Abstract
FLASH has been shown to be required for S phase progression and to interact with a nuclear protein, ataxia-telangiectasia locus (NPAT), a component of Cajal bodies in the nucleus and an activator of histone transcription. We investigated the role of human FLASH by using an inducible FLASH knockdown system in the presence or absence of various mutant forms of mouse FLASH. While carboxyl-terminal deletion mutants of FLASH, which do not interact with NPAT, can support S phase progression, its amino-terminal deletion mutants, which are unable to self associate, cannot support S phase progression, replication-dependent histone transcription, or the formation of Cajal bodies. Furthermore, FLASH was shown to be associated with arsenite resistance protein 2 (ARS2) through its central region, which is composed of only 13 amino acids. The expression of ARS2 and the interaction between FLASH and ARS2 are required for S phase progression. Taking these results together, FLASH functions in S phase progression through interaction with ARS2.
FADD-like interleukin-1β-converting enzyme/caspase-8-associated huge protein (FLASH)/CASP8AP2, originally identified as a component of the Fas-caspase-8-mediated apoptosis-inducing pathway (13), was reported to be involved in Fas-mediated apoptotic signaling through translocation from the nucleus to the cytoplasm (20). FLASH also was shown to act in the nucleus as an enhancer and/or repressor of steroid hormone receptor-mediated transcription (14, 15, 21). On the other hand, FLASH was found to be essential for cell division by the high-throughput screening of a genome-scale library of short interfering RNAs (16). FLASH also was reported to be a potential prognostic marker in cases of acute lymphoblastic leukemia (10). Recently, it was shown that FLASH is an essential component of Cajal bodies in the nucleus, playing an important role in histone transcription and S phase progression (3, 4). In addition, these reports indicated that FLASH interacts with a nuclear protein, ataxia-telangiectasia locus (p220NPAT [NPAT]), a component of Cajal bodies. The phosphorylation of NPAT by cyclin E/Cdk2 was reported to regulate the activation of histone gene transcription in S phase and to be necessary to maintain the structure of Cajal bodies during the cell cycle (7, 19, 27, 29, 30). Therefore, FLASH has been thought to play an essential role in S phase progression, probably through interaction with NPAT. However, the molecular mechanisms underlying the role of FLASH in cell cycle progression largely have remained unknown.
Cajal bodies are small subnuclear organelles, originally described by Santiago Ramon Y Cajal in 1903, that are involved in several nuclear functions, including the biogenesis and trafficking of small nuclear and nucleolar ribonucleoprotein particles (snRNPs and snoRNPs, respectively) and the processing of pre-rRNA and replication-dependent histone mRNA (6, 11). Recently, a number of Cajal bodies were found to contain FLASH and NPAT but not coilin, a conventional marker of such bodies, suggesting that Cajal bodies can be classified into at least two types, those containing coilin and those containing both FLASH and NPAT (5). In addition, variability in the number of Cajal bodies was reported to be relevant to cell ploidy in cancer cell lines (5).
In this article, we describe a novel factor interacting with FLASH, namely arsenite resistance protein 2 (ARS2; originally called ASR2). ARS2 first was identified as a gene product conferring resistance to arsenite on an arsenite-hypersensitive cell line (24). ARS2 was reported to be highly conserved in mammals, zebrafish, and plants (2, 12, 23, 28). Recently, ARS2 was shown to be essential for early development in mammals by an analysis of ARS2-null embryos derived from ARS2 knockout mice (28). However, the biological function of ARS2 is largely unknown, especially at the molecular level.
Here, we investigated the role of FLASH in cell cycle progression by using an inducible FLASH knockdown system. We show that both FLASH and ARS2 are essential for S phase progression, and the interaction of FLASH with ARS2, but not with NPAT, is required for normal S phase progression.
MATERIALS AND METHODS
Cell lines, culture conditions, and transient transfection.
Human pharyngeal carcinoma-derived KB and human embryonic kidney-derived 293T cells were cultured in Dulbecco's modified Eagle's medium (Nacalai Tesque) supplemented with 10% fetal calf serum (Sigma), 100 U/ml penicillin, and 100 μg/ml streptomycin (Nacalai Tesque) at 37°C in 5% CO2. For transient transfection, 293T cells were transfected with expression vectors using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions.
Lentiviral expression vectors.
Lentiviral vectors, provided by H. Miyoshi (Riken), were prepared as described earlier (22). Human ARS2 (hARS2) cDNA was amplified by reverse transcription-PCR (RT-PCR) from human T lymphoma-derived Jurkat cell mRNA using 5′-ATGGGTGACAGTGATG-3′ as a forward primer and 5′-TCAAAAGAAATCAACATCGT-3′ as a reverse primer, and then it was cloned in frame with hemagglutinin (HA), DsRed-Monomer, or FLAG3 tag into a lentiviral vector, CSII-EF-MCS-IRES-Hyg (22), by using its unique restriction sites XhoI and BamHI. Mouse FLASH (mFLASH) cDNA (13) was subcloned in frame with enhanced green fluorescent protein (EGFP) or FLAG3 tags into CSII-EF-MCS. For the construction of various deletion mutants of mFLASH and hARS2, mutant cDNAs were amplified from wild-type cDNA by PCR with specific primers and cloned into CSII-EF-MCS or CSII-EF-MCS-IRES-Hyg. All PCR-generated plasmid constructs were verified by DNA sequencing.
Tetracycline-inducible shRNA expression system.
To induce specific gene silencing, the tetracycline-inducible short hairpin RNA (shRNA) expression system (Tet-On shRNA system) with lentivirus-based vectors was utilized as described previously (17). shRNA expression was induced by the treatment of cells with 1 μg/ml doxycycline (Dox) (Nacalai Tesque). The oligonucleotides of shRNA for human FLASH (shFLASH) and hARS2 (shARS2) were generated from nucleotides (nt) 755 to 777 of FLASH cDNA (GenBank no. NM_012115) and nt 380 to 400 of ARS2 cDNA (GenBank no. NM_015908), respectively. The oligonucleotides of shRNA for silencing lacZ (shlacZ) were generated from nt 407 to 425 of lacZ cDNA (GenBank no. V00296).
Flow cytometry.
For analyses of DNA content (cell cycle analysis), cells were fixed with ice-cold 70% ethanol in phosphate-buffered saline (PBS) and incubated with PBS containing 50 μg/ml propidium iodide (PI) for 30 min at 4°C after treatment with 250 μg/ml RNase for 15 min at 37°C. For the bromodeoxyuridine (BrdU) incorporation assay, cells were labeled with 10 μM BrdU for 30 min and fixed in PBS containing both 70% ethanol and 50 mM glycine, pH 2.0, for 30 min at 4°C. After treatment with 4 N HCl for 20 min at room temperature, the incorporated BrdU was determined by staining with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody (Ab) (BD Biosciences) or allophycocyanin-conjugated anti-BrdU Ab (BD Pharmingen) for 45 min at 37°C. Before the analysis, cells were incubated in PBS containing 10 μg/ml PI for 30 min at 4°C. Cells were analyzed with an EPICS XL flow cytometer (Beckman Coulter) or a FACSCanto II (Becton Dickinson). The data were analyzed with EXPO32 software (Beckman Coulter) or BD FACSDiva software (Becton Dickinson).
Immunoprecipitation and Western blotting.
