng/ml. (A) The diameter of myotubes was measured under a high-power field with WinRoof software. Scale bar, 50 μm. Data are means ± standard deviations (n = 4). *, P < 0.05 compared to results of Dex treatment in mock vector-transfected C2C12 cells; #, P < 0.05 compared to results of Dex and IGF-1 treatment in mock vector-transfected C2C12 cells. veh, vehicle. (B) Levels of atrogin-1, MuRF-1, and GAPDH were measured by real-time RT-PCR. Data are means ± standard deviations (n = 4). *, P < 0.05 compared to results for the Dex treatment of mock vector-transfected C2C12 cells; #, P < 0.05 compared to results for the Dex and IGF-1 treatment of mock vector-transfected C2C12 cells. (C) Mock vector- or Cbl-b-HA-transfected C2C12 myotube lysates were subjected to SDS-PAGE, followed by immunoblotting (IB) for the indicated proteins. Data are means ± standard deviations (n = 4). *, P < 0.05 compared to results for the Dex treatment of mock vector-transfected C2C12 cells; #, P < 0.05 compared to results for the Dex and IGF-1 treatment of mock vector-transfected C2C12 cells. (D) Mock vector- or Cbl-b-HA-transfected C2C12 myotube lysates and their immunoprecipitates (IP) by anti-HA antibody were subjected to IB for the indicated proteins. Three independent experiments showed similar results. MMSTD, molecular mass standards.
Overexpression of Cbl-b ubiquitinates and degrades IRS-1 in COS7 cells and skeletal muscle.
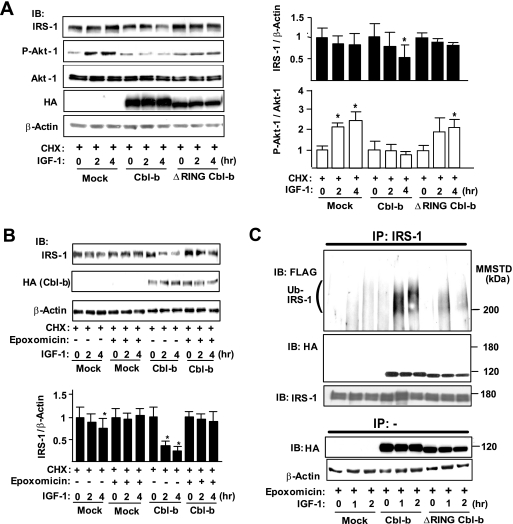
To further examine the mechanisms of Cbl-b-mediated degradation of IRS-1, we expressed Cbl-b and ΔRING-Cbl-b, a deleted mutant that lacks E3 ligase activity, in COS7 cells. Under normal conditions, COS7 cells expressed negligible levels of Cbl-b proteins (see Fig. S2A in the supplemental material). The expression of Cbl-b significantly reduced the amount of IRS-1 after IGF-1 treatment (Fig. 3A), although the rates of the decrease of IRS-1 levels were very low after IGF-1 treatment in mock vector- and ΔRING-Cbl-b-transfected cells. Moreover, Cbl-b inhibited the phosphorylation of Akt-1 but did not alter the levels of Akt-1 protein (Fig. 3A). The transfection of mock vector and ΔRING-Cbl-b did not affect the IGF-1-mediated phosphorylation of Akt-1.
FIG. 3.
Transient expression of Cbl-b induces ubiquitination and degradation of IRS-1 in vitro. (A) COS7 cells were transfected with mock vector/pCEFL-Cbl-b-HA/pCEFL-ΔRING-Cbl-b-HA, pcDNA3-FLAG-Ubiquitin, and pLX2-IRS-1. To analyze the degradation rates of IGF-1 signaling molecules, COS7 cells were treated with 100 μg/ml cycloheximide (CHX) 3 h before treatment with recombinant human long-R3 IGF-I (10 ng/ml). Whole-cell lysates before or after IGF-1 treatment were subjected to immunoblotting (IB) for the indicated proteins. Densitometric ratios of IRS-1 to β-actin or P-Akt-1 to Akt-1 in the indicated samples are expressed relative to values at time zero of IGF-1 treatment. Data are means ± standard deviations (n = 3). *, P < 0.05 compared to the values at time zero. (B) Mock vector- or pCEFL-Cbl-b-HA-transfected COS7 cells were treated with vehicle (dimethylsulfoxide) or 100 nM epoxomicin in addition to CHX 3 h before IGF-1 treatment. Whole-cell lysates were subjected to IB for the indicated proteins. Data are the densitometric ratios of IRS-1 to β-actin in vehicle- and epoxomicin-treated cells. Data are means ± standard deviations (n = 3). *, P < 0.05 compared to the values at time zero. (C) Epoxomicin was added to all cells 3 h before IGF-1 treatment. Lysates from IRS-1-, FLAG-ubiquitin-, and Cbl-b-HA/ΔRING-Cbl-b-HA-expressing COS7 cells treated with IGF-1 for the indicated time intervals were immunoprecipitated (IP) with an anti-IRS-1 antibody. The immunoprecipitates were subjected to IB for the indicated proteins. Three independent experiments showed similar results. MMSTD, molecular mass standards.
IRS-1 degradation in CHO cells and adipocytes is regulated by the proteasomal pathway (9, 26). Consistently with previous reports (26), ubiquitin expression enhanced the Cbl-b-mediated degradation of IRS-1 (see Fig. S2C in the supplemental material). Therefore, we examined whether Cbl-b-mediated IRS-1 degradation is also proteasomal using epoxomicin, a proteasome inhibitor. In the presence of IGF-1, epoxomicin prevented both Cbl-b-induced IRS-1 degradation in COS7 cells and IRS-1 degradation in mock transfection controls (Fig. 3B), indicating that Cbl-b-independent basal IRS-1 degradation in COS7 cells also is mediated by the proteasome. Anti-IRS-1 immunoprecipitates from Cbl-b-expressing cells showed that the overexpression of Cbl-b induced the ubiquitination of IRS-1 in a time-dependent manner after IGF-1 treatment compared to that of mock transfection controls (Fig. 3C). The inactive mutant ΔRING-Cbl-b failed to induce the ubiquitination of IRS-1, although it bound IRS-1 (Fig. 3C). Consistently with a specific role for Cbl-b in the regulation of IRS-1, the atrophy-associated ubiquitin ligase Siah-1A, which also is induced by microgravity unloading (30), failed to induce the ubiquitination of IRS-1 in COS7 cells (see Fig. S2D in the supplemental material).
