Abstract
Complete inhibition of E protein transcription factors by Id1 blocks the developmental transition of CD4/CD8 double-negative 1 (DN1; CD44+ CD25−) thymocytes to the DN2 (CD44+ CD25+) stage. To understand the underlying mechanisms, we observed that mRNA levels of Deltex1, as well as Deltex4, were dramatically elevated in Id1-expressing thymocytes, which could result in developmental arrest by attenuating Notch function. In support of this hypothesis, we found that Deltex1 ablation enabled Id1-expressing progenitors to differentiate to the DN3 (CD44− CD25+) stage, which was accompanied by enhanced Notch1 expression in T-cell progenitors. Consistently, constitutive activation of Notch1 drove the differentiation of Id1-expressing progenitors to the DN3 stage. Furthermore, we showed that Gfi1b levels decreased, whereas GATA3 levels increased in Id1 transgenic thymocytes. When overexpressed, GATA3 was able to upregulate Deltex1 transcription. Thus, T-cell commitment may be controlled by the interplay among E proteins, Gfi1b, and GATA3 transcription regulators, which influence Notch function through the expression of Deltex1.
T-lineage cells differentiate from multipotent progenitors that migrate from the bone marrow to the thymus (40, 41, 52). These progenitors exhibit a phenotype of CD4− CD8− c-kit+ CD44+ CD25− and are generally classified into the CD4 and CD8 double-negative 1 subset (DN1) (3, 14). Upon T-lineage commitment, DN1 cells progress to the DN2 stage by expressing CD25 on the cell surface, and this process is thought to be independent of pre-T-cell receptor (pre-TCR) function. Rearrangement of the TCR β-chain gene takes place at the DN2 (CD44+ CD25+) and DN3 (CD44− CD25+) stages and allows the formation of pre-TCRs, consisting of TCRβ and pre-TCRα (13, 33). The survival and expansion of DN3 and DN4 (CD44− CD25−) T cells and their subsequent differentiation to the CD4 and CD8 double-positive stage (DP) are driven by pre-TCR signaling, whereas the maturation of DP T cells to CD4 or CD8 single-positive (SP) stages is controlled by receptors made of TCRα and TCRβ (17, 26).
Although TCR signaling plays crucial roles in T-cell development, signaling from Notch receptors is essential at the initial stages of T-cell commitment (2, 30). The Notch family consists of four members, Notch1 to -4. Upon binding to their ligands, Delta-like (DL) and Jagged, Notch receptors are activated by proteolytic cleavages that result in the release of their intracellular domain, which translocates into the nucleus and acts as a transcription coactivator by binding to its DNA-binding partner, RBP-J (5, 47). Among the members of the Notch family, Notch1 appears to play a nonredundant function at the early stages of T-cell development. Upregulation of Notch1 occurs in a subset of DN1 cells, classified as ETP or DN1a/-b, which are thought to be robust T-cell progenitors (37, 42). Notch1 levels peak at the DN3 stage and decrease in the DP or SP stages (41, 42, 49). Consistent with this pattern of expression, the loss of Notch1 results in the arrest of T-cell development at the DN1 stage (39). Likewise, disruption of the gene encoding its DNA-binding partner, RBP-J, causes a similar block in differentiation (15).
The molecular mechanism by which Notch signaling governs T-cell commitment and differentiation remains largely elusive and probably involves the coordinated expression of a growing number of genes that have been identified as targets of Notch (31). Among these genes, Hes1 and Deltex1 are widely used as indicators of Notch activity (11, 22). While ablation of Hes1 has effects similar to those of Notch1 deficiency (48), mutation of the Deltex genes does not cause any abnormalities in T-cell development (28, 44). However, overexpression of Deltex1 has been shown to antagonize the function of Notch1 with regard to T-cell development (21, 56). Deltex1 expression blocks T-cell differentiation in fetal organ cultures and in bone marrow transplant recipients, while allowing B cells to develop. These consequences are analogous to the situation when Notch1 is inducibly deleted (39, 51). The inhibitory effect of Deltex1 is thought to be mediated by its ability to interfere with the transcriptional activity of Notch (21). Therefore, it appears that a negative feedback loop is in place to modulate Notch function, i.e., upregulation of Deltex1 by Notch signaling could eventually lead to inhibition of Notch function. This would also prevent a positive feedback control of the transcription of the Notch1 gene, which itself is stimulated by Notch signaling (12, 50). In addition, Deltex1 expression might be influenced by other regulatory mechanisms, and this could have a direct impact on Notch function (19).
The early stages of T-cell development are also regulated by a variety of transcription factors. For example, the function of GATA3 is essential for the earliest stages of T-cell development, because GATA3 deficiency prevents the formation of any T-lineage cells (16, 46). However, overexpression of GATA3 also blocks the initiation of T-cell differentiation (4), suggesting that an optimal level of GATA3 activity is crucial for T-cell commitment. A cross talk between GATA3 and the Notch signaling pathway has been exemplified by the effect of GATA3 on the expression of several Notch target genes, such as Hes1 and Notch1 (41).
Another family of transcription factors involved in the early stages of T-cell development consists of the basic helix-loop-helix E proteins, including the products of the E2A and HEB genes (27, 34, 45). Germ line mutation of the E2A or HEB gene leads to partial developmental blocks beginning at the DN1-to-DN2 transition, as well as additional abnormalities during T-cell maturation (6, 9, 54). Moreover, when the function of both E2A and HEB proteins is abolished by Id1, which inhibits the DNA-binding activity of E proteins, the differentiation of DN1 cells is almost completely arrested, suggesting that E2A and HEB are indispensable for T-cell commitment (24, 25, 55). Although E2A has been suggested to regulate the expression of Notch1, it is not clear if E2A directly activates Notch1 transcription at the DN1 stage (20). Interestingly, E2A is also shown to be responsible for the expression of a transcriptional repressor, Gfi1b, which prevents GATA3 expression (53). Therefore, it is possible that aberrant upregulation of GATA3 in the absence of E protein function may contribute to the developmental arrest at the DN1-to-DN2 transition.
Here, we provide evidence to suggest that loss of E protein function as a result of Id1 expression leads to precocious expression of Deltex1, which could interfere with the function of Notch1 in promoting T-cell commitment and differentiation. In support of this notion, null mutation of the Deltex1 gene in Id1 transgenic mice facilitated the differentiation of T-cell progenitors in vivo and in culture, which was accompanied by an upregulation of Notch1 expression in these cells. Consistently, the expression of a constitutively active form of Notch1 enables Id1 transgenic cells to differentiate to the DN3 stage. In exploring the mechanism of aberrant Deltex1 expression, we found that Gfi1b levels were reduced, whereas GATA3 levels were elevated, in T-cell progenitors of Id1 transgenic mice and that GATA3 overexpression was capable of increasing the levels of Deltex1 mRNA in multipotent progenitors. Taking these results together, we propose a regulatory network that controls T-cell commitment and differentiation. This involves E protein-mediated modulation of Gfi1b and GATA3 levels, which in turn prevents precocious expression of Deltex1, thus ensuring optimal Notch function for T-cell commitment.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were originally obtained from Jackson Laboratories (Bar Harbor, ME). Id1 transgenic mice in the FVB/N background, called Id1-28, in which expression of the Id1 cDNA is driven by a T-cell-specific proximal promoter of the lck gene, were previously described (24). These mice were also backcrossed onto the C57BL/6 background for at least 10 generations. Deltex1−/− mice were kind gifts from M. J. Bevan (Department of Immunology and Howard Hughes Medical Institute, University of Washington, Seattle, WA) (28).