Total cell lysate was prepared with ice-cold lysis buffer (20 mM Tris-HCl [pH 7.4], 10% glycerol, 1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, and 1 mM EGTA) or radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 0.5% sodium deoxycholate) supplemented with complete protease inhibitor cocktails (Roche Molecular Biochemicals). After immunoprecipitation was carried out, the cell lysate and eluted immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting as described earlier (18). For the detection of FLASH, 4% acrylamide was used as a separate gel to attain sufficient sensitivity, since the efficiency with which FLASH was transferred to the nylon membrane was very low. The Abs used were anti-FLASH monoclonal Ab (MAb) (1:1,000; generated in our laboratory against a peptide of amino acids [aa] 1260 to 1575 of mFLASH and with an ability to react with both mouse and human FLASH), anti-Flag M2-agarose resin (Sigma), anti-GFP (1:1,000; 1E4 MAb or polyclonal Ab; MBL), anti-NPAT (1:100; BD Biosciences), anti-HA (1:2,000; 16B12; Covance), anti-Flag M2 (1:1,000; Sigma), anti-ARS2 (1:500; Novus Biologicals), and antiactin (1:5,000; Chemicon).
Immunofluorescence staining.
Cells in chamber slides were fixed with 3% formaldehyde in PBS at 4°C for 20 min and permeabilized with 0.5% Triton buffer (0.5% Triton X-100, 20 mM HEPES-KOH [pH 7.4], 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose) for 10 min at room temperature. After being blocking with 10% bovine serum albumin (BSA) in PBS for 1 h, cells were treated with a primary Ab overnight and incubated with an Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary Ab (1:1,000; Molecular Probes) for 1 h. DNA was stained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) in PBS for 20 min. Fluorescence images were acquired under a confocal microscope (TCS SP2 AOBS; Leica). For counting the number of nuclear foci in cells, images were acquired with a DAS microscope (Leica). The following primary Abs were used: anti-NPAT (1:120; BD Pharmingen) and anticoilin (1:125; ab11822; Abcam).
Mass spectrometry.
For affinity purification of the protein complex with ΔC2 mFLASH-FLAG3, KB cells with Tet-On shFLASH expressing ΔC2 mFLASH-FLAG3 in 10 150-mm dishes were treated with 1 μg/ml Dox for 3 days, and then cell lysate was prepared using ice-cold RIPA buffer. The soluble fraction was recovered by centrifugation for 50 min at 9,400 × g and precleared with 100 μl of protein G beads for 1 h at 4°C. The supernatants then were further incubated with 100 μl of anti-Flag M2-agarose resin (Sigma) for 5 h at 4°C. The resin was washed seven times with lysis buffer. For elution, sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol) was added and incubated for 10 min at room temperature after 5 min at 37°C. The eluted fractions were concentrated, separated by SDS-PAGE (5 to 15% gradient acrylamide gels), and visualized by silver staining. For the mass spectrometric analysis, the separated gel was cut into 13 slices and digested with trypsin (Promega). The peptides extracted from each slice were analyzed by a hybrid mass spectrometer (QSTAR XL; Applied Biosystems) equipped with a nanoelectrospray ion source (MDS Protana).
Northern blotting and real-time RT-PCR.
Total mRNA for Northern blotting and real-time RT-PCR was prepared using ISOGEN (Nippon Gene) and an RNeasy Mini kit (Qiagen), respectively, according to the manufacturers' instructions. For Northern blotting, total RNA (20 μg) was loaded on a 1.0% formaldehyde agarose gel for electrophoresis and transferred to a Hybond N+ nylon membrane (GE healthcare). The membrane was hybridized with alkaline phosphate-labeled DNA probes for histones and human eEF1A1 (hEF1α) by using AlkPhos Direct (GE Healthcare). For probes for histone mRNAs, the following coding region of each histone was amplified by PCR: H1c (nt 44 to 685; GenBank no. NM_005319) for H1, H2ad (nt 1 to 393; GenBank no. NM_021065) for H2A, H2bf (nt 1 to 381; GenBank no. NM_003522) for H2B, H3a (nt 1 to 411; GenBank no. NM_003529) for H3, and H4a (nt 1 to 312; GenBank no. NM_003538) for H4. The probe for human eEF1A1 (hEF1α) cDNA was used as a standard.
For checking the knockdown efficiency by shRNA, RT products were prepared with a high-capacity cDNA RT kit (Applied Biosystems) and analyzed by quantitative PCR using the StepOne real-time PCR system (Applied Biosystems) with the following primer sets: 5′-CATTGTCAAGATGCTGGATG-3′ and 5′-CTCATCCTTGTCGTTGGTTT-3′ for ARS2 and 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-GACAAGCTTCCCGTTCTCAG-3′ for glyceraldehyde-3-phosphate dehydrogenase (as a standard).
RESULTS
N-terminal region of FLASH is required for S phase progression.
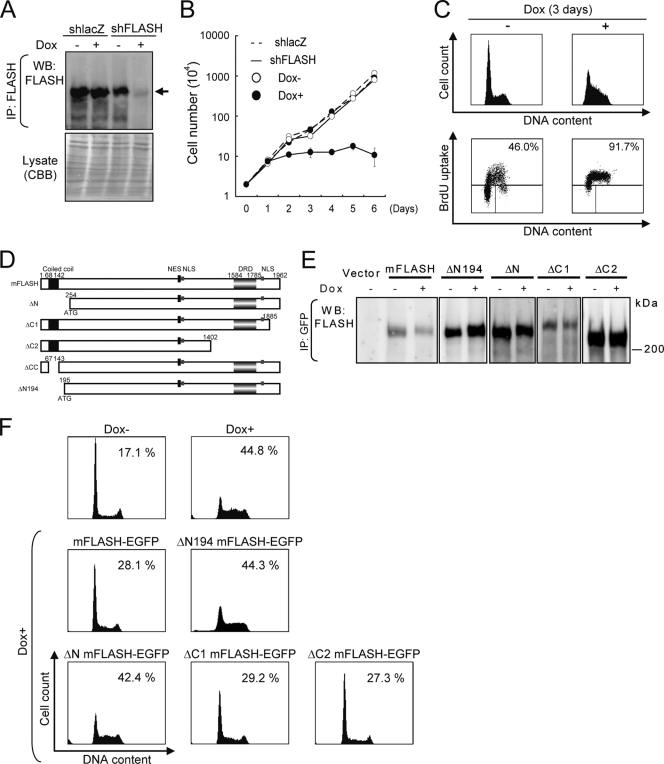
FLASH previously was shown to play a crucial role in S phase progression by analyses of cells transiently expressing shFLASH (1, 3, 4). To investigate the function of FLASH in S phase progression in detail, we utilized a previously established Tet-On shFLASH expression system using lentivirus-based expression vectors (17). In KB cells with Tet-On shFLASH, FLASH expression was downregulated on treatment with Dox for 3 days (Fig. 1A). Simultaneously, an analysis using phase-contrast microscopy indicated that FLASH knockdown KB cells were moderately detached from culture dishes and seemed to be larger than normal cells (see Fig. S1A in the supplemental material). An increase in cell volume in the downregulation of FLASH expression was confirmed by flow cytometry (see Fig. S1B in the supplemental material). In addition, cell proliferation was inhibited by the downregulation (Fig. 1B). A cell cycle analysis of shFLASH-expressing KB cells pulse-labeled with BrdU showed that the cells accumulated in S phase, although they continuously incorporated BrdU at low levels (Fig. 1C). Similar effects of the downregulation of FLASH expression were observed in U2OS cells (data not shown).
FIG. 1.