To extend our findings to an animal system, we first used plasmid gene delivery to determine whether Cbl-b is involved in skeletal muscle atrophy in vivo. Compared to that with mock vector administration, the injection of plasmids encoding Cbl-b into the rat tibialis anterior muscle resulted in increased muscle protein ubiquitination, indicating that Cbl-b functions as a ubiquitin ligase in vivo (Fig. 4A). The direct measurement of muscle fiber size showed a 65% decrease in size upon Cbl-b expression (Fig. 4A and B). Moreover, consistently with the results of the cell-free ubiquitination assay (see Fig. S2E in the supplemental material), the expression of Cbl-b in the tibialis anterior muscle increased the ubiquitination and degradation of IRS-1, but Cbl-b did not undergo significant autoubiquitination (Fig. 4C).
FIG. 4.
Overexpression of Cbl-b induces muscle atrophy in vivo. (A) Mock vector or pCEFL-Cbl-b-HA was injected directly into the rat tibialis anterior muscle. Two weeks later, muscle sections were stained with anti-HA and anti-ubiquitin (Ub) antibodies and counterstained with H&E. Arrows represent injection points, which were indicated by bromophenol blue. Scale bar, 100 μm. (B) Cross-sectional areas of myofibers expressing Cbl-b-HA (from the boxed area of panel A, containing 50 myofibers) were calculated with WinRoof software. We also measured the area of 50 myofibers in uninjected or mock vector-injected myofibers as controls. Data are means ± standard deviations (n = 4). *, P < 0.05 for results for injected myofibers compared to those for control myofibers. (C) Homogenates from muscle blocks expressing Cbl-b or injected with mock vector were immunoprecipitated (IP) with an anti-IRS-1 or anti-HA antibody, followed by immunoblotting (IB) for the indicated proteins. Results shown in this figure are representative of four experiments. MMSTD, molecular mass standards.
Cbl-b-deficient mice are protected from unloading-induced skeletal muscle atrophy and dysfunction.
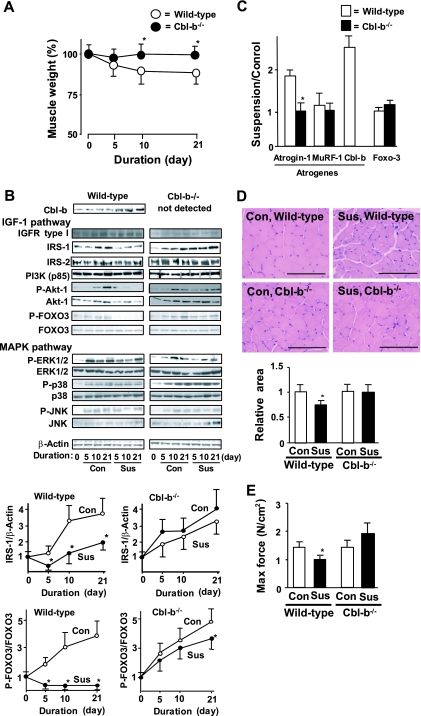
We used Cbl-b-deficient (Cbl-b−/−) mice (5) to further examine the role of Cbl-b in unloading-induced muscle atrophy in vivo. Wild-type (C57BL/6) and Cbl-b−/− mice (8 weeks old) were subjected to hind limb unloading stress induced by long-term tail suspension. In wild-type mice, this suspension-unloading paradigm reduced the wet weight (per kilogram of body mass) of gastrocnemius muscle by 20% compared to that of untreated animals (Fig. 5A). In marked contrast, the long-term tail suspension of Cbl-b−/− mice failed to induce a significant decrease in muscle mass. Moreover, tail suspension significantly induced the ubiquitination of IRS-1 (data not shown) and decreased its protein levels in a time-dependent manner but did not decrease those of IGF-1 receptor, IRS-2, and PI3K in gastrocnemius muscle of wild-type mice (Fig. 5B). However, tail suspension in Cbl-b−/− mice did not decrease IRS-1 levels, confirming that IRS-1 is a potent substrate for Cbl-b in vivo.
FIG. 5.
Animals deficient in Cbl-b are protected from unloading-induced decreases in muscle mass and IRS-1 protein levels in vivo. (A) Wild-type and Cbl-b−/− mice (8 weeks old) were subjected to unloading by tail suspension for 21 days. Muscle wet weight per kilogram of body weight in suspended animals at the indicated times is expressed as the percent weight reduction relative to that of nonsuspended (sham-operated, control) animals. The results are representative of three independent experiments and are expressed as means ± standard deviations (n = 5). *, P < 0.05 for results for Cbl-b−/− mice compared to those for the wild-type mice. (B) Levels/phosphorylation of molecules in the IGF-1 pathway, including IGFR-1, IRS-1, IRS-2, PI3K, Akt-1, and FOXO3, and those in the MAPK pathway, such as ERK1/2, p38, and JNK, in gastrocnemius muscle were determined by immunoblotting (IB). Densitometric ratios of IRS-1 to β-actin and phosphorylated to total FOXO3 in wild-type and Cbl-b−/− mice are shown. Data are means ± standard deviations (n = 5). *, P < 0.05 for results for Cbl-b−/− mice compared to those for the wild-type mice. Sus, tail suspension; Con, control. (C) The mRNA levels of atrogin-1, MuRF-1, Cbl-b, FOXO3, and GAPDH in gastrocnemius muscle were measured by real-time RT-PCR. Ratios of target transcript to GAPDH mRNA in suspended mice are expressed relative to those of control animals. Results are means ± standard deviations (n = 5). *, P < 0.05 for results for Cbl-b−/− mice compared to those for the wild-type mice. (D) Sections of gastrocnemius muscle were stained with H&E, and cross-sectional myofiber areas were determined with WinRoof software. Scale bar, 100 μm. Data are means ± standard deviations (n = 5). *, P < 0.05 for results for Cbl-b−/− mice compared to those for the wild-type mice. (E) Soleus muscles isolated from suspended and control mice were subjected to contraction analysis. Maximal specific tetanic force was determined as described in Materials and Methods. Results are means ± standard deviations (n = 3). *, P < 0.05 for results for Cbl-b−/− mice compared to those for the wild-type mice.