Transplant assays.
Transplantation experiments were conducted as previously described (36). Briefly, B6-CD45.1 Thy1.1 recipient mice were preconditioned with 6.5 Gy in a Mark I gamma irradiator using a 137Cs source (J. L. Shepard & Associates, Glendale, CA). One and 10 million whole bone marrow cells from wild-type B6-CD45.2 Thy1.1 and homozygous Id1 transgenic (Id1tg/tg) B6-CD45.2 Thy1.2 mice, respectively, were mixed and injected intravenously into irradiated recipients. Three weeks after transplantation, thymocytes from the recipients were evaluated by gating on donor marker CD45.2, as well as different Thy1 markers specific for wild-type and Id1 transgenic donors.
Cell enrichment, analytical flow cytometry, and cell sorting.
For all experiments, cell collection and staining were conducted using Hanks balanced salt solution supplemented with 5% fetal calf serum. After staining with appropriate antibodies, dead cells were excluded based on propidium iodide incorporation (Molecular Probes, Eugene, OR). Analysis of cell surface phenotypes was performed on an LSRII cytometer (BD Immunocytometry, San Jose, CA). Cell sorting was performed using a MoFlo (Beckman Coulter, Fullerton, CA) or FACSAria flow cytometer (BD Immunocytometry), and the purity was generally over 95% based on postsort analyses. The results were analyzed using the FlowJo program (Tree Star, Inc., San Carlos, CA). Lineage-negative thymocytes were enriched through immunomagnetic depletion as previously described (36). Briefly, thymocytes were first labeled with rat immunoglobulin against CD4, CD8, and B220, which was followed by incubation with BioMag anti-rat immunoglobulin-conjugated magnetic particles (Qiagen, Valencia, CA).
Antibodies purchased from BD Pharmingen (San Diego, CA) included anti-CD3ɛ (145-2C11), anti-CD4 (GK1.5), anti-CD8 (53-7.3), anti-TCRγδ (GL3), anti-B220 (RA3-6B2), anti-CD11b (Mac-1; M1/70), anti-Ly6G (1A8), anti-CD11c (HL3), anti-NK1.1 (PK136), anti-CD19 (1D3), anti-CD45.2 (104), anti-Thy1.1 (OX-7), anti-Thy1.2 (53-2.1), and anti-CD49b (DX5). These antibodies were conjugated with phycoerythrin (PE), fluorescein isothiocyanate, allophycocyanin (APC), APC-Cy7, or biotin. Biotin-conjugated antibodies were revealed with either PE-Texas red-streptavidin or APC-Cy7-streptavidin. Additionally, PE-Cy5.5-conjugated anti-CD44 (1M7), PE-Cy5-conjugated anti-CD24 (M1/69), and APC-Cy7-conjugated anti-CD25 (PC61), used to define the subpopulations within DN thymocytes, were from eBioscience, San Diego. CA.
Coculture of progenitor and stromal cells.
OP9 stromal cells transduced with vector or DL1-expressing retroviruses were maintained as previously published (43). Sorted DN1 or DN1′ cells (see Results) were seeded into 24-well tissue culture plates containing a near-confluent monolayer of stromal cells. Alpha medium (Invitrogen, Carlsbad, CA) was supplemented with 1 ng/ml of interleukin-7 (IL-7) and 5 ng/ml of Flt-3 ligand (Flt-3L) (R&D Systems, Minneapolis, MN). The coculture system was maintained by refreshing half the volume of the medium every 3 to 4 days and transferring cells to a new stromal monolayer every 6 to 7 days. Cocultured cells were harvested at desired time points by forcefully pipetting and filtering the resulting cell suspension. Contaminated stromal cells, usually representing less than 5% of the total, were excluded based on forward and side scatter characteristics in flow cytometric analyses.
Retroviral transduction.
The Phoenix-E packaging cell line was cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and transfected with retroviral constructs using the calcium phosphate precipitation method in the presence of 25 μM chloroquine. The Notch1 intracellular domain (Notch1-IC) and GATA3-expressing retroviral constructs were kind gifts from Warren S. Pear (University of Pennsylvania) and José Alberola-Ila (Oklahoma Medical Research Foundation), respectively. The cultures were given fresh medium 8 and 20 h after transfection. Viral stocks were collected by harvesting culture supernatants 44 h after transfection. To transduce Id1tg/tg DN1′ cells, cells were stimulated with 5 ng/ml of both Flt-3L and IL-7 in Alpha medium overnight. Spin infection was performed by centrifugation at 2,000 rpm for 90 min at room temperature in retroviral stocks supplemented with polybrene at a final concentration of 4 μg/ml. After incubation at 37°C for 6 h, cells were seeded with stromal cells in fresh medium.
For bone marrow progenitors from 5-fluorouracil (5-FU)-treated mice, cells were cultured in Alpha medium containing 20 ng/ml of each stem cell factor, Flt-3L and thrombopoietin (R&D Systems), for 3 h. Spin infection was then performed in the presence of 8 μg/ml Polybrene. After incubation at 37°C for 12 h, a second round of spin infection was carried out and followed by incubation at 37°C for 8 h. Transduced progenitor cells were sorted 24 h after the second infection.
Real-time quantitative reverse transcriptase PCR (RT-PCR).
Total RNA was extracted from sorted cells by using TRIzol reagent (Invitrogen). DNase-I-treated total RNA samples were used to produce cDNA templates with Moloney murine leukemia virus RT (Invitrogen). PCR amplification was performed using SYBR green PCR master mix and an ABI Prism 7500 machine (Applied Biosystems, Foster City, CA). The primer sets for Deltex1, Deltex4, Hes1, Notch1, and Id2 were from Qiagen QuantiTect primer assays. The primers for Gfi1b were ATGCCACGGTCCTTTCTAGTG and GGAAGGCTCTGGTTCAGCAA, and the primers for IL-2 receptor β (IL-2rb) were TGGAGCCTGTCCCTCTACG and TCCACATGCAAGAGACATTGG. Those for GATA3 were as described previously (53).
RESULTS
Id1 impairs T-cell development in transgenic mice at the DN1 to DN2 transition.
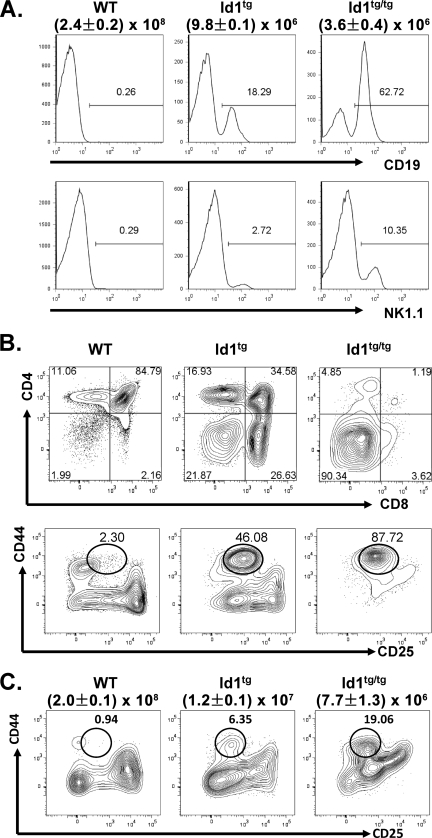
We have previously shown that T-cell development is severely impaired in Id1 transgenic mice, which exhibit dramatically reduced thymic cellularity and massive apoptosis of developing thymocytes (24, 38). Substantial fractions of live thymocytes in the rudimentary thymuses of heterozygous Id1 transgenic (Id1tg) and Id1tg/tg mice in the FVB/N background were CD19+ B cells or NK1.1+ NK cells, though the absolute numbers of these cells per thymus did not increase compared to their levels in wild-type mice (Fig. 1A). Even when CD19+ cells were gated out, Id1 transgenic thymocytes were found to consist of much-higher percentages of CD4 and CD8 DN cells, and Id1tg/tg mice had very few cells that developed beyond the DN stage. Within the DN compartment, a distinct DN1-like population characterized as CD4− CD8− CD44+ CD25lo was detected in Id1 transgenic mice, and we termed this subset DN1′ (Fig. 1B). Since the CD4 and CD8 DN compartment was analyzed after gating out cells expressing TCRγδ, B220, Mac-1, NK1.1, and DX5, the DN1′ population does not include cells differentiated along the γδ T-cell, B-cell, myeloid, and NK lineages. The percentage of DN1′ cells corresponded to levels of exogenous Id1 expressed in the thymus (Id1tg versus Id1tg/tg). Likewise, thymocytes of Id1 transgenic mice on the C57BL/6 background exhibited similar profiles of CD4 and CD8 expression (data not shown). However, the developmental block at the DN1′ stage was less dramatic (Fig. 1C). Nevertheless, the accumulation of DN1′ cells thus represented a significant developmental block in Id1 transgenic mice, which is also seen in E2A-deficient mice (6, 54).