N-terminal region of FLASH is required for S phase progression. (A) KB cells with Tet-On shFLASH or shlacZ were cultured with or without 1 μg/ml Dox for 3 days. Cell lysate was immunoprecipitated (IP) with anti-FLASH MAb and analyzed by Western blotting (WB) using the same anti-FLASH MAb. To confirm the amounts of total protein, total cell lysates were resolved by SDS-PAGE, and the gels were stained with Coomassie brilliant blue (CBB). Arrows indicate full-length FLASH. Some degraded bands of FLASH, which disappeared after the expression of shFLASH, were observed. (B) KB cells with Tet-On shlacZ or shFLASH were treated with 1 μg/ml Dox for the period indicated. Cell numbers were quantified by the trypan blue exclusion method using a hemocytometer. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means. (C) Cell cycle analysis using cells pulse-labeled with BrdU. After treatment with 1 μg/ml Dox for 3 days, KB cells with Tet-On shFLASH were pulse-labeled with 10 μM BrdU for 30 min, stained with FITC-conjugated anti-BrdU and PI, and analyzed by flow cytometry. Percentages of BrdU-positive cells in S phase are indicated. Representative data from three independent experiments are shown. (D) A graphical overview of full-length mFLASH and various deletion mutants. The numbers of amino acid residues in mFLASH are indicated. NLS, nuclear localization signal; NES, nuclear export signal; DRD, DED-recruiting domain. (E) KB cells with Tet-On shFLASH expressing various deletion mutants of EGFP-fused mFLASH were treated with Dox for 3 days. Cell lysates were immunoprecipitated with anti-GFP Ab and analyzed by Western blotting with anti-FLASH MAb. (F) KB cells depicted in panels A and E were treated with Dox for 3 days and analyzed by flow cytometry after being stained with PI. The percentages of cells in S phase are indicated.
We next analyzed the region of FLASH required for S phase progression. The expression of FLASH was restored in KB cells with Tet-On shFLASH by the infection of lentivirus-based expression vectors encoding various deletion mutants of mFLASH fused with EGFP at their C-terminal ends (Fig. 1D and E). In this lentivirus-based expression system, almost all cells were shown to express EGFP (data not shown), indicating that the transfection efficiency is nearly 100%. The defect of S phase progression in shFLASH-expressing KB cells was recovered by the expression of EGFP-fused full-length mFLASH (mFLASH-EGFP) (Fig. 1F). The expression of the deletion mutant missing the N-terminal aa 1 to 253 (ΔN) or N-terminal aa 1 to 194 (ΔN194) could not restore the function of FLASH in S phase progression. In contrast, the deletion of the C-terminal aa 1886 to 1962 (ΔC1) and 1403 to 1962 (ΔC2) did restore this function of FLASH (Fig. 1F). Thus, the N-terminal region containing a coiled-coil domain (aa 68 to 142) was required for FLASH to function in S phase progression.
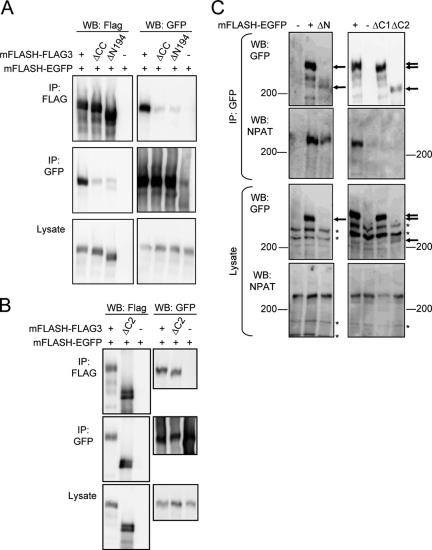
To examine further the roles of the N-terminal region of FLASH including the coiled-coil domain, we carried out immunoprecipitation experiments using various deletion mutants of mFLASH together with full-length mFLASH. 293T cells were transfected with mFLASH-EGFP together with full-length mFLASH, mFLASH without its N-terminal coiled-coil domain (aa 68 to 142), or mFLASH without its N-terminal 194 aa, all of which were tagged with three copies of FLAG (FLAG3) at the C terminus (mFLASH-FLAG3, ΔCC mFLASH-FLAG3, and ΔN194 mFLASH-FLAG3, respectively) (Fig. 1D). mFLASH-EGFP was coimmunoprecipitated with mFLASH-FLAG3 but not with ΔCC mFLASH-FLAG3 or ΔN194 mFLASH-FLAG3 (Fig. 2A). In contrast, mFLASH-EGFP was coimmunoprecipitated with ΔC2 mFLASH-FLAG3 (Fig. 2B). These results suggest that FLASH protein self associates through its N-terminal region including the coiled-coil domain, and that the self-association activity of FLASH is important for S phase progression.
FIG. 2.
Characterization of C-terminal and N-terminal deletion mutants of FLASH. (A and B) 293T cells were transfected with an empty vector (−) or an expression vector encoding mFLASH-FLAG3, ΔCC mFLASH-FLAG3, or ΔN194 mFLASH-FLAG3 together with that encoding mFLASH-EGFP. After 48 h of cultivation, mFLASH immunoprecipitates (IP) obtained with anti-Flag agarose resin or anti-GFP Ab were analyzed by Western blotting (WB) with anti-Flag and anti-GFP Abs. (C) 293T cells were transfected with empty vector (−) or an expression vector encoding mFLASH-EGFP, ΔN mFLASH-EGFP, ΔC1 mFLASH-EGFP, or ΔC2 mFLASH-EGFP. After 48 h of cultivation, immunoprecipitates obtained with anti-GFP Ab were analyzed by Western blotting with anti-GFP or anti-NPAT Ab. Representative results from three independent experiments are shown. Arrows indicate various deletion mutants of FLASH, and asterisks indicate nonspecific bands.
Interaction of FLASH with NPAT is not required for S phase progression.
FLASH was reported to interact with NPAT, and this interaction has been speculated to be important in S phase progression (3). To identify the region of FLASH interacting with NPAT, 293T cells were transiently transfected with an expression vector encoding mFLASH-EGFP, ΔN mFLASH-EGFP, ΔC1 mFLASH-EGFP, or ΔC2 mFLASH-EGFP, and a coimmunoprecipitation analysis was performed with endogenous NPAT and the deletion mutants of mFLASH. While the expression level of ΔN mFLASH was too low to be detected by anti-GFP Ab, it could be observed in the immunoprecipitate with anti-GFP Ab. mFLASH and ΔN mFLASH were shown to be coimmunoprecipitated with NPAT; however, ΔC1 and ΔC2 mFLASH did not (Fig. 2C). Thus, FLASH was shown to interact with NPAT through its C-terminal region. Meanwhile, ΔC1 and ΔC2 mFLASH without activity to bind NPAT could support S phase progression similarly to full-length mFLASH. Taken together, the interaction of FLASH with NPAT is not correlated with the function of FLASH in supporting S phase progression.
Formation of Cajal bodies containing coilin is correlated with FLASH-dependent S phase progression.