The unloading-induced loss of IRS-1 in wild-type mice resulted in the impairment of IGF-1 signaling in vivo. Tail suspension significantly prevented Akt-1 activation, while the amount of Akt-1 remained constant until day 10. In this case, the amount of Akt-1 in skeletal muscle was significantly decreased on day 21 after tail suspension, suggesting that the prolonged disturbance of IGF-1 signaling affects the synthesis and/or degradation of Akt-1. Moreover, the suppression of IRS-1-dependent Akt-1 activity led to the dephosphorylation/activation of FOXO3 (Fig. 5B) and the induction of atrogin-1 expression (Fig. 5C). These results are consistent with the finding of a previous study that showed the induction of atrogin-1 expression by an unphosphorylated FOXO3 mutant (38). The deficiency of Cbl-b maintained FOXO3 in its phosphorylated inactive state during tail suspension and significantly attenuated the unloading-induced expression of atrogin-1. Unexpectedly, tail suspension did not alter MuRF-1 expression in either wild-type or Cbl-b−/− mice (Fig. 5C).
Since Cbl-b interacts with EGFR (12), we also tested the effect of Cbl-b deficiency on the MAPK pathway. Tail suspension and Cbl-b deficiency hardly changed the levels of the signaling molecules, such as ERK1/2, p38, and JNK (Fig. 5B). However, tail suspension and sham treatment induced the phosphorylation of all tested signaling molecules; there was no significant difference in phosphorylation between Cbl-b−/− and wild-type mice.
The sparing of muscle mass and IGF-1 signaling after 21-day tail suspension also was reflected in the preservation of the mean cross-sectional area of the gastrocnemius muscle fiber of Cbl-b−/− mice compared to that of suspended wild-type mice (Fig. 5D). Remarkably, no significant reduction in muscle fiber size was apparent after a 21-day tail suspension in Cbl-b−/− mice compared to that of nonsuspended controls (Fig. 5D). The effect of Cbl-b on muscle mass also was associated with muscle dysfunction in response to unloading; tail suspension for 21 days significantly decreased the maximal value of the tetanic force (specific force) of isolated mouse soleus muscle to 70% of the level for control muscle (Fig. 5E). In Cbl-b−/− mice, tail suspension did not result in a decrease of the force development. Thus, Cbl-b is an important mediator of the loss of muscle mass and function in response to unloading. The increased stability of IRS-1 in Cbl-b−/− animals contributed, at least in part, to their resistance to tail suspension-induced muscle atrophy. In contrast, tail suspension significantly decreased the time required to achieve the peak force of soleus muscle in wild-type mice, indicating that tail suspension changes the slow-twitch type of myofibers in soleus muscle to the fast-twitch type (see Fig. S4A in the supplemental material). However, fiber type switching induced by unloading was not suppressed in Cbl-b−/− mice. Immunohistochemistry for slow- or fast-type MyHC also showed that tail suspension increased the ratio of fast- to slow-type fibers in the soleus muscle of both wild-type and Cbl-b−/− mice (see Fig. S4B in the supplemental material). These findings suggest that Cbl-b plays an important role in regulating the size of myocytes but not the type of myofibers.
Synthetic oligopeptides inhibit Cbl-b-mediated IRS-1 ubiquitination and degradation.
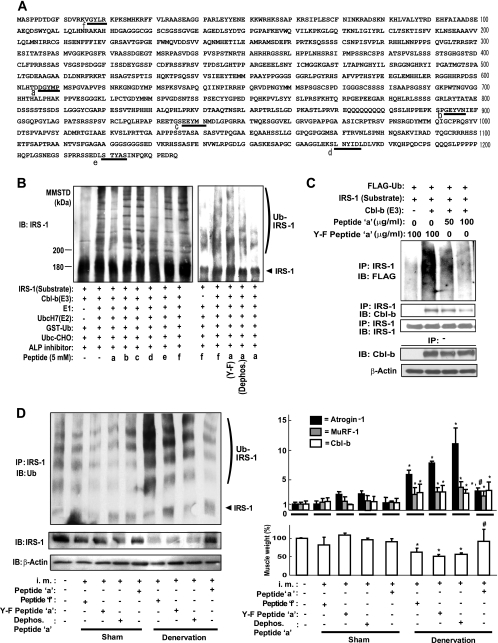
The observation that unloading failed to induce muscle atrophy and dysfunction in Cbl-b−/− mice (Fig. 5) suggests that the inhibition of Cbl-b-mediated IRS-1 ubiquitination is critical for the development of therapeutic strategies for unloading-mediated muscle atrophy. Furthermore, the phosphorylation of IRS-1 on tyrosine residues is indispensable for insulin/IGF-1-stimulated responses (16, 17). Cbl-b preferentially binds to phosphorylated tyrosine residues of substrates (45). Based on these findings, we screened the inhibitory activities of the synthetic pentapeptides consisting of the tyrosine phosphorylation domains of IRS-1 (Fig. 6A) against Cbl-b-mediated ubiquitination using a cell-free ubiquitination system.
FIG. 6.