FIG. 1.
T-cell development is arrested at the DN1-to-DN2 transition in Id1 transgenic mice. (A) Total thymocytes from wild-type (WT), Id1tg and Id1tg/tg mice in the FVB/N background were analyzed for expression of CD19 (top) and NK1.1 (bottom). Numbers show the percentages of CD19+ or NK1.1+ cells within propidium iodide-negative live-cell gates. Average total thymocyte counts ± standard deviations are shown beneath the genotypes of the mice. (B) CD4 and CD8 expression on CD19− thymocytes from mice as described for panel A. Numbers show percentages of cells in each quadrant. Levels of CD44 and CD25 expression on thymocytes negative for B220, CD4, CD8, TCRγδ, DX5, NK1.1, and Mac1 staining (Lin−) are shown at the bottom. The DN1′ cell gate is shown as a circle, and percentage of cells within the gate are indicated. (C) CD44 and CD25 expression on thymocytes of mice in the C57BL/6 background. Average total thymocyte counts ± standard deviations are shown beneath the genotypes of the mice.
The developmental block is T-cell intrinsic.
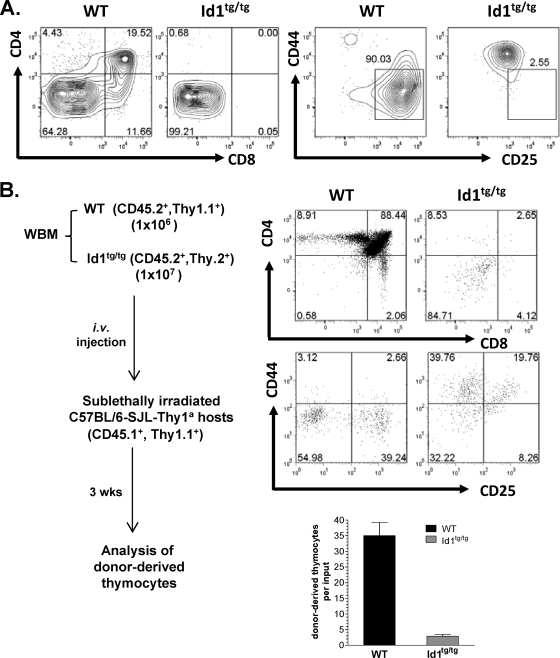
To determine whether the defect in T-cell development is an intrinsic feature of the progenitors, DN1′ cells were isolated from Id1tg/tg mice and cocultured with OP9-DL1 stromal cells. As a control, wild-type DN1 cells were cultured in parallel. After 19 days, wild-type DN1 cells had already differentiated to the DN3 stage and some had reached the DP stage (Fig. 2A). In contrast, Id1 transgenic DN1′ cells did not progress. Although the wild-type cells expanded 919-fold, the numbers of Id1 transgenic cells increased by only 47-fold.
FIG. 2.
The DN1-to-DN2 block in Id1 transgenic mice is T-cell intrinsic. (A) Sorted DN1 cells from wild-type (WT) or DN1′ cells from Id1tg/tg mice were seeded onto OP9-DL1 stromal cells in medium supplemented with 1 ng/ml of IL-7 and 5 ng/ml of Flt-3L. After 19 days, T-cell differentiation was assessed by examining surface expression of CD4 and CD8 or CD44 and CD25. The CD4/CD8 plots were gated on live cells, and the CD44/CD25 plots were gated on Lin− live cells. The numbers on the plots represent the percentage of cells in each gate. (B) Sublethally irradiated recipients were transplanted with a 1:10 mixture of WT mouse and Id1tg/tg mouse whole bone marrow (WBM). Donor-derived thymocytes were analyzed after 3 weeks as described for Fig. 1B. The numbers on the plots represent the percentage of cells in each gate. Total numbers of thymocytes developed from WT and Id1tg/tg donors are shown in a bar graph with averages from six recipients and standard deviations. i.v., intravenous.
Furthermore, we performed a competitive repopulation assay by cotransplanting whole bone marrow cells from wild-type and Id1tg/tg mice into sublethally irradiated recipient mice (Fig. 2B). Even at a 1-to-10 ratio of wild-type to transgenic bone marrow cells transplanted, wild-type cells developed normally and outgrew the transgenic cells by more than 10-fold. However, the transgenic cells failed to develop beyond the DN1′ stage. These results suggest that the developmental defect in Id1 transgenic thymocytes is T-cell intrinsic. Moreover, Id1 transgenic thymocytes did not exert any detectable negative effect on wild-type cells under this experimental condition, because the level of recovery of wild-type cells from the mixed transplant was similar to that from recipients receiving wild-type bone marrow alone (data not shown).
Aberrant gene expression in Id1 transgenic mice.