Earlier reports indicated that FLASH is colocalized with NPAT in Cajal bodies, and the downregulation of FLASH expression results in the disruption of nuclear foci of NPAT and those of coilin, a major component of Cajal bodies (3, 4). We examined the effects of mFLASH-EGFP and deletion mutants similarly fused to EGFP on nuclear foci of NPAT or coilin in shFLASH-expressing KB cells. Signals for both NPAT and coilin foci disappeared in KB cells with Tet-On shFLASH after treatment with Dox for 3 days (Fig. 3A and B; also see Fig. S2A and B in the supplemental material). Full-length mFLASH-EGFP was observed to form nuclear foci in shFLASH-expressing KB cells, and FLASH-EGFP foci were completely colocalized with NPAT foci in the nucleus (Fig. 3A; also see Fig. S2A in the supplemental material). These observations are consistent with earlier reports (3, 4). On the other hand, about 20% of FLASH foci and 21% of coilin foci were colocalized with coilin and FLASH foci, respectively (Fig. 3B and data not shown; also see Fig. S2B in the supplemental material). Neither ΔN, ΔC1, nor ΔC2 mFLASH formed foci in the nucleus, and the formation of NPAT foci was not observed in FLASH knockdown KB cells expressing either ΔN, ΔC1, or ΔC2 mFLASH-EGFP (Fig. 3C and Table 1). Meanwhile, the formation of coilin foci was restored in FLASH knockdown KB cells expressing either mFLASH-EGFP, ΔC1 mFLASH-EGFP, or ΔC2 mFLASH-EGFP but not in cells expressing ΔN mFLASH-EGFP (Fig. 3D and Table 1). Collectively, FLASH and NPAT colocalized in the same foci, and the formation of foci by FLASH, which is dependent on both the N- and C-terminal regions of FLASH, correlated well with that of NPAT. However, the formation of neither FLASH nor NPAT foci correlated with the ability of FLASH to support S phase progression (Table 1). On the other hand, foci of coilin, which depend not on foci of FLASH but on the expression of mFLASH constructs with the ability to promote S phase progression (full-length, ΔC1, or ΔC2 mFLASH), seem to be required for S phase progression.
FIG. 3.
Interaction of FLASH with NPAT is not required for S phase progression. (A and B) KB cells with Tet-On shFLASH expressing or not expressing mFLASH-EGFP were cultured with (+) or without (−) 1 μg/ml Dox for 3 days, and NPAT (A) or coilin (B) was stained with anti-NPAT and anticoilin Ab, respectively. NPAT or coilin (red) and EGFP (green) were observed under a confocal fluorescence microscope. DNA was stained with DAPI (blue). Scale bars, 5 μm. (C and D) The graphs on the left show the percentages of cells (n > 100) with NPAT foci (C) and coilin foci (D), which were determined in KB cells as described for panels A and B, respectively. The graphs on the right show the percentages of cells with NPAT foci or coilin foci together with FLASH foci (EGFP foci), which were determined in KB cells as described in the legend to Fig. 1E. Representative results from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means.
TABLE 1.
Characterization of various mutants of FLASHa
| mFLASH type | Presence of Dox | Nuclear focus formation
|
Expression of histone mRNA | Cell cycle progression | ||
|---|---|---|---|---|---|---|
| FLASH | NPAT | Coilin | ||||
| Exogenous mFLASH | − | ++ | ++ | ++ | ++ | |
| Exogenous mFLASH | + | − | − | ± | − | |
| mFLASH | + | ++ | ++ | ++ | ++ | ++ |
| ΔCC mFLASH | + | − | − | − | ± | − |
| ΔC1 mFLASH | + | − | − | + | + or ++ | ++ |
| ΔC2 mFLASH | + | − | − | + | + or ++ | ++ |
| ΔFARB mFLASH | + | + | + | + | ± | ± |
Various biological functions of exogenously expressed FLASH and its mutants are summarized. KB cells with Tet-On shFLASH were cultured with (+) or without (−) 1 μg/ml Dox for 3 days, and then nuclear foci of FLASH, NPAT, and coilin, the expression level of histone mRNAs, and cell cycle progression were examined as described in the legends to Fig. 1 and 3A to D (also see Fig. S1C in the supplemental material). ++, normal; +, weak but significant; ±, nearly undetectable; −, undetectable.
Expression of ARS2 is required for S phase progression.
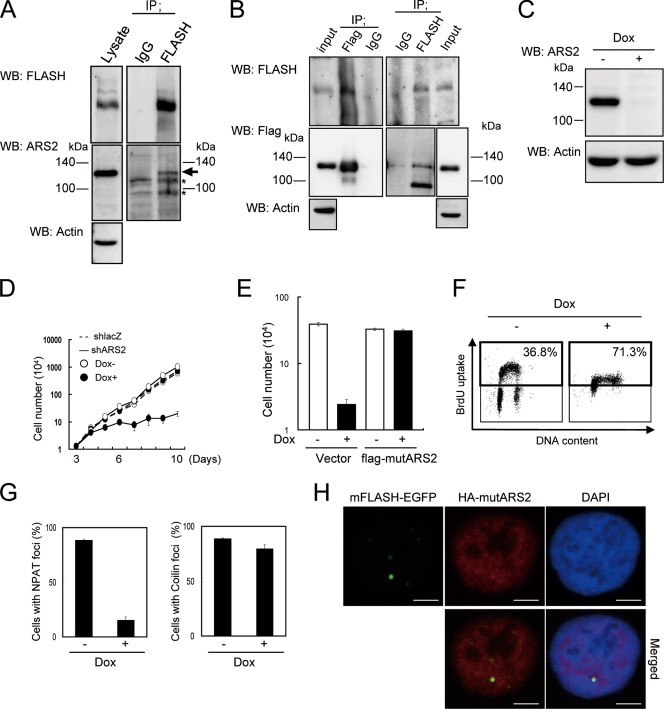
To elucidate how ΔC2 mFLASH functions in S phase progression, we purified proteins associated with ΔC2 mFLASH-FLAG3 in shFLASH-expressing KB cells. We identified ARS2 by the mass spectrometry of ΔC2 mFLASH immunoprecipitates. The physical association between endogenous FLASH and endogenous ARS2 in 293T cells was confirmed by immunoprecipitation and Western blotting (Fig. 4A). We also confirmed the coimmunoprecipitation of exogenously expressed ARS2 and endogenous FLASH in KB cells (Fig. 4B).
FIG. 4.
ARS2 required for S phase progression interacts with FLASH. (A) Immunoprecipitates (IP) obtained with anti-FLASH Ab or control immunoglobulin G (IgG) from 293T cell lysate were analyzed by Western blotting (WB) with anti-FLASH MAb or anti-ARS2 polyclonal Ab. Actin also was detected as a loading control. (B) KB cells with Tet-On shARS2, infected with a lentiviral vector encoding FLAG3-mutARS2 (ARS2 with silent mutations), were treated with 1 μg/ml Dox for 6 days. Immunoprecipitation was carried out with anti-FLASH, anti-FLAG agarose resin, or control IgG, and immunoprecipitates were analyzed by Western blotting with anti-FLASH or anti-Flag M2 MAb. (C) KB cells with Tet-On shARS2 were cultured with or without 1 μg/ml Dox for 6 days, and the knockdown of ARS2 was confirmed by Western blotting with anti-ARS2 polyclonal Ab. Actin also was detected as a loading control. Representative results from three independent experiments are shown. (D) KB cells with Tet-On shlacZ or shARS2 were treated with 1 μg/ml Dox for the period indicated. Cell numbers were quantified by the trypan blue exclusion method using a hemocytometer. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means. (E) KB cells with Tet-On shARS2 were infected with an expression vector encoding flag-mutARS2 or an empty vector (Vector), plated at 1.0 × 103 cells/well in 24-well plates, and cultured with 1 μg/ml Dox for 9 days. Cell numbers were quantified by the trypan blue exclusion method using a hemocytometer. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means. (F) Cell cycle analysis using cells pulse-labeled with BrdU. After treatment with 1 μg/ml Dox for 6 days, KB cells with Tet-On shARS2 were pulse labeled with 10 μM BrdU for 30 min, stained with FITC-conjugated anti-BrdU and PI, and analyzed by flow cytometry. The percentages of BrdU-positive cells in S phase are indicated. Representative data from three independent experiments are shown. (G) Cells shown in panel E were stained with anti-NPAT or anti-Coilin Ab, and NPAT foci (upper) and Coilin foci (lower) were enumerated (n > 100). Error bars denote ± standard errors of the means. (H) KB cells with Tet-On shARS2 expressing both mFLASH-EGFP and HA-mutARS2 were treated with 1 μg/ml Dox for 6 days, stained with anti-HA Ab (red), and observed under a confocal fluorescence microscope. DNA was stained with DAPI (blue). Scale bars, 5 μm.