Synthetic peptides inhibit IRS-1 ubiquitination and degradation. (A) Primary structure of rat IRS-1. Underlined letters a to f indicate the synthetic peptides a to f, respectively, that were used in this experiment as potential inhibitors of Cbl-b. (B) The Cbl-b-mediated ubiquitination of IRS-1 was performed in the presence of synthetic peptides a to f using a cell-free ubiquitination assay. Data are representative of three experiments. (C) COS7 cells were transfected with combinations of pCAG-Cbl-b-HA, pcDNA3-FLAG-Ubiquitin, and pLX2-IRS-1. Twenty hours later, cells were treated with 100 nM epoxomicin and peptide a (DGpYMP) or Y-F-mutated peptide a (DGFMP) at 50 or 100 μg/ml. Immunoprecipitates (IP) from whole-cell lysates incubated with an anti-IRS-1 antibody were subjected to immunoblotting (IB) for the indicated protein. Data are representative of three experiments. (D) C57BL/6 mice were subjected to denervation or sham operation (control). Six milligrams of peptide a, peptide f, Y-F-mutated peptide a, or dephosphorylated peptide a were injected into their gastrocnemius muscle every 2 days for 10 days after operation. Gastrocnemius muscles were isolated 6 h after the last injection, and their wet weight was measured. Immunoprecipitates (IP) from the muscle homogenates with an anti-IRS1 antibody were subjected to IB for ubiquitin (left). Data are representative of three experiments. The right upper panel shows the levels of atrogin-1, MuRF-1, and Cbl-b mRNA in gastrocnemius muscle measured by real-time RT-PCR. Muscle wet weight per kilogram of body weight is expressed as the percentage relative to that of control animals (lower right). Results are means ± standard deviations (n = 3). *, P < 0.05 for results for injected animals compared to those of noninjected animals; #, P < 0.05 for results with peptide a compared to those of control peptide f treatment in denervated mice. Data are representative of three experiments. i.m., intramuscular injection.
In this system, the interaction between Cbl-b and IRS-1 resulted in the Cbl-b-dependent ubiquitination of IRS-1 along with the expected electrophoretic pattern of IRS-1-ubiquitinated products (see Fig. S2E in the supplemental material). Among the various pentapeptides employed, peptide a (DGpYMP) and peptide d (LNpYID) significantly inhibited Cbl-b-mediated IRS-1 ubiquitination (Fig. 6B) in the cell-free system. Interestingly, the replacement of tyrosine with phenyalanine (Y-F mutation) and the dephosphorylation of peptide a reduced its inhibitory action on Cbl-b-mediated IRS-1 ubiquitination (Fig. 6B). ΔRING Cbl-b failed to induce IRS-1 ubiquitination in a cell-free ubiquitination system (see Fig. S2F in the supplemental material) as well as in COS7 cells (Fig. 3C). c-Cbl, another member of the Cbl family protein, also did not mediate the ubiquitination of IRS-1 (see Fig. S2F in the supplemental material). The replacement of tyrosine608 with phenylalanine in rat IRS-1 also induced resistance to Cbl-b-mediated ubiquitination, while the replacement of tyrosine18 with phenylalanine failed to prevent the ubiquitination (see Fig. S2F in the supplemental material). Furthermore, peptide a inhibited IRS-1 ubiquitination in COS7 cells expressing Cbl-b and IRS-1, but the mutated peptide a did not (Fig. 6C). Anti-IRS-1-IP from COS7 cells treated with peptide a contained small amounts of Cbl-b (Fig. 6C), suggesting that peptide a significantly prevented the interaction between Cbl-b and IRS-1. Peptide d showed the loss of inhibitory activity in cells (data not shown).
Finally, we examined the inhibitory effects of peptide a in vivo. Denervation stimulated the ubiquitination of IRS-1 and the expression of Cbl-b, atrogin-1, and MuRF-1 transcripts in gastrocnemius muscle, resulting in reduced muscle wet weight (Fig. 6D). In mice, the intramuscular injection of a high dose of peptide a significantly prevented the denervation-induced ubiquitination of IRS-1, a fall in the amount of IRS-1, and the expression of atrogin-1 compared to results of the injection of control peptide f, Y-F-mutated peptide a, and dephosphorylated peptide a, resulting in the restoration of decreased muscle weight (Fig. 6D). The injection of peptides did not change the increased amount of Cbl-b and MuRF-1 mRNAs (Fig. 6D). Neither denervation nor the injection of peptides affected plasma IGF-1 concentrations (see Fig. S5C in the supplemental material).
DISCUSSION
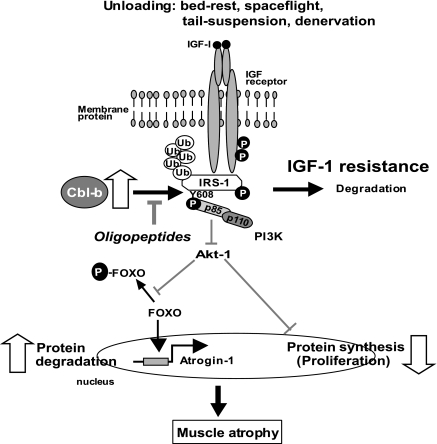
Our results indicate that the ubiquitin ligase Cbl-b plays a major role in skeletal muscle atrophy induced by unloading. The mechanism of Cbl-b-induced muscle atrophy is unique, in that it does not appear to involve the degradation of structural components of the muscle; rather, it impairs muscular trophic signals in response to unloading conditions. Recent studies of the molecular mechanisms of muscle atrophy have focused on the role of the IGF-1/PI3K/Akt-1 signaling cascade as a vital pathway in the regulation of the balance between hypertrophy and atrophy (38, 41). These studies indicate that under muscle-wasting conditions, such as disuse, diabetes, and fasting, decreased IGF-1/PI3K/Akt-1 signaling augments the expression of atrogin-1, resulting in muscle atrophy. However, these studies did not address the mechanisms of the unloading-induced impairment of growth factor signaling. In the present study, we found that under both in vitro and in vivo experimental conditions, Cbl-b ubiquitinated and induced the specific degradation of IRS-1, a key intermediate of skeletal muscle growth regulated by IGF-1/insulin and growth hormone, resulting in the inactivation of Akt-1 (Fig. 1, 3, and 4). The inactivation of Akt-1 led to the upregulation of atrogin-1 (Fig. 2) through the dephosphorylation (activation) of FOXO3, as well as a reduced mitogen response in skeletal muscle. Thus, the activation of Cbl-b may be an important mechanism underlying the failure of atrophic muscle to respond to growth factor-based treatments such as IGF-1 (Fig. 7).