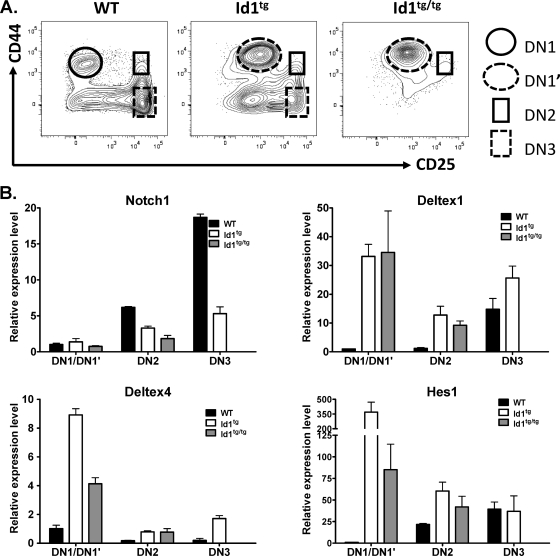
To investigate the molecular mechanisms underlying the defect in T-cell differentiation in Id1 transgenic mice, we examined the expression levels of several genes involved in early stages of T-cell development. Different DN populations, DN1 or DN1′, DN2, and DN3, were purified from wild-type, Id1tg, and Id1tg/tg mice for quantitative RT-PCR assays (Fig. 3A). Since Notch1 is essential for T-cell commitment and development, we measured Notch1 transcript levels in these cells. Notch1 levels in Id1 transgenic DN1′ cells were similar to the levels in DN1 cells of wild-type mice but corresponded to only one-fifth of the level found in wild-type DN2 cells. The levels of expression of Notch1 in Id1-expressing DN2 and DN3 cells were also reduced by two- to threefold compared to its levels in their wild-type counterparts (Fig. 3B). These data suggested that Notch signaling might be minimized. Surprisingly, the expression level of several genes known to be the downstream targets of Notch signaling was found to be elevated rather than diminished in Id1 transgenic cells. These genes include Deltex1 and -4, as well as Hes1. In DN1′ cells, the levels of Deltex1 were over 30-fold higher than the levels in wild-type DN1 cells (Fig. 3B). The Deltex1 levels in Id1-expressing DN2 cells were 10 times higher than in wild-type controls. Despite the sharp increase in Deltex1 expression in wild-type DN3 thymocytes, DN3 cells from Id1tg mice still expressed higher levels of Deltex1. Id1tg/tg mice did not have sufficient DN3 cells for this measurement. Deltex4 expression followed a similar trend, although the magnitude of elevation was smaller than that of Deltex1. In contrast, Deltex2 expression was not altered by Id1 (data not shown). Moreover, the level of transcription of another downstream target of Notch, Hes1, increased dramatically in DN1′ and DN2 cells of Id1 transgenic mice compared to its transcription level in wild-type controls. Taken together, these results indicated that elevated expression of Deltex and Hes1 in Id1 transgenic thymocytes occurred prior to upregulation of Notch1 transcription during the DN1-to-DN3 transition, thus implying Notch-independent mechanisms. In contrast, the reduction in levels of the Notch1 transcript in DN2 and DN3 cells of Id1 transgenic mice might result from high levels of Deltex1, which could compromise Notch function and, in turn, diminish the autoregulation of Notch1 transcription.
FIG. 3.
Analysis of gene expression. (A) Illustration of cell populations from wild-type, Id1tg, and Id1tg/tg mice that were used for real-time RT-PCR assay. (B) The expression levels of indicated genes by purified DN subsets. Levels of transcripts were normalized against the level of β-actin by calculating ΔCT, where CT is the threshold cycle. Expression levels relative to those of wild-type DN1 cells were determined by using the 2−ΔΔCT formula. Data show the averages ± standard deviations, which were calculated as described previously (10). WT, wild type.
Deltex1 deficiency rescues T-cell commitment in Id1 transgenic mice.
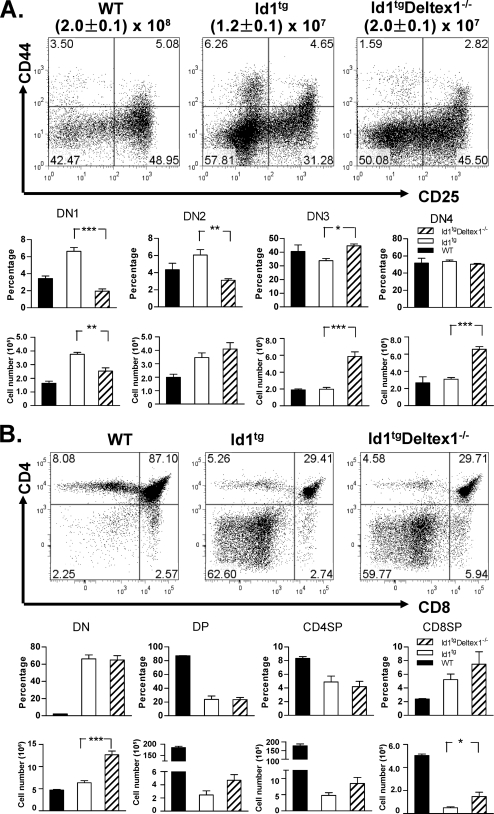
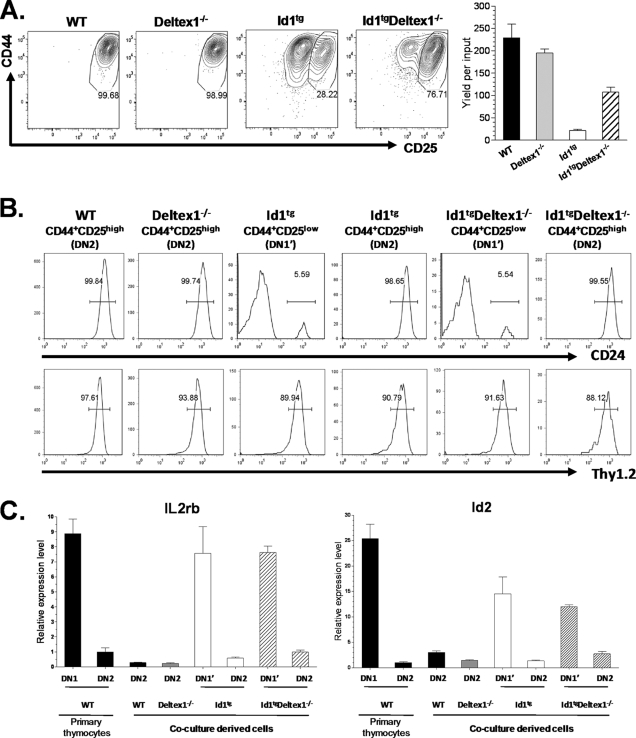
To test the hypothesis that high levels of Deltex1 are, at least in part, responsible for the developmental block at the DN1-to-DN2 transition, we determined the effect of Deltex1 ablation on T-cell commitment in Id1 transgenic mice. Deltex1 has been shown to be dispensable for T-cell development (28, 44). We also found that the developmental profile of Deltex1−/− thymocytes was indistinguishable from that of wild-type mice (data not shown). Since the Deltex1−/− mice were in the C57BL/6 background, we crossed these mice with our Id1 transgenic mice that were backcrossed into the C57BL/6 background. Id1 transgenic mice in the C57BL/6 background have slightly milder defects (Fig. 1C) than those in the FVB/N background (Fig. 1B), but their expression profiles for the Notch1 and Deltex1 genes were similar to those found in FVB/N mice (data not shown). In comparing Id1tg Deltex1−/− to Id1tg mice, we observed a twofold increase overall in thymic cellularity (Fig. 4A). Examination of the developmental profile of DN thymocytes using CD44 and CD25 markers revealed that Deltex1 ablation in Id1 transgenic mice eliminated the block at the DN1′ stage, reducing the percentage and number of DN1′ cells compared to these parameters in Id1tg mice with an intact Deltex1 gene (Fig. 4A). Since the total thymocyte counts increased as a result of Deltex1 ablation, the average numbers of DN2 cells did not change markedly even though the percentages of these cells decreased. Deltex1 deficiency also led to a small but statistically significant increase in the average percentage of DN3 cells compared to that in Id1 transgenic mice, which is reflected by a more dramatic increase in the number of DN3 thymocytes in Id1tg Deltex1−/− mice. The average number of DN4 thymocytes also increased (Fig. 4A). In contrast, T-cell development beyond the DN stages was not significantly affected by loss of Deltex1 (Fig. 4B). These results suggest that aberrant elevation of Deltex1 levels is likely responsible for the block at early stages of T-cell commitment and differentiation and that additional developmental defects exist at the DN3 and subsequent stages, as previously noted by several groups (1, 7-9, 25, 38, 55).
FIG. 4.