To examine the physiological functions of ARS2, ARS2 was knocked down using the Tet-On shRNA expression system in KB cells. The expression of ARS2 was downregulated by treatment with Dox for 6 days (Fig. 4C). The expression of shARS2 induced cell growth arrest (Fig. 4D), but the growth was recovered by the expression of exogenous flag-tagged ARS2 with silent mutations in the target nucleotides of shARS2 (flag-mutARS2) (Fig. 4E; also see Fig. S4A in the supplemental material). Intriguingly, an analysis of shARS2-expressing cells pulse-labeled with BrdU indicated that the knockdown of ARS2 induced an S phase arrest similar to that observed in FLASH knockdown cells (Fig. 4F).
ARS2 is involved in both replication-dependent transcription of histone and formation of NPAT foci.
When cells enter S phase, the amounts of replication-dependent histone mRNAs increase, which are rapidly degraded at the end of S phase (26). Replication-dependent histone mRNAs are the only metazoan mRNAs that are not polyadenylated, while replacement (replication-independent) histone mRNAs are polyadenylated (8). To examine the levels of histone mRNA in shARS2-expressing KB cells, Northern blotting was performed, and mRNAs for whole variants of replication-dependent and -independent histones were detected as different bands at the same time. The mRNA levels of the replication-dependent linker histone (H1) and core histones (H2A, H2B, H3, and H4) were decreased in shARS2-expressing KB cells (see Fig. S3A in the supplemental material). A similar decrease in the expression of histone mRNAs was observed in shFLASH-expressing KB cells (see Fig. S1C in the supplemental material), as reported previously (3). Thus, the downregulation of ARS2 expression as well as FLASH expression was shown to be correlated with the suppression of the transcription of replication-dependent histone genes.
We analyzed nuclear foci of NPAT and coilin after the expression of shARS2 in KB cells, finding that signals for NPAT, but not coilin, were disrupted by the downregulation of ARS2 expression (Fig. 4G; see Fig. S3B in the supplemental material).
We next analyzed the subcellular distribution of ARS2 and FLASH. KB cells with Tet-On shARS2 were infected with lentiviral vectors encoding mFLASH-EGFP and HA-tagged mutARS2 (HA-mutARS2). The immunofluorescent staining of HA-mutARS2 by anti-HA Ab showed that ARS2 was located throughout the nucleus but formed nuclear foci that colocalized with FLASH foci (Fig. 4H).
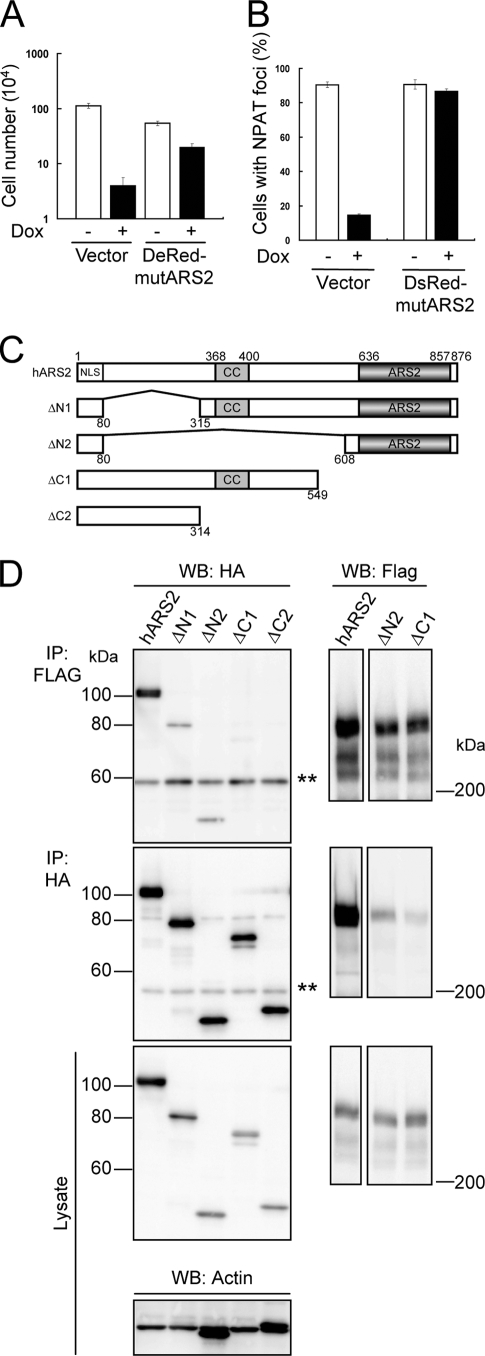
To confirm that ARS2 is required for the formation of NPAT foci, KB cells with Tet-On shARS2 were infected with lentiviral vectors encoding mutARS2 tagged by DsRed-Monomer at its N terminus (DsRed-mutARS2). Nuclear foci of NPAT were observed under the confocal immunofluorescence microscope after treatment with Dox for 6 days. The expression of DsRed-mutARS2, which could recover cell-proliferating activity in ARS2 knockdown cells (Fig. 5A), was shown to restore the ability to form foci of NPAT in shARS2-expressing KB cells (Fig. 5B; also see Fig. S4B in the supplemental material). Therefore, ARS2 is required for nuclear foci of NPAT, which are identical to those of FLASH.
FIG. 5.
ARS2 interacts with FLASH through its ARS2 domain. (A) KB cells with Tet-On shARS2 were infected with an expression vector encoding DsRed-Monomer-mutARS2 (DsRed-mutARS2) or an empty vector (Vector), plated at 1.25 × 103 cells/well in 24-well plates, and cultured with 1 μg/ml Dox for 9 days. Cell numbers were quantified by the trypan blue exclusion method using a hemocytometer. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means. (B) KB cells described in panel A were treated with (+) or without (−) 1 μg/ml Dox for 6 days, and NPAT was stained with anti-NPAT Ab. Cells were observed under a confocal fluorescence microscope, and the percentages of cells with NPAT foci (n > 100) were determined. Representative results from three independent experiments are shown. Error bars denote ± standard errors of the means. (C) A graphical overview of full-length hARS2 and various deletion mutants of hARS2. Numbers of amino acid residues in hARS2 are indicated. CC, coiled-coil domain; ARS2, ARS2 domain. (D) 293T cells were transfected with an expression vector encoding HA-hARS2, HA-ΔN1 ARS2, HA-ΔN2 ARS2, HA-ΔC1 ARS2, or HA-ΔC2 ARS2 together with that encoding mFLASH-FLAG3. After 48 h, ARS2 and mFLASH immunoprecipitates (IP) obtained with anti-HA Ab and anti-Flag agarose resin, respectively, were analyzed by Western blotting (WB) with anti-HA and anti-Flag Abs. Actin also was detected as a loading control. Representative results from three independent experiments are shown. Double asterisks indicate the heavy chain of Ab.