FIG. 7.
Mechanistic model of unloading-mediated muscle atrophy. Unloading induces ubiquitin ligase Cbl-b in myocytes. Cbl-b stimulates ubiquitination and the degradation of IRS-1, an important intermediate in IGF-1 signaling pathway, resulting in IGF-1 resistance in myocytes during unloading. IGF-1 resistance induces impaired protein synthesis and enhances protein degradation in muscle, leading to muscle atrophy. Cbl-b and PI3K may interact with phosphotyrosine608 of IRS-1. The inhibition of Cbl-b and IRS-1 interaction by oligopeptides may restore this impairment of IGF-1 signaling (see Discussion for more details). GSK3, glycogen synthase kinase 3; mTOR, mammalian target of rapamycin; S6K, p70 S6 kinase; Ub, ubiquitin.
Recently, several studies reported that SOCS1/3 and Cul7 are ubiquitin ligases for IRS-1 (35, 49). SOCS1/3 is associated with insulin resistance and glucose intolerance mainly in the liver by promoting the ubiquitination and degradation of IRS-1 and IRS-2. In contrast, Cul7 plays a role in IRS-1 degradation in a process that may contribute to the regulation of cellular senescence. In the present study, the expression of SOCS1 and Cul7 in skeletal muscle did not change following tail suspension (see Fig. S1B in the supplemental material). Moreover, although SOCS1 deficiency does not prevent diet-induced insulin resistance (10), the deficiency of the Cbl-b gene enhances the infiltration of macrophages in adipose tissue and causes peripheral insulin resistance in mice (21), suggesting that Cbl-b is involved in insulin resistance through the regulation of macrophage function. It is likely that the expression of ubiquitin ligase for IRS-1 is distinctly regulated in various cells.
A near-universal property of diverse muscle-wasting diseases is the presence of IGF-1 resistance. Attempts to overcome this barrier clinically have led to the use of high doses of IGF-1 sufficient to produce numerous and, in many cases, prohibitively undesirable side effects (2). Our study revealed the molecular basis of this treatment barrier. Our data indicate that Cbl-b is required for unloading-induced IRS-1 degradation and the resultant loss of muscle mass and function; in its absence, muscle IGF-1 responsiveness was restored (Fig. 4 and 5). Furthermore, we found that synthetic peptide a, DGpYMP, blocked IRS-1 ubiquitination both in vitro and in vivo, resulting in the restoration, at least in part, of denervation-induced muscle atrophy. Since peptides are rapidly degraded by aminopeptidase in muscle, our studies indicate that a high dose of peptide a and frequent intramuscular injection were required to prevent the IRS-1 ubiquitination. To circumvent these issues, we are synthesizing and testing low-molecular-weight chemical mimetics to peptide a for their efficacy in the inhibition of Cbl-b.
Peptide a is a small pentapeptide corresponding to amino acids 606 to 610 of rat IRS-1. Interestingly, tyrosine608 of IRS-1 is phosphorylated in response to insulin/IGF-1 and becomes a major docking site for PI3K (11). The substitution or dephosphorylation of tyrosine in peptide a did not prevent Cbl-b-mediated IRS-1 ubiquitination (Fig. 6B and D). The replacement of tyrosine608 with phenylalanine in rat IRS-1 also induced resistance to Cbl-b-mediated ubiquitination (see Fig. S2F in the supplemental material). These findings suggest that peptide a inhibited the interaction between Cbl-b and IRS-1 through phosphotyrosine608 of IRS-1 (Fig. 7). Previous biochemical and structural studies (29, 50) indicated that SDGpYTPEPA, a phosphopeptide of ZAP-70, interacted with the TKB domain of c-Cbl, which is highly homologous to Cbl-b. The residues that interact with the ZAP-70 peptide (50) also are highly conserved in Cbl ligases. Interestingly, the ZAP-70 peptide is very similar to peptide a. Furthermore, the homology modeling of the Cbl-b TKB domain using the complex structure of the c-Cbl and ZAP-70 peptide as the template predicted that the peptide binding surface in the TKB domain is highly conserved in the two Cbl ligases (data not shown). The superimposition of the model structure onto the complex structure of c-Cbl and the ZAP-70 peptide shows a lack of steric hindrance between Cbl-b and the ZAP-70 peptide (data not shown). Based on these observations, it is conceivable that Cbl-b binds to the ZAP-70 peptide and peptide a. It is likely that peptide a inhibits the interaction between Cbl-b and IRS-1 at this phosphorylated site, since peptide a could competitively inhibit the binding of Cbl-b to IRS-1, thereby leading to a significant inhibition of Cbl-b-mediated IRS-1 ubiquitination and muscle mass loss.
The unloading-related reduced muscle strength correlated with the reduction in muscle fiber size. IGF-1 is involved in the stimulation of the IRS-1/PI3K/Akt-1 kinase cascade, which affects muscle size composition (25, 32), while the switching of the muscle fiber type may be regulated by other factors, such as peroxisome-proliferator-activated receptor-γ coactivator −1α, which regulates the expression of slow-type fiber-associated genes (28). Interestingly, tail suspension was not associated with a decreased maximal specific tetanic force of the soleus muscle of Cbl-b−/− mice (Fig. 5E). These findings support our hypothesis that Cbl-b-targeted therapy also preserves muscle mass and strength by maintaining the IGF-1 signaling. In contrast, the myofiber type affects the endurance of muscle power. In the present study, Cbl-b deficiency failed to restore the decline in the endurance of muscle contraction (see Fig. S4A in the supplemental material) and the switching of muscle fiber type during unloading (see Fig. S4B in the supplemental material). Based on these results, approaches other than Cbl-b targeting are necessary to prevent fiber type switching caused by unloading.