Disruption of Deltex1 rescues T-cell development from DN1 to DN3 in Id1 transgenic mice. (A) Thymocytes from wild-type, Id1tg, and Id1tg Deltex1−/− mice were analyzed for expression of CD44 and CD25. Plots show expression in live Lin− cells, and the numbers within each quadrant represent the percentage of cells. Average percentages and total numbers of DN1 through DN4 subpopulations are shown in bar graphs, with standard deviations. (B) The same samples were analyzed for expression of CD4 and CD8. Average percentages and total numbers of CD4 and CD8 DN, DP, and SP (CD4SP or CD8SP) cells are shown in the bar graphs, with standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The numbers of age-matched mice analyzed in the same experiment were four, six, and three for wild-type, Id1tg, and Id1tg Deltex1−/− mice, respectively. The data are representative of two independent experiments. WT, wild type.
To further evaluate the effect of Deltex1 ablation on T-cell commitment, we tested the ability of Id1tg Deltex1−/− DN1′ cells to differentiate in OP9-DL1 stromal cell cocultures (Fig. 5A). After 7 days in culture, the majority of wild-type DN1 cells progressed to a DN2-like (CD44+ CD25high) stage. In contrast, only 28% of Id1tg DN1′ cells progressed to this stage. Deltex1 deficiency significantly enhanced the differentiation of DN1′ cells taken from Id1tg Deltex1−/− mice (28% versus 77%). The yield of CD44+ CD25high cells produced from ID1tg Deltex1−/− DN1′ cells was over fivefold higher than that from Id1tg DN1′ cells (Fig. 5A).
FIG. 5.
Deltex1 deficiency facilitates differentiation of DN1′ cells in culture. (A) Sorted DN1 cells from wild-type (WT) or Deltex1−/− mice and DN1′ cells from Id1tg or Id1tg Deltex1−/− mice were plated onto OP9-DL1 stromal cells in medium supplemented with 1 ng/ml of IL-7 and 5 ng/ml of Flt-3L. After 7 days, cells were assayed for surface expression of CD44 and CD25 by flow cytometry. Plots show the analysis of live cells and are representative of four wells for each genotype. Numbers show the percentages of differentiated cells. The average yield of CD44+ CD25high cells per input is shown in a bar graph, with standard deviations. The data are representative of at least three independent experiments. (B) CD24 and Thy1.2 expression in cells produced as described for panel A. Undifferentiated CD44+ CD25low (DN1′) and differentiated CD44+ CD25high (DN2) cells were gated and assessed separately for the expression of CD24 and Thy1. Numbers show the percentages of CD24+ or Thy1.2+ cells. (C) Transcription levels of indicated genes in coculture-derived cells, with standard deviations. Cell populations described for panel B were sorted, along with primary WT DN1 and DN2 thymocytes. Levels of IL-2rb and Id2 transcripts were measured using real-time RT-PCR and normalized against the level of β-actin. Expression levels in primary WT DN1 and DN2 cells served as controls.
To further verify the identity of the DN2-like cells, we examined additional characteristics of T-cell differentiation in the cells derived from the cocultures. Since E proteins are known to be involved in the rearrangement of TCRβ and -γ genes and in the transcription of the pre-TCRα gene, these common features of T-cell differentiation could not be used. Therefore, we monitored CD24 expression on the cell surface, which is known to be upregulated as T cells progress from the DN1 to the DN2 stage (41). Indeed, the DN2-like cells were exclusively CD24+, whereas the remaining DN1′-like cells were largely CD24− (Fig. 5B). Moreover, we also found these cultured cells to be positive for the pan-T-cell marker, Thy1.2 (Fig. 5B). T-cell differentiation was further evaluated by examining changes in gene expression. As shown in Fig. 5C, expression of the IL-2rb and Id2 genes was dramatically reduced in wild-type DN2 primary thymocytes compared to their expression in DN1 cells (41). Similarly, all DN2-like cells produced in culture had low levels of IL-2rb and Id2, in contrast to the high levels seen in residual DN1′ cells from Id1tg and Id1tg Deltex1−/− mice (Fig. 5C). Collectively, the results from both surface marker staining and gene expression profiles strongly suggested that ablation of Deltex1 enabled DN1′ cells to commit to the T lineage and progress to the DN2 stage. However, these DN2-like cells from Id1 transgenic cells with or without Deltex1 were unable to progress further and eventually disappeared, suggesting that additional factors are needed to facilitate their differentiation in the culture system. Nevertheless, these in vitro culture results, together with the results obtained in vivo, support the notion that aberrant levels of Deltex1 in Id1 transgenic DN1′ cells hinder T-cell commitment, at least in part, and prevent their transition to the DN2 stage.
Enhanced Notch signaling enables DN1′ cells to differentiate.
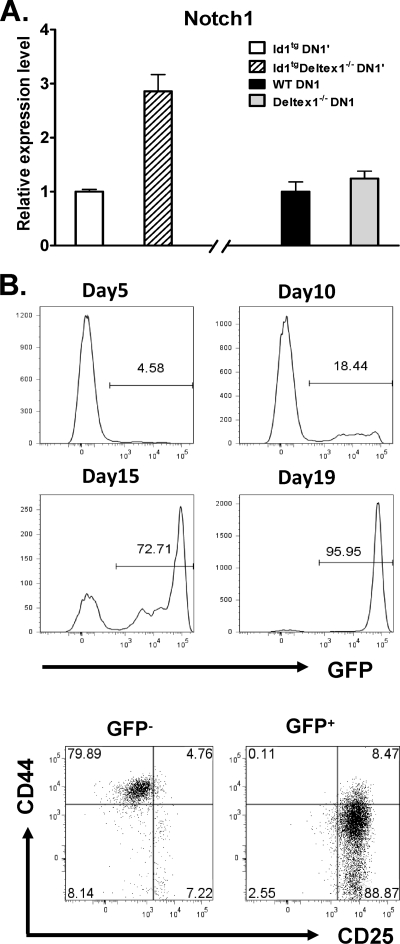
Our hypothesis stated that aberrant Deltex1 expression tampers with Notch signaling, which in turn diminishes transcription of the Notch1 gene itself. If so, one would expect that deletion of the Deltex1 gene leads to an elevation of Notch1 levels. Indeed, we found that Notch1 expression increased nearly threefold in Id1 transgenic DN1′ cells lacking Deltex1 compared to its expression in DN1′ cells with Deltex1 (Fig. 6A). In contrast, Deltex1 deficiency alone did not significantly alter Notch1 expression in DN1 cells compared to its expression in their wild-type counterparts (Fig. 6A). The increase in Notch levels could be interpreted to mean that Notch activity was enhanced by the elimination of Deltex1, which then facilitated the differentiation of DN1′ cells as seen in Fig. 4 and 5.
FIG. 6.
Notch signaling potentiates differentiation of DN1′ cells. (A) DN1′ cells from Id1tg and Id1tg Deltex1−/− mice or DN1 cells from wild-type (WT) and Deltex1−/− mice were isolated by cell sorting and used to analyze Notch1 expression. The level of Notch1 mRNA was determined by using real-time RT-PCR and normalized against the level of β-actin mRNA. Data are presented as the relative levels of expression in Id1tg DN1′ or wild-type DN1 cells in the absence or presence of Deltex1 and are shown as averages ± standard deviations from triplicates. The data are representative of two separate experi- ments. (B) DN1′ cells, sorted from Id1tg/tg mice, were transduced with the retrovirus encoding Notch1-IC and GFP. The cells were then plated onto a 24-well plate containing OP9 stromal cells and cultured in medium supplemented with 1 ng/ml of IL-7 and 5 ng/ml of Flt-3L. At different time points of the culture, GFP expression was analyzed, and the percentages of live, GFP+ cells are shown in the histograms. Cells from day-19 cultures were stained with antibodies against CD44 and CD25 and analyzed by gating on live, GFP− and GFP+ populations. Numbers indicate percentage of cells in each quadrant. Data are representative of at least two independent experiments.