ARS2 interacts with FLASH through its ARS2 domain.
To identify the region of ARS2 binding to FLASH, we constructed various deletion mutants of hARS2, as shown in Fig. 5C. These ARS2 mutants were tagged with HA at their N-terminal ends and expressed in 293T cells together with mFLASH-FLAG3. The mutants containing the ARS2 domain (ΔN1 and ΔN2) as well as full-length ARS2 were coimmunoprecipitated with FLASH, while the mutants without the ARS2 domain (ΔC1 and ΔC2) failed to interact with FLASH (Fig. 5D). These results indicate that the ARS2 domain is required for ARS2 to interact with FLASH.
FLASH interacts with ARS2 through its central region composed of 13 amino acids.
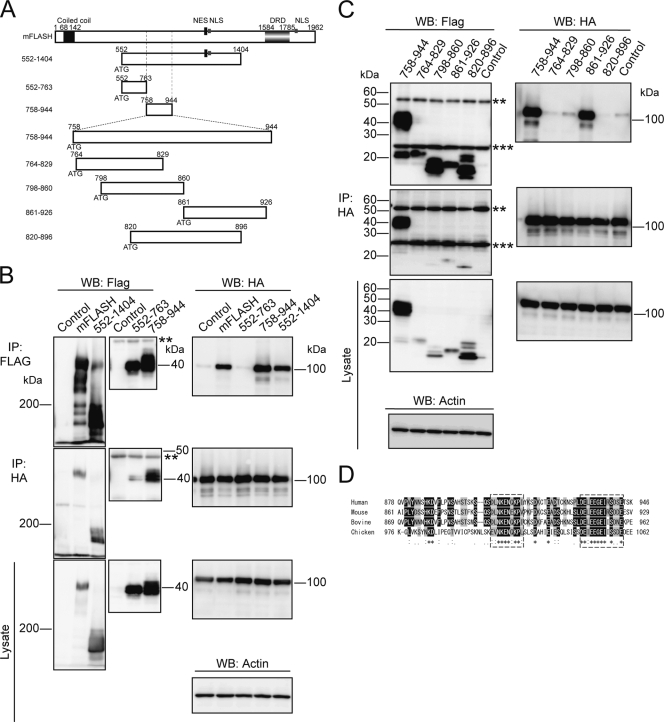
To identify the region of FLASH binding to ARS2, coimmunoprecipitation experiments were carried out using 293T cells transiently expressing HA-tagged ARS2 together with various deletion mutants of mFLASH (Fig. 6A; also see Fig. S5A in the supplemental material). These mFLASH deletion mutants were tagged with three copies of FLAG at the C terminus. Full-length mFLASH and ΔC2 mFLASH were confirmed to coimmunoprecipitate with HA-ARS2 (see Fig. S5B in the supplemental material). Coiled-coil domain-deleted mFLASH (ΔCC) and two N-terminal deletion mutants, ΔN194 and ΔN551 (comprising aa 195 to 1962 and 552 to 1962 of mFLASH, respectively), also interacted with HA-ARS2 (see Fig. S5B and C in the supplemental material). However, ΔN943 mFLASH, composed of aa 944 to 1962, could not interact (see Fig. S5D in the supplemental material), suggesting that aa 552 to 943 of mFLASH are important for the interaction with ARS2. To further examine this region, we constructed two deletion mutants of mFLASH consisting of aa 552 to 763 and aa 758 to 944 (Fig. 6A). ARS2 was shown to coimmunoprecipitate with aa 758 to 944 of mFLASH but not with aa 552 to 763 of mFLASH (Fig. 6B). All results indicated that the central region of mFLASH composed of aa 758 to 944 is required for interaction with ARS2.
FIG. 6.
FLASH interacts with ARS2 thorough its central region. (A) A graphical overview of mFLASH and its various deletion mutants. Numbers of amino acid residues in mFLASH are indicated. (B and C) 293T cells were transiently transfected with an empty vector (Control) or an expression vector encoding mFLASH-FLAG3 or the indicated deletion mutant of mFLASH-FLAG3 together with that encoding HA-ARS2. After 48 h, various mutants of mFLASH-FLAG3 or HA-ARS2 were immunoprecipitated (IP) with anti-FLAG resin and anti-HA Ab, respectively. Immunoprecipitates were analyzed by Western blotting (WB). Actin also was detected as a loading control. Representative results from three independent experiments are shown. **, heavy chain of immunoglobulin G (IgG); ***, light chain of IgG. (D) Alignment of human, mouse, bovine, and chicken FLASH in their center region. Identical and similar amino acids are indicated in black and shaded boxes, respectively. Highly conserved regions are surrounded by dotted lines.
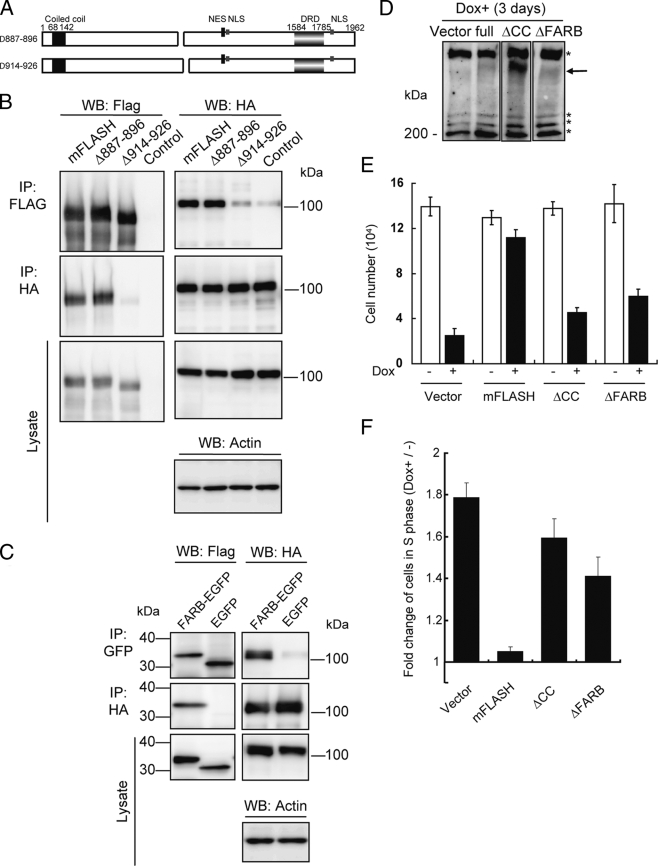
We next constructed mFLASH deletion mutants composed of aa 764 to 829, 798 to 860, 861 to 926, and 820 to 896 of mFLASH, as shown in Fig. 6A. Only aa 861 to 926 of mFLASH could interact with ARS2 (Fig. 6C). This region of mFLASH (aa 861 to 926) was aligned against the corresponding region of human, bovine, and chicken FLASH (Fig. 6D), revealing the two regions of mFLASH composed of aa 887 to 896 and 914 to 926 to be highly conserved. We then constructed two types of FLASH deletion mutants without aa 887 to 896 and 914 to 926 of mFLASH, as shown in Fig. 7A. Δ887-896 mFLASH bound to ARS2, while Δ914-926 mFLASH did not (Fig. 7B). We designated the region composed of aa 914 to 926 of mFLASH FLASH-ARS2 binding (FARB) region. The mouse and human FARB sequences are DELEEGEIRSDDE and DELEEGEIRSDSE, respectively.
FIG. 7.