Cbl-b interacts with various proteins. Among them, EGFR is one of the most-characterized Cbl-b-interacting proteins (12). The present study showed no interaction between Cbl-b and EGFR (Fig. 1C) and little effect of Cbl-b deficiency on the MAPK pathway (Fig. 5B). Based on these findings, we suggest that Cbl-b affects the MAPK pathway less than the IGF-1 signaling pathway in skeletal muscle during unloading. In skeletal muscles, Cbl-b preferentially interacted with IRS-1 among the tested signaling molecules, such as Vav and C3G. However, these findings cannot exclude the possible interaction of Cbl-b with other unidentified proteins involved in atrophic signals in skeletal muscle. Further studies are necessary to explore this possibility.
Both atrogin-1 and MuRF-1 are important ubiquitin ligases during muscle atrophy and are regulated by the IGF-1/PI3K/Akt-1 pathway. In this study, the expression of MuRF-1 mRNA did not respond to changes in the IGF-1 signaling compared to that of atrogin-1 mRNA (Fig. 2B, 5C, and 6D). This is consistent with the previous report of Sacheck et al. (36) that treatment with IGF-1 did not markedly suppress the dexamethasone-induced expression of MuRF-1 in C2C12 myotubes compared to that of atrogin-1. They suggested that the expression of MuRF-1 in response to IGF-1 treatment is different from that of atrogin-1; atrogin-1 expression is much more responsive to hormonal signals. The different sensitivity to IGF-1 signaling may explain the different response to Cbl-b deficiency between MuRF-1 and atrogin-1.
Because Cbl-b and c-Cbl share a high degree of structural similarity (23, 46), they may play functionally redundant roles as ubiquitin ligases. In other systems, both Cbl-b and c-Cbl appear to regulate equally the ubiquitination of the same binding partner, such as the adaptor protein CIN85 (44) and the tyrosine kinase Syk (8, 40). However, the complete lack of IRS-1 degradation in Cbl-b−/− mice (Fig. 5B) suggests that IRS-1 is a more physiologically relevant substrate for Cbl-b than for c-Cbl in skeletal muscle atrophied by unloading. Furthermore, the specific expression of Cbl-b during unloading in muscle (Fig. 1A; also see Fig. S1A and B in the supplemental material) and the failure of IRS-1 ubiquitination by c-Cbl (see Fig. S2F in the supplemental material) suggest that the regulatory system for Cbl-b expression is distinct from that for c-Cbl expression. On the other hand, a recent study showed that Cbl-b, but not c-Cbl, acts as a negative regulator of tumor necrosis factor α and interleukin-6 produced by bone-marrow-derived mast cells (18) or macrophages infiltrated into white adipose tissue (21). Thus, Cbl-b may play a role in protein metabolism in skeletal muscle that is distinct from that of c-Cbl.
The Cbl-b gene is highly sensitive to mechanical stress (unloading) (30, 31). Therefore, we hypothesized that some of the components of the signal transduction pathways that regulate the expression of Cbl-b are important sensors of mechanical stress. We identified transcription factors that regulate Cbl-b expression using rat L6 myoblasts and differentiated myotubes (unpublished data). The elucidation of the biological relevance of Cbl-b expression as a sensor of unloading is a critical next step for defining the molecular mechanism through which mechanical unloading is transduced into biochemical signaling in skeletal muscle.
The disturbance of bone/muscle functions is a key factor in unloading-mediated diseases. To conquer these bone/muscle-wasting diseases, the identification of common genetic targets in bone and muscle is critical. We reported previously that denervation in mice induces high levels of Cbl-b expression in osteoblasts (43). Moreover, skeletal muscle recently has been reported to regulate bone maintenance through the release of IGF-1 (1). Thus, treatment with peptide a targeting Cbl-b may be a more efficient therapeutic option for simultaneously attenuating bone and muscle atrophy during unloading.
In summary, we propose a role for Cbl-b in the downregulation of IGF-1 signaling in skeletal muscle under unloading conditions. Our data also suggest that the selective inhibition of Cbl-b-mediated IRS-1 ubiquitination represents a novel therapeutic strategy for wasting diseases in the musculoskeletal locomotor system.
Supplementary Material
Acknowledgments
We thank K. Kishi for pLX2-IRS-1, S. Lipkowitz for pCEFL-Cbl-b-HA, T. Suzuki for pcDNA3-FLAG-Ubiquitin, and J. Miyazaki for pCAG. We are grateful to H. Gu and T. Inazu for providing Cbl-b−/− mice. We also thank Otsuka Pharmaceutical Co. and Ajinomoto Co. for providing synthetic peptides.
This study was carried out as part of the Ground Research Announcement for Space Utilization, promoted by the Japan Aerospace Exploration Agency (JAXA) and the Japan Space Forum (to T.N.), a grant in aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.N.), Seed Production by the Japan Science and Technology Agency (to T.N.), and the Promotion of Basic Research Activities for Innovative Biosciences from the Bio-Oriented Technology Research Advancement Institution of Japan (to T.N.). This study also was supported in part by a high-technology research center grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan to the Juntendo University Research Institute for Disease of Old Ages, Tokyo, Japan.