To further test this idea, we transduced Id1tg/tg DN1′ cells with the retrovirus producing Notch1-IC, a constitutively active form of Notch1, and green fluorescent protein (GFP) (35). The cells were then cocultured with OP9 stromal cells. Notch1-IC-expressing cells were identified by GFP expression. Although GFP-positive (GFP+) cells constituted only about 5% at the beginning of the culture, these cells became progressively enriched over time (Fig. 6B). A 350-fold expansion of Notch1-IC-expressing cells without any increase in the number of GFP-negative (GFP−) cells led to an enrichment of GFP+ cells to 96% by day 19. When these cells were analyzed using flow cytometry, it was revealed that GFP+ cells differentiated into DN3-like cells, whereas GFP− cells remained at the DN1′ stage (Fig. 5C). As a control, we also performed parallel experiments by transducing Id1tg/tg DN1′ cells with vector control retrovirus and did not observe any effect on the differentiation and proliferation of DN1′ cells (data not shown). Thus, this result supports the notion that Notch signaling is essential for the differentiation of Id1 transgenic DN1′ cells.
Potential mechanisms underlying aberrant Deltex1 expression.
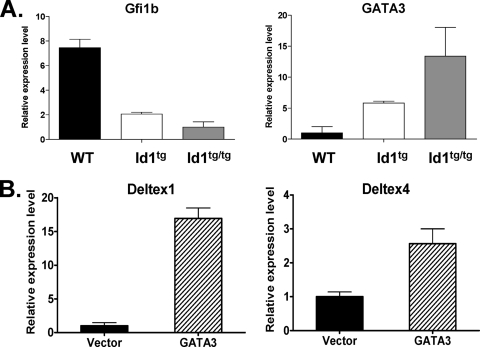
To understand the mechanism by which Deltex1 was aberrantly expressed independently of Notch signaling in Id1 transgenic mice, we examined the promoter sequence of the Deltex1 gene. Strikingly, we discovered a 70-bp sequence containing at least 17 GATA-binding sites located 1.6 kb upstream of the transcriptional start site of Deltex1. This immediately begged the question of whether GATA3 expression in DN1′ cells may be responsible for Deltex1 upregulation. Xu and Kee (53) have shown that a transcriptional repressor, Gfi1b, is a target of E2A and represses GATA3 expression in T-cell lines. If so, one might expect Gfi1b expression to decrease while GATA3 levels increase in Id1 transgenic thymocytes where E protein functions are inhibited. We therefore compared the levels of Gfi1b and GATA3 in subsets of Id1 transgenic thymocytes with their levels in wild-type counterparts. Indeed, the levels of Gfi1b were found to be reduced by four- and eightfold in DN1′ cells of Id1tg and Id1tg/tg mice, respectively (Fig. 7A). Correspondingly, GATA3 expression was dramatically elevated in DN1′ cells compared to its level of expression in wild-type DN1 cells (Fig. 7A). Thus, this increase in GATA3 levels could be responsible for activating Deltex1 transcription.
FIG. 7.
GATA3 upregulates Deltex1 expression. (A) Gfi1b and GATA3 expression in DN1 or DN1′ populations of cells isolated from wild-type (WT), Id1tg, and Id1tg/tg mice. Levels of indicated transcripts were normalized against the level of β-actin mRNA. Data shown are Gfi1b levels relative to the level in Id1tg/tg DN1′ cells and GATA3 levels relative to the level in WT DN1 cells. They are presented as averages ± standard deviations from triplicates. (B) Bone marrow cells from 5-FU-treated WT mice were transduced with vector or GATA3-expressing retrovirus. Transduced cells were isolated based on GFP expression from the same viral constructs and used to analyze Deltex1 and Deltex4 expression. Levels of indicated mRNA, determined by using real-time RT-PCR, were normalized against the level of β-actin. Data are presented as the average expression levels ± standard deviations relative to the levels in vector-transduced cells and are calculated from triplicates. Representatives of two experiments are shown.
To test whether ectopic expression of GATA3 is capable of stimulating Deltex1 expression, we expressed GATA3 in bone marrow progenitor cells obtained from 5-FU-treated wild-type mice, because DN1 thymocytes are difficult to transduce with retrovirus. We reasoned that these bone marrow-derived multipotent progenitors may be more comparable to the uncommitted thymic seeding progenitors in the DN1 population than any differentiated thymocyte subsets. Retrovirally transduced cells were sorted based on GFP expression, and Deltex1 levels were measured using quantitative RT-PCR assays (Fig. 7B). Compared to the results for cells transduced with the control virus, GATA3 expression led to a 15-fold increase in Deltex1 expression, indicating that GATA3 can indeed activate Deltex1 transcription. In contrast, the Deltex4 gene does not contain a cluster of GATA-binding sites within 3 kb of its 5′ flanking sequence, and its expression increased by less than threefold in the same assay (Fig. 6B). Taking these results together, it appears that elevated levels of the GATA3 transcription factor in Id1 transgenic mice may play a significant role in the aberrant expression of Deltex1.
DISCUSSION
Overexpression of Id1 in transgenic thymocytes inhibits the function of both E2A and HEB proteins in a dose-dependent manner and results in severe blocks in T-cell development (24). One of the most striking phenotypes is the accumulation of cells displaying characteristics of DN1 thymocytes, except that these cells express slightly higher levels of CD25. Therefore, we referred to these cells as DN1′ cells, which might represent cells arrested at the DN1-to-DN2 transition. These DN1′ cells do not express any specific markers for NK, myeloid, B, and γδ T-lineage cells. However, they had a high propensity to differentiate into DX5+ NK1.1− cells when cocultured with OP9 stromal cells with or without the Notch ligand, DL1 (data not shown). It remains to be understood whether the DN1′ population consists of multipotent progenitors or whether it is a mixture of progenitors with different cell fates. Nevertheless, in the presence of Id1, T cells fail to develop.
To understand the molecular mechanism underlying the block at the earliest stage of T-cell development, we examined gene expression profiles in these DN1′ cells of Id1 transgenic mice in comparison to the gene expression profiles in wild-type DN1 cells. We discovered that levels of Deltex1 were dramatically increased. Furthermore, we provided evidence to suggest that elevated expression of Deltex1 impairs Notch signaling and, consequently, impedes T-cell commitment and developmental progression from the DN1 to the DN2 stage. This block was alleviated by Deltex1 deficiency or by enforced Notch signaling, suggesting that Deltex1 indeed had a negative effect on Notch function. This notion is consistent with previous findings that overexpression of Deltex1 in multipotent progenitors inhibited T-cell development while allowing B cells to be produced in the thymus (21, 56). These effects of Deltex1 are similar to those observed in Notch1 conditional-knockout mice (39). Paradoxically, Deltex1 is a mammalian homolog of Drosophila melanogaster Deltex, which was found through genetic approaches to play a positive role in Notch signaling (32). Whether Deltex1 has any positive function in Notch pathways is a complicated issue and likely depends on cellular contexts (18, 29). Although it is reasonable to expect Deltex to facilitate Notch function, the negative effect of various Deltex proteins on Notch function may be viewed as a feedback control mechanism that could be useful for maintaining proper levels of Notch signaling (28). Incidentally, Deltex1 appears dispensable for T-cell differentiation, but high levels of Deltex1 can be harmful (21, 28).