Interaction of FLASH with ARS2 through its FARB region is required for S phase progression. (A) A graphical overview of two types of mFLASH deletion mutants. (B) 293T cells were transiently transfected with an empty vector (Control) or an expression vector encoding mFLASH-FLAG3, Δ887-896 mFLASH-FLAG3, or Δ914-926 mFLASH-FLAG3 together with that encoding HA-ARS2. After 48 h, mFLASH or ARS2 was immunoprecipitated (IP) with anti-FLAG resin and anti-HA Ab, respectively. Immunoprecipitates were analyzed by Western blotting (WB). Actin also was detected as a loading control. Representative results from three independent experiments are shown. (C) 293T cells were transiently transfected with an expression vector encoding an EGFP-fused FARB region of mFLASH or EGFP together with that encoding HA-ARS2. After 48 h, ARS2 and EGFP or FARB-EGFP were immunoprecipitated with anti-HA Ab and anti-GFP Ab, respectively. Immunoprecipitates were analyzed by Western blotting. Actin also was detected as a loading control. Representative results from three independent experiments are shown. (D) KB cells with Tet-On shFLASH were infected with an empty vector (Vector) or a lentiviral expression vector encoding mFLASH-EGFP, ΔCC mFLASH-EGFP, or ΔFARB mFLASH-EGFP. Cells were cultured with 1 μg/ml Dox for 3 days and lysed for Western blotting using anti-FLASH MAb. Arrows, EGFP-fused FLASH; *, nonspecific bands. (E) KB cells indicated in panel D were plated at 2.5 × 103 cells/well in 24-well plates and cultured with 1 μg/ml Dox for 3 days. Cell numbers were quantified by the trypan blue exclusion method using a hemocytometer. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means. (F) Cells described in panel E were pulse-labeled with BrdU for 30 min, and the percentage of cells in S phase (BrdU-positive cells) was determined by flow cytometry. The change (n-fold) in the percentage of cells expressing shFLASH (Dox+) in S phase against that of cells not expressing shFLASH (Dox−) in S phase was calculated. Representative data from three independent experiments, all performed in triplicate, are shown. Error bars denote ± standard errors of the means.
We then fused EGFP with FARB at its N-terminal end. FARB-EGFP was able to interact with ARS2 (Fig. 7C). All results indicate that the FARB region of FLASH is required and sufficient for interaction with ARS2.
Interaction of FLASH and ARS2 is necessary for S phase progression.
To confirm that the interaction between FLASH and ARS2 is essential for S phase progression, we analyzed the proliferation of shFLASH-transfected KB cells expressing mFLASH, ΔCC mFLASH, and ΔFARB mFLASH (Δ914-926 mFLASH) (Fig. 7D). ΔFARB mFLASH showed a reduced ability to promote the proliferation of cells, as did ΔCC mFLASH, compared to the ability of full-length mFLASH (Fig. 7E). To examine the number of cells in S phase, a cell cycle analysis was performed with BrdU-incorporated cells. The ratio of cells in S phase was increased in shFLASH-transfected KB cells expressing ΔFARB mFLASH as well as ΔCC mFLASH compared to that of cells expressing full-length mFLASH (Fig. 7F). To confirm that ΔFARB mFLASH-EGFP still was capable of interacting with NPAT, 293T cells were transiently transfected with an expression vector encoding mFLASH-EGFP or ΔFARB mFLASH-EGFP, and coimmunoprecipitation analysis indicated that both mFLASH and ΔFARB mFLASH were coimmunoprecipitated with NPAT (see Fig. S5E in the supplemental material). These results are consistent with preceding data (Fig. 2C). All of the results indicate that the interaction between FLASH and ARS2, but not between FLASH and NAPT, is involved in FLASH-dependent S phase progression.
We examined the ability of ΔFARB mFLASH to form nuclear foci of FLASH, NPAT, or coilin in shFLASH-expressing KB cells. Confocal immunofluorescence microscopy revealed that ΔFARB FLASH retains a weak but significant ability to form nuclear foci composed of FLASH, NPAT, and coilin (Table 1).
To examine the levels of histone mRNAs in shFLASH-transfected KB cells expressing ΔFARB mFLASH, Northern blotting was performed, indicating that the mRNA levels of all linker and core histones were markedly decreased in the ΔFARB mFLASH-expressing cells (see Table S1 in the supplemental material). These results show that the interaction between FLASH and ARS2 is important to maintain the histone mRNA levels as well as to promote S phase progression.
DISCUSSION
Here, we showed that not only the expression of FLASH but also the expression of ARS2 and the interaction between FLASH and ARS2 play important roles in S phase progression. Previously, the inhibition of the transcription of replication-dependent histone genes was suggested to be responsible for the accumulation of FLASH knockdown cells in S phase, and the interaction of FLASH with NPAT, an activator of histone transcription, also was suggested to play a role in S phase progression (3). However, we showed here that the interaction of FLASH with ARS2, not NPAT, is necessary for S phase progression. Moreover, although NPAT was reported to be required for S phase entry (7, 19, 27, 29, 30), FLASH, ARS2 and their interaction are necessary for S phase progression but not S phase entry (Fig. 1C, 4F, and 7F). Taking these results together, ARS2 may play an important role in FLASH-dependent S phase progression. However, it is still unclear whether the inhibition of the transcription of replication-dependent histone is responsible for or a consequence of the accumulation of FLASH knockdown and ARS2 knockdown cells in S phase.
ARS2 was identified as one of the gene products conferring resistance to arsenite on arsenite-sensitive CHO cells by expression cloning using a cDNA library from arsenite-resistant cells (24). hARS2 has been reported to interact with RNA binding protein S1 (RNPS1) in a high-throughput study combining immunoprecipitation and mass spectrometry (9). ARS2 also has been shown to be essential for early mammalian development (28). However, the precise biological functions of ARS2 protein have remained unclear. We suppose that the functions of ARS2 other than the activation of histone transcription are involved in FLASH-dependent S phase progression. While ARS2 was shown to be distributed throughout the nucleus, it partly colocalized with FLASH in Cajal bodies (Fig. 4H). FLASH may act as an anchor for ARS2 in Cajal bodies, allowing it to function properly. To explain the molecular mechanism of the FLASH- and ARS2-dependent S phase progression, the precise biological functions of ARS2 need to be clarified.
As reported recently (5), Cajal bodies can be classified into at least two types, those containing coilin but not FLASH or NPAT and those with FLASH and NPAT but not coilin (Fig. 3A and B). Interestingly, the conventional type containing coilin seems not to merge but to associate with those containing FLASH and NPAT (Fig. 3B; also see Fig. S2B in the supplemental material), while FLASH was indicated to be involved in the formation of not only Cajal bodies with FLASH and NPAT but also those with coilin (Table 1). Taking these results together, the association of the complex of FLASH and ARS2 with Cajal bodies containing coilin might be involved in S phase progression. Thus, the biological functions of Cajal bodies containing coilin and those containing FLASH and NPAT need to be clarified in detail.
The N-terminal region of FLASH including the coiled-coil domain was shown to be essential to the biological functions of FLASH, including S phase progression and the formation of Cajal bodies with coilin and with both FLASH and NPAT (Table 1; also see Table S1 in the supplemental material). The N-terminal region of FLASH including the coiled-coil domain also is suggested to be involved in the self association of FLASH (Fig. 2A), suggesting that the self association of FLASH plays an important role in its various biological functions. To understand the molecular mechanism behind both the formation of Cajal bodies and the promotion of S phase progression, it may be necessary to determine whether FLASH forms a dimer or oligomer through the coiled-coil domain.