Footnotes
Published ahead of print on 22 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alzghoul, M. B., D. Gerrard, B. A. Watkins, and K. Hannon. 2004. Ectopic expression of IGF-1 and Shh by skeletal muscle inhibits disuse-mediated skeletal muscle atrophy and bone osteopenia in vivo. FASEB J. 18221-223. [DOI] [PubMed] [Google Scholar]
- 2.Bach, L. A. 1999. The insulin-like growth factor system: basic and clinical aspects. Aust. N.Z. J. Med. 29355-361. [DOI] [PubMed] [Google Scholar]
- 3.Bodine, S. C., E. Latres, S. Baumhueter, V. K. Lai, L. Nunez, B. A. Clarke, W. T. Poueymirou, F. J. Panaro, E. Na, K. Dharmarajan, et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2941704-1708. [DOI] [PubMed] [Google Scholar]
- 4.Bustelo, X. R., P. Crespo, M. Lopez-Barahona, J. S. Gutkind, and M. Barbacid. 1997. Cbl-b, a member of the Sli-1/c-Cbl protein family, inhibits Vav-mediated c-Jun N-terminal kinase activation. Oncogene 152511-2520. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, Y. J., H. K. Kole, K. Brown, M. Naramura, S. Fukuhara, R. J. Hu, I. K. Jang, J. S. Gutkind, E. Shevach, and H. Gu. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403216-220. [DOI] [PubMed] [Google Scholar]
- 6.Costelli, P., M. Muscaritoli, M. Bossola, F. Penna, P. Reffo, A. Bonetto, S. Busquets, G. Bonelli, F. J. Lopez-Soriano, G. B. Doglietto, et al. 2006. IGF-1 is downregulated in experimental cancer cachexia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291R674-R683. [DOI] [PubMed] [Google Scholar]
- 7.Criswell, D. S., F. W. Booth, F. DeMayo, R. J. Schwartz, S. E. Gordon, and M. L. Fiorotto. 1998. Overexpression of IGF-1 in skeletal muscle of transgenic mice does not prevent unloading-induced atrophy. Am. J. Physiol. Endocrinol. Metab. 275E373-E379. [DOI] [PubMed] [Google Scholar]
- 8.Dangelmaier, C. A., P. G. Quinter, J. Jin, A. Y. Tsygankov, S. P. Kunapuli, and J. L. Daniel. 2005. Rapid ubiquitination of Syk following GPVI activation in platelets. Blood 1053918-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egawa, K., N. Nakashima, P. M. Sharma, H. Maegawa, Y. Nagai, A. Kashiwagi, R. Kikkawa, and J. M. Olefsky. 2000. Persistent activation of phosphatidylinositol 3-kinase causes insulin resistance due to accelerated insulin-induced insulin receptor substrate-1 degradation in 3T3-L1 adipocytes. Endocrinology 1411930-1935. [DOI] [PubMed] [Google Scholar]
- 10.Emanuelli, B., Y. Macotela, J. Boucher, and K. C. Ronald. 2008. SOCS-1 deficiency does not prevent diet-induced insulin resistance. Biochem. Biophys. Res. Commun. 377447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito, D. L., Y. Li, A. Cama, and M. J. Quon. 2001. Tyr(612) and Tyr(632) in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 1422833-2840. [DOI] [PubMed] [Google Scholar]
- 12.Ettenberg, S. A., M. M. Keane, M. M. Nau, M. Frankel, L. M. Wang, J. H. Pierce, and S. Lipkowitz. 1999. cbl-b inhibits epidermal growth factor receptor signaling. Oncogene 181855-1866. [DOI] [PubMed] [Google Scholar]
- 13.Goldspink, G. 2006. Impairment of IGF-I gene splicing and MGF expression associated with muscle wasting. Int. J. Biochem. Cell Biol. 38481-489. [DOI] [PubMed] [Google Scholar]
- 14.Gomes, M. D., S. H. Lecker, R. T. Jagoe, A. Navon, and A. L. Goldberg. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 9814440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González, E., M. L. Messi, and O. Delbono. 2000. The specific force of single intact extensor digitorum longus and soleus mouse muscle fibers declines with aging. J. Membr. Biol. 178175-183. [DOI] [PubMed] [Google Scholar]
- 16.Greene, M. W., N. Morrice, R. S. Garofalo, and R. A. Roth. 2004. Modulation of human insulin receptor substrate-1 tyrosine phosphorylation by protein kinase Cδ. Biochem. J. 378105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gual, P., Y. Le Marchand-Brustel, and J. F. Tanti. 2005. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 8799-109. [DOI] [PubMed] [Google Scholar]
- 18.Gustin, S. E., C. B. Thien, and W. Y. Langdon. 2006. Cbl-b is a negative regulator of inflammatory cytokines produced by IgE-activated mast cells. J. Immunol. 1775980-5989. [DOI] [PubMed] [Google Scholar]
- 19.Harman, S. M., and M. R. Blackman. 2003. The effects of growth hormone and sex steroid on lean body mass, fat mass, muscle strength, cardiovascular endurance and adverse events in healthy elderly women and men. Horm. Res. 60121-124. [DOI] [PubMed] [Google Scholar]
- 20.Hirasaka, K., T. Nikawa, L. Yuge, I. Ishihara, A. Higashibata, N. Ishioka, A. Okubo, T. Miyashita, N. Suzue, T. Ogawa, et al. 2005. Clinorotation prevents differentiation of rat myoblastic L6 cells in association with reduced NF-κB signaling. Biochim. Biophys. Acta 743130-140. [DOI] [PubMed] [Google Scholar]
- 21.Hirasaka, K., S. Kohno, J. Goto, H. Furochi, K. Mawatari, N. Harada, T. Hosaka, Y. Nakaya, K. Ishidoh, T. Obata, Y. Ebina, H. Gu, S. Takeda, K. Kishi, and T. Nikawa. 2007. Deficiency of Cbl-b gene enhances infiltration and activation of macrophages in adipose tissue and causes peripheral insulin resistance in mice. Diabetes 562511-2522. [DOI] [PubMed] [Google Scholar]
- 22.Ikemoto, M., T. Nikawa, S. Takeda, C. Watanabe, T. Kitano, K. M. Baldwin, R. Izumi, I. Nonaka, T. Towatari, S. Teshima, K. Rokutan, and K. Kishi. 2001. Space shuttle flight (STS-90) enhances degradation of rat myosin heavy chain in association with activation of ubiquitin-proteasome pathway. FASEB J. 151279-1281. [DOI] [PubMed] [Google Scholar]
- 23.Keane, M. M., O. M. Rivero-Lezcano, J. A. Mitchell, K. C. Robbins, and S. Lipkowitz. 1995. Cloning and characterization of cbl-b: a SH3 binding protein with homology to the c-cbl proto-oncogene. Oncogene 102367-2377. [PubMed] [Google Scholar]