Deltex1 deficiency facilitates the commitment of Id1 transgenic progenitors to the T lineage by reducing the percentages and numbers of DN1′ cells while increasing the numbers of DN3 and DN4 cells compared to these parameters in Id1 transgenic mice with the Deltex1 gene intact. However, loss of Deltex1 boosted the total number of thymocytes by only twofold and did not have a significant impact on T-cell development beyond the DN stages. This is likely due to additional defects at later stages of development as a result of the loss of E protein function. For example, we did not detect any VDJ rearrangement events at the TCRβ or -γ loci in Id1 transgenic DN3 cells that developed as a result of Notch1-IC expression (data not shown). This notion is supported by observations made by using E2A-deficient thymocytes, where TCRβ gene rearrangement is severely impaired (1). Additionally, very low levels of pre-TCRα transcripts were detected in Id1 transgenic DN3 cells compared to the levels in wild-type counterparts (data not shown). Consequently, an inability to signal through pre-TCR would not allow the tremendous expansion of thymocytes and developmental progression to later stages. Furthermore, we have previously shown that developing Id1 transgenic thymocytes undergo apoptosis (25, 38, 55). These defects may not be corrected by Deltex1 ablation.
Moreover, enhanced Deltex1 expression is only one of the abnormalities found in Id1 transgenic DN1′ cells. Deltex4 levels were also elevated, albeit less dramatically, and could have contributed to the inhibitory effects on Notch signaling together with Deltex1 (28). It would be interesting to determine the combined role of Deltex1 and -4 in preventing T-cell commitment in Id1 transgenic mice. However, it is technically difficult to ablate both genes simultaneously in Id1 transgenic mice. Deltex4-deficient mice have not been reported. Curiously, Hes1 expression also increased dramatically in Id1 transgenic DN1′ cells. However, whether high levels of Hes1 are detrimental to T-cell commitment is not clear. Unlike the results for Deltex1, retrovirus-mediated expression of Hes1 in bone marrow progenitors did not block T-cell development (23). On the contrary, analyses of Hes1-deficient mice suggested that Hes1 was required for T-cell progenitors to progress to the DN2 stage (48).
Although Deltex1 has been well recognized as a Notch1 target gene, evidence has also emerged to suggest that it can be regulated by additional factors (19). We showed in this report that overexpression of GATA3 increased transcription of the endogenous Deltex1 gene by 15-fold. GATA3 probably binds to a cluster of GATA sites located 1.6 kb upstream of the Deltex1 gene. However, GATA3 may not act alone, because it was unable to stimulate the expression of a luciferase reporter gene driven by a 1.9-kb Deltex1 promoter sequence that included these GATA sites (data not shown). This promoter construct was also not responsive to activated Notch1. Despite the fact that Deltex1 is a well-documented downstream target gene of Notch signaling, its transcriptional regulation is poorly understood. The regulation of Deltex1 transcription appears to involve complicated schemes in which GATA3 cooperates with additional transcription factors. Alternatively, transcription of the Deltex1 gene may depend on its proper chromatin configuration in its native locus. As we have demonstrated here, a tight control of Deltex1 expression is necessary for the proper development of mammalian systems such as T lymphopoiesis, thus representing an interesting subject for future investigation.
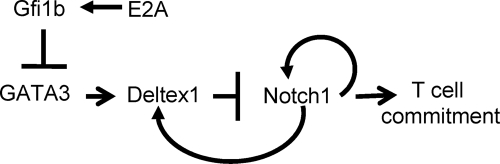
The elevation of Deltex1 expression possibly results from a chain of events originating from the loss of E protein function (Fig. 8). In a recent study, GATA3 levels were found to be elevated in E2A-deficient cell lines due to the downregulation of a transcriptional repressor, Gfi1b (53). Gfi1b is thought to be a target gene of E2A transcription factors. In Id1 transgenic mice, we found that Gfi1b levels were significantly lower and GATA3 levels were higher in DN1′ cells than in their wild-type counterparts. Previous studies have shown that overexpression of GATA3 blocks T-cell development even though a lack of GATA3 also arrests T-cell differentiation (4, 16, 46). It is therefore important to maintain an optimal level of GATA3 during T-cell development. One of the crucial roles of E proteins at early stages of T lymphopoiesis may be to suppress GATA3 expression, as well as Deltex1 expression, through upregulation of the Gfi1b transcriptional repressor (Fig. 7). This is important for ensuring proper Notch signaling, which is essential for T-cell differentiation. E proteins have also been proposed to regulate Notch1 transcription itself, a conclusion largely based on studies using E2A-deficient cell lines and fetal thymuses (20). While E proteins may play a direct role in Notch1 transcription at later stages, no evidence is available to suggest a direct effect of E proteins on Notch1 transcription in uncommitted T-cell progenitors. The findings from this study illustrate how cellular differentiation processes are normally controlled by a network of transcriptional regulators. This control mechanism offers flexibility and fine tuning but also vulnerability to multiple pathological agents. On the other hand, the interdependence of multiple regulators may protect cells from any single pathogenic assault, thus safeguarding the differentiation processes.
FIG. 8.
Role of E proteins in controlling Notch-mediated T-cell commitment. E protein function, which is inhibited by Id1, controls the transcription of the Gfi1b gene, encoding a known repressor of GATA3 transcription. As a result, elevated levels of GATA3 in T-cell progenitors of Id1 transgenic mice lead to an increase in Deltex1 levels, which interferes with Notch signaling in directing T-cell commitment, as well as Notch autoregulation.
Acknowledgments
We thank Ying Zhao for technical assistance and members of the Sun laboratory for advice. We are grateful to Jose Alberola-Ila and Flora Ling for critical reading of the manuscript.
We wish to dedicate the manuscript to the memory of Wei-Feng Chen.
This work was supported by grants from the National Institute of Health to X.-H.S. (CA77553 and AI56129). X.-H.S. holds the Eli Lilly Distinguished Chair in Biomedical Research.