FLASH was indicated to bind ARS2 through its FARB sequence composed of only 13 amino acids. ΔFARB FLASH lost the ability to bind ARS2, but FARB-fused EGFP could bind to ARS2. In addition, ΔFARB-FLASH lost the functions of FLASH in S phase progression and the transcription of histones. The regulation of the interaction between FLASH and ARS2, possibly achieved by the expression of FARB peptide within the cells or the treatment of cells with a low-molecular-weight component binding to the FARB region, would be useful in the treatment of malignant diseases by controlling the proliferation of malignant cells.
In conclusion, we revealed a novel mechanism supporting the progression of S phase involving FLASH, ARS2, and interaction between the two. Recently, nuclear FLASH foci were reported to be colocalized with c-Myb in active RNA polymerase II foci in the nucleus (1), and hARS2 has been shown to interact with RNPS1 (9), which is concerned with the regulation of pre-mRNA splicing in vivo (25). Thus, the system to support S phase progression by FLASH and ARS2 also may be involved in regulating the function of several transcription factors, and these two different activities may be tightly intercorrelated. The determination of the precise biological functions of both FLASH and ARS2 at the molecular level is necessary to completely understand this novel system regulating S phase progression.
Supplementary Material
Acknowledgments
We thank E. Nishida and all the members of our laboratory for discussions and help.
This study was supported by the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and grants in aid from MEXT. M.K. is a research fellow of the Japan Society for the Promotion of Science.
There is no conflict of financial interest.
Footnotes
Published ahead of print on 22 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alm-Kristiansen, A. H., T. Saether, V. Matre, S. Gilfillan, O. Dahle, and O. S. Gabrielsen. 2008. FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene 274644-4656. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam, A., R. M. Nissen, Z. Sun, E. C. Swindell, S. Farrington, and N. Hopkins. 2004. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 10112792-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcaroli, D., L. Bongiorno-Borbone, A. Terrinoni, T. G. Hofmann, M. Rossi, R. A. Knight, A. G. Matera, G. Melino, and V. De Laurenzi. 2006. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA 10314808-14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcaroli, D., D. Dinsdale, M. H. Neale, L. Bongiorno-Borbone, M. Ranalli, E. Munarriz, A. E. Sayan, J. M. McWilliam, T. M. Smith, E. Fava, R. A. Knight, G. Melino, and V. De Laurenzi. 2006. FLASH is an essential component of Cajal bodies. Proc. Natl. Acad. Sci. USA 10314802-14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bongiorno-Borbone, L., A. De Cola, P. Vernole, L. Finos, D. Barcaroli, R. A. Knight, G. Melino, and V. De Laurenzi. 2008. FLASH and NPAT positive but not Coilin positive Cajal bodies correlate with cell ploidy. Cell Cycle 72357-2367. [DOI] [PubMed] [Google Scholar]
- 6.Cioce, M., and A. I. Lamond. 2005. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21105-131. [DOI] [PubMed] [Google Scholar]
- 7.DeRan, M., M. Pulvino, E. Greene, C. Su, and J. Zhao. 2008. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 28435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 2391-14. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, R. M., P. Chu, F. Elisma, H. Li, P. Taylor, S. Climie, L. McBroom-Cerajewski, M. D. Robinson, L. O'Connor, M. Li, R. Taylor, M. Dharsee, Y. Ho, A. Heilbut, L. Moore, S. Zhang, O. Ornatsky, Y. V. Bukhman, M. Ethier, Y. Sheng, J. Vasilescu, M. Abu-Farha, J. P. Lambert, H. S. Duewel, I. I. Stewart, B. Kuehl, K. Hogue, K. Colwill, K. Gladwish, B. Muskat, R. Kinach, S. L. Adams, M. F. Moran, G. B. Morin, T. Topaloglou, and D. Figeys. 2007. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flotho, C., E. Coustan-Smith, D. Pei, S. Iwamoto, G. Song, C. Cheng, C. H. Pui, J. R. Downing, and D. Campana. 2006. Genes contributing to minimal residual disease in childhood acute lymphoblastic leukemia: prognostic significance of CASP8AP2. Blood 1081050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall, J. G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16273-300. [DOI] [PubMed] [Google Scholar]
- 12.Golling, G., A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S. Y. Lin, R. M. Nissen, and N. Hopkins. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31135-140. [DOI] [PubMed] [Google Scholar]
- 13.Imai, Y., T. Kimura, A. Murakami, N. Yajima, K. Sakamaki, and S. Yonehara. 1999. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature 398777-785. [DOI] [PubMed] [Google Scholar]
- 14.Kino, T., and G. P. Chrousos. 2003. Tumor necrosis factor alpha receptor- and Fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J. Biol. Chem. 2783023-3029. [DOI] [PubMed] [Google Scholar]
- 15.Kino, T., T. Ichijo, and G. P. Chrousos. 2004. FLASH interacts with p160 coactivator subtypes and differentially suppresses transcriptional activity of steroid hormone receptors. J. Steroid Biochem. Mol. Biol. 92357-363. [DOI] [PubMed] [Google Scholar]
- 16.Kittler, R., G. Putz, L. Pelletier, I. Poser, A. K. Heninger, D. Drechsel, S. Fischer, I. Konstantinova, B. Habermann, H. Grabner, M. L. Yaspo, H. Himmelbauer, B. Korn, K. Neugebauer, M. T. Pisabarro, and F. Buchholz. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 4321036-1040. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, Y., and S. Yonehara. 2009. Novel cell death by downregulation of eEF1A1 expression in tetraploids. Cell Death Differ. 16139-150. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K. K., and S. Yonehara. 2002. Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST). J. Biol. Chem. 27712351-12358. [DOI] [PubMed] [Google Scholar]
- 19.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 142298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milovic-Holm, K., E. Krieghoff, K. Jensen, H. Will, and T. G. Hofmann. 2007. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 26391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obradoviæ, D., M. Tirard, Z. Nemethy, O. Hirsch, H. Gronemeyer, and O. F. Almeida. 2004. DAXX, FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells—toward a basis for the opposite actions elicited by two nuclear receptors? Mol. Pharmacol. 65761-769. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, K., J. Fujisawa, M. Reth, and S. Yonehara. 2006. Human T-cell leukemia virus type-I oncoprotein Tax inhibits Fas-mediated apoptosis by inducing cellular FLIP through activation of NF-κB. Genes Cells 11177-191. [DOI] [PubMed] [Google Scholar]
- 23.Prigge, M. J., and D. R. Wagner. 2001. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 131263-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossman, T. G., and Z. Wang. 1999. Expression cloning for arsenite-resistance resulted in isolation of tumor-suppressor fau cDNA: possible involvement of the ubiquitin system in arsenic carcinogenesis. Carcinogenesis 20311-316. [DOI] [PubMed] [Google Scholar]
- 25.Sakashita, E., S. Tatsumi, D. Werner, H. Endo, and A. Mayeda. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 241174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sittman, D. B., R. A. Graves, and W. F. Marzluff. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 801849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei, Y., J. Jin, and J. W. Harper. 2003. The cyclin E/Cdk2 substrate and Cajal body component p220NPAT activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 233669-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson, M. D., D. Wang, R. Wagner, H. Breyssens, M. Gertsenstein, C. Lobe, X. Lu, A. Nagy, R. D. Burke, B. F. Koop, and P. L. Howard. 2008. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol. Cell. Biol. 281503-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye, X., Y. Wei, G. Nalepa, and J. W. Harper. 2003. The cyclin E/Cdk2 substrate p220NPAT is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 238586-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 142283-2297. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.