- 24.Kedar, V., H. McDonough, R. Arya, H. H. Li, H. A. Rockman, and C. Patterson. 2004. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA 10118135-18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latres, E., A. R. Amini, A. A. Amini, J. Griffiths, F. J. Martin, Y. Wei, H. C. Lin, G. D. Yancopoulos, and D. J. Glass. 2005. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 2802737-2744. [DOI] [PubMed] [Google Scholar]
- 26.Lee, A. V., J. L. Gooch, S. Oesterreich, R. L. Guler, and D. Yee. 2000. Insulin-like growth factor I-induced degradation of insulin receptor substrate 1 is mediated by the 26S proteasome and blocked by phosphatidylinositol 3′-kinase inhibition. Mol. Cell. Biol. 201489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, H. H., V. Kedar, C. Zhang, H. McDonough, R. Arya, D. Z. Wang, and C. Patterson. 2004. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 1141058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J., H. Wu, P. T. Tarr, C. Y. Zhang, Z. Wu, O. Boss, L. F. Michael, P. Puigserver, E. Isotani, E. N. Olson, B. B. Lowell, R. Bassel-Duby, and B. M. Spiegelman. 2002. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418797-801. [DOI] [PubMed] [Google Scholar]
- 29.Meng, W., S. Sawasdikosol, S. E. Shoelson, and M. J. Eck. 1999. Structure of the amino-terminal domain of Cbl complexed to its binding site of ZAP-70 kinase. Nature 39884-90. [DOI] [PubMed] [Google Scholar]
- 30.Nikawa, T., K. Ishidoh, K. Hirasaka, I. Ishihara, M. Ikemoto, M. Kano, E. Kominami, I. Nonaka, T. Ogawa, G. R. Adams, K. M. Baldwin, N. Yasui, K. Kishi, and S. Takeda. 2004. Skeletal muscle gene expression in space-flown rats. FASEB J. 18522-524. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, T., H. Furochi, M. Mameoka, K. Hirasaka, Y. Onishi, N. Suzue, M. Oarada, M. Akamatsu, H. Akima, T. Fukunaga, K. Kishi, N. Yasui, K. Ishido, H. Fukuoka, and T. Nikawa. 2006. Ubiquitin ligase gene expression in healthy volunteers with 20-day bed rest. Muscle Nerve 34463-469. [DOI] [PubMed] [Google Scholar]
- 32.Pallafacchina, G., E. Calabria, A. L. Serrano, J. M. Kalhovde, and S. A. Schiaffino. 2002. Protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc. Natl. Acad. Sci. USA 999213-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Primig, M., T. H. Chang, and M. Buckingham. 1998. A novel GFP-neo vector designed for the isolation and analysis of enhancer elements in transfected mammalian cells. Gene 215181-189. [DOI] [PubMed] [Google Scholar]
- 34.Rennie, M. J. 2003. Claims for the anabolic effects of growth hormone: a case of the emperor's new clothes? Br. J. Sports Med. 37100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rui, L., M. Yuan, D. Frantz, S. Shoelson, and M. F. White. 2002. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 27742394-42398. [DOI] [PubMed] [Google Scholar]
- 36.Sacheck, J. M., A. Ohtsuka, S. C. McLary, and A. L. Goldberg. 2004. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am. J. Physiol. Endocrinol. Metab. 287E591-E601. [DOI] [PubMed] [Google Scholar]
- 37.Sacheck, J. M., J. P. Hyatt, A. Raffaello, R. T. Jagoe, R. R. Roy, V. R. Edgerton, S. H. Lecker, and A. L. Goldberg. 2007. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 21140-155. [DOI] [PubMed] [Google Scholar]
- 38.Sandri, M., C. Sandri, A. Gilbert, C. Skurk, E. Calabria, A. Picard, K. Walsh, S. Schiaffino, S. H. Lecker, and A. L. Goldberg. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimura, H., N. Hattori, S. Kubo, Y. Mizuno, S. Asakawa, S. Minoshima, N. Shimizu, K. Iwai, T. Chiba, K. Tanaka, and T. Suzuki. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25302-305. [DOI] [PubMed] [Google Scholar]
- 40.Sohn, H. W., H. Gu, and S. K. Pierce. 2003. Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. J. Exp. Med. 1971511-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stitt, T. N., D. Drujan, B. A. Clarke, F. Panaro, Y. Timofeyva, W. O. Kline, M. Gonzalez, G. D. Yancopoulos, and D. J. Glass. 2004. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14395-403. [DOI] [PubMed] [Google Scholar]
- 42.Sun, X. J., D. L. Crimmins, M. G. Myers, Jr., M. Miralpeix, and M. F. White. 1993. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol. Cell. Biol. 137418-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzue, N., T. Nikawa, Y. Onishi, C. Yamada, K. Hirasaka, T. Ogawa, H. Furochi, H. Kosaka, K. Ishidoh, H. Gu, S. Takeda, N. Ishimaru, Y. Hayashi, H. Yamamoto, K. Kishi, and N. Yasui. 2006. Ubiquitin ligase Cbl-b downregulates bone formation through suppression of IGF-1 signaling in osteoblasts during denervation. J. Bone Miner. Res. 21722-734. [DOI] [PubMed] [Google Scholar]
- 44.Szymkiewicz, I., K. Kowanetz, P. Soubeyran, A. Dinarina, S. Lipkowitz, and I. Dikic. 2002. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J. Biol. Chem. 27739666-39672. [DOI] [PubMed] [Google Scholar]
- 45.Thien, C. B., and W. Y. Langdon. 2001. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2294-307. [DOI] [PubMed] [Google Scholar]
- 46.Thien, C. B., and W. Y. Langdon. 2005. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem. J. 391153-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tintignac, L. A., J. Lagirand, S. Batonnet, V. Sirri, M. P. Leibovitch, and S. A. Leibovitch. 2005. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 2802847-2856. [DOI] [PubMed] [Google Scholar]
- 48.Wells, D. J. 1993. Improved gene transfer by direct plasmid injection associated with regeneration in mouse skeletal muscle. FEBS Lett. 332179-182. [DOI] [PubMed] [Google Scholar]
- 49.Xu, X., A. Sarikas, D. C. Dias-Santagata, G. Dolios, P. J. Lafontant, S. C. Tsai, W. Zhu, H. Nakajima, H. O. Nakajima, L. J. Field, R. Wang, and Z. Q. Pan. 2008. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 30403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102533-539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.