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Agata, Y., N. Tamaki, S. Sakamoto, T. Ikawa, K. Masuda, H. Kawamoto, and C. Murre. 2007. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 27871-884. [DOI] [PubMed] [Google Scholar]
- 2.Allman, D., J. A. Punt, D. J. Izon, J. C. Aster, and W. S. Pear. 2002. An invitation to T and more: notch signaling in lymphopoiesis. Cell 109 Suppl.:S1-S11. [DOI] [PubMed] [Google Scholar]
- 3.Allman, D., A. Sambandam, S. Kim, J. P. Miller, A. Pagan, D. Well, A. Meraz, and A. Bhandoola. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4168-174. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. K., G. Hernandez-Hoyos, C. J. Dionne, A. M. Arias, D. Chen, and E. V. Rothenberg. 2002. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev. Biol. 246103-121. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas, S., K. Matsuno, and M. E. Fortini. 1995. Notch signaling. Science 268225-232. [DOI] [PubMed] [Google Scholar]
- 6.Bain, G., I. Engel, E. C. Robanus Maandag, H. P. te Riele, J. R. Voland, L. L. Sharp, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 174782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bain, G., M. W. Quong, R. S. Soloff, S. M. Hedrick, and C. Murre. 1999. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J. Exp. Med. 1901605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barndt, R., M. F. Dai, and Y. Zhuang. 1999. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J. Immunol. 1633331-3343. [PubMed] [Google Scholar]
- 9.Barndt, R. J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 206677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bookout, A. L., and D. J. Mangelsdorf. 2003. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 1e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J. W., C. Pampeno, S. Vukmanovic, and D. Meruelo. 2002. Characterization of the transcriptional expression of Notch-1 signaling pathway members, Deltex and HES-1, in developing mouse thymocytes. Dev. Comp. Immunol. 26575-588. [DOI] [PubMed] [Google Scholar]
- 12.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 1373-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley, E. C., H. T. Petrie, L. M. Shah, M. J. Owen, and A. C. Hayday. 1994. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity 183-93. [DOI] [PubMed] [Google Scholar]
- 14.Godfrey, D. I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 1504244-4252. [PubMed] [Google Scholar]
- 15.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14637-645. [DOI] [PubMed] [Google Scholar]
- 16.Ho, I. C., and S. Y. Pai. 2007. GATA-3—not just for Th2 cells anymore. Cell. Mol. Immunol. 415-29. [PubMed] [Google Scholar]
- 17.Hoffman, E. S., L. Passoni, T. Crompton, T. M. Leu, D. G. Schatz, A. Koff, M. J. Owen, and A. C. Hayday. 1996. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10948-962. [DOI] [PubMed] [Google Scholar]
- 18.Hori, K., M. Fostier, M. Ito, T. J. Fuwa, M. J. Go, H. Okano, M. Baron, and K. Matsuno. 2004. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 1315527-5537. [DOI] [PubMed] [Google Scholar]
- 19.Hu, X., A. Y. Chung, I. Wu, J. Foldi, J. Chen, J. D. Ji, T. Tateya, Y. J. Kang, J. Han, M. Gessler, R. Kageyama, and L. B. Ivashkiv. 2008. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity 29691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikawa, T., H. Kawamoto, A. W. Goldrath, and C. Murre. 2006. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 2031329-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izon, D. J., J. C. Aster, Y. He, A. Weng, F. G. Karnell, V. Patriub, L. Xu, S. Bakkour, C. Rodriguez, D. Allman, and W. S. Pear. 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16231-243. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama, R., and T. Ohtsuka. 1999. The Notch-Hes pathway in mammalian neural development. Cell Res. 9179-188. [DOI] [PubMed] [Google Scholar]
- 23.Kageyama, R., T. Ohtsuka, and K. Tomita. 2000. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cells 101-7. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D., X. C. Peng, and X. H. Sun. 1999. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol. Cell. Biol. 198240-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, D., M. Xu, L. Nie, X. C. Peng, E. Jimi, R. E. Voll, T. Nguyen, S. Ghosh, and X. H. Sun. 2002. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-kappaB activities. Immunity 169-21. [DOI] [PubMed] [Google Scholar]
- 26.Kisielow, P., and H. von Boehmer. 1995. Development and selection of T cells: facts and puzzles. Adv. Immunol. 5887-209. [DOI] [PubMed] [Google Scholar]
- 27.Lazorchak, A., M. E. Jones, and Y. Zhuang. 2005. New insights into E-protein function in lymphocyte development. Trends Immunol. 26334-338. [DOI] [PubMed] [Google Scholar]
- 28.Lehar, S. M., and M. J. Bevan. 2006. T cells develop normally in the absence of both Deltex1 and Deltex2. Mol. Cell. Biol. 267358-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W. H., and M. Z. Lai. 2005. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell. Biol. 251367-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald, H. R., A. Wilson, and F. Radtke. 2001. Notch1 and T-cell development: insights from conditional knockout mice. Trends Immunol. 22155-160. [DOI] [PubMed] [Google Scholar]
- 31.Maillard, I., T. Fang, and W. S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23945-974. [DOI] [PubMed] [Google Scholar]
- 32.Matsuno, K., R. J. Diederich, M. J. Go, C. M. Blaumueller, and S. Artavanis-Tsakonas. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 1212633-2644. [DOI] [PubMed] [Google Scholar]
- 33.Michie, A. M., and J. C. Zuniga-Pflucker. 2002. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin. Immunol. 14311-323. [DOI] [PubMed] [Google Scholar]
- 34.Murre, C. 2005. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 61079-1086. [DOI] [PubMed] [Google Scholar]
- 35.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 1832283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, S. S., R. S. Welner, T. Kouro, P. W. Kincade, and X. H. Sun. 2006. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J. Immunol. 1772880-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porritt, H. E., L. L. Rumfelt, S. Tabrizifard, T. M. Schmitt, J. C. Zuniga-Pflucker, and H. T. Petrie. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20735-745. [DOI] [PubMed] [Google Scholar]
- 38.Qi, Z., and X. H. Sun. 2004. Hyperresponse to T-cell receptor signaling and apoptosis of Id1 transgenic thymocytes. Mol. Cell. Biol. 247313-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10547-558. [DOI] [PubMed] [Google Scholar]
- 40.Rolink, A. G., S. Massa, G. Balciunaite, and R. Ceredig. 2007. Early lymphocyte development in bone marrow and thymus. Swiss Med. Wkly. 137(Suppl. 155)20S-24S. [DOI] [PubMed] [Google Scholar]
- 41.Rothenberg, E. V., J. E. Moore, and M. A. Yui. 2008. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 89-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambandam, A., I. Maillard, V. P. Zediak, L. Xu, R. M. Gerstein, J. C. Aster, W. S. Pear, and A. Bhandoola. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 6663-670. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt, T. M., and J. C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17749-756. [DOI] [PubMed] [Google Scholar]
- 44.Storck, S., F. Delbos, N. Stadler, C. Thirion-Delalande, F. Bernex, C. Verthuy, P. Ferrier, J. C. Weill, and C. A. Reynaud. 2005. Normal immune system development in mice lacking the Deltex-1 RING finger domain. Mol. Cell. Biol. 251437-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, X. H. 2004. Multitasking of helix-loop-helix proteins in lymphopoiesis. Adv. Immunol. 8443-77. [DOI] [PubMed] [Google Scholar]
- 46.Taghon, T., M. A. Yui, and E. V. Rothenberg. 2007. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 8845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanigaki, K., and T. Honjo. 2007. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 8451-456. [DOI] [PubMed] [Google Scholar]
- 48.Tomita, K., M. Hattori, E. Nakamura, S. Nakanishi, N. Minato, and R. Kageyama. 1999. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 131203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visan, I., J. B. Tan, J. S. Yuan, J. A. Harper, U. Koch, and C. J. Guidos. 2006. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 7634-643. [DOI] [PubMed] [Google Scholar]
- 50.Weng, A. P., J. M. Millholland, Y. Yashiro-Ohtani, M. L. Arcangeli, A. Lau, C. Wai, C. Del Bianco, C. G. Rodriguez, H. Sai, J. Tobias, Y. Li, M. S. Wolfe, C. Shachaf, D. Felsher, S. C. Blacklow, W. S. Pear, and J. C. Aster. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 202096-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, A., H. R. MacDonald, and F. Radtke. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 1941003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L. 2006. T lineage progenitors: the earliest steps en route to T lymphocytes. Curr. Opin. Immunol. 18121-126. [DOI] [PubMed] [Google Scholar]
- 53.Xu, W., and B. L. Kee. 2007. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood 1094406-4414. [DOI] [PubMed] [Google Scholar]
- 54.Yan, W., A. Z. Young, V. C. Soares, R. Kelley, R. Benezra, and Y. Zhuang. 1997. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol. Cell. Biol. 177317-7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, Y., H. C. Liou, and X. H. Sun. 2006. Id1 potentiates NF-kappaB activation upon T cell receptor signaling. J. Biol. Chem. 28134989-34996. [DOI] [PubMed] [Google Scholar]
- 56.Yun, T. J., and M. J. Bevan. 2003. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 1705834-5841. [DOI] [PubMed] [Google Scholar]