Abstract
Accurate chromosome segregation requires the capture of sister kinetochores by microtubules from opposite spindle poles prior to the initiation of anaphase, a state termed chromosome biorientation. In the budding yeast Saccharomyces cerevisiae, the conserved protein kinase Ipl1 (Aurora B in metazoans) is critical for ensuring correct chromosomal alignment. Ipl1 associates with its activators Sli15 (INCENP), Nbl1 (Borealin), and Bir1 (Survivin), but while Sli15 clearly functions with Ipl1 to promote chromosome biorientation, the role of Bir1 has been uncertain. Using a temperature-sensitive bir1 mutant (bir1-17), we show that Bir1 is needed to permit efficient chromosome biorientation. However, once established, chromosome biorientation is maintained in bir1-17 cells at the restrictive temperature. Ipl1 is partially delocalized in bir1-17 cells, and its protein kinase activity is markedly reduced under nonpermissive conditions. bir1-17 cells arrest normally in response to microtubule depolymerization but fail to delay anaphase when sister kinetochore tension is reduced. Thus, Bir1 is required for the tension checkpoint. Despite their robust mitotic arrest in response to nocodazole, bir1-17 cells are hypersensitive to microtubule-depolymerizing drugs and show a more severe biorientation defect on recovery from nocodazole treatment. The role of Bir1 therefore may become more critical when spindle formation is delayed.
Accurate chromosome segregation during anaphase is vital for ensuring the maintenance of genome integrity during cell division and, in turn, depends critically on the correct attachment of sister chromatids to kinetochore microtubules. For high-fidelity chromosome segregation, kinetochores must capture spindle microtubules such that sister chromatids are connected to opposite spindle poles (termed amphitelic attachment or chromosome biorientation), ensuring that they are pulled in opposite directions during the subsequent anaphase.
In the budding yeast Saccharomyces cerevisiae, the majority of sister chromatids remain attached to microtubules from a single pole (mono-oriented) without the intervention of a correction mechanism to promote amphitelic attachment (36), a key element of which is the Ipl1 protein kinase. Ipl1 has been proposed to promote the detachment of incorrect microtubule-kinetochore connections so that correct attachments subsequently can form (35). In the absence of Ipl1 function, at the point of anaphase onset around two-thirds of sister chromatids remain mono-oriented, attached to microtubules originating from a single pole to which they then cosegregate (35). Kinetochore proteins such as Dam1 and Ndc80 have been proposed as key Ipl1 substrates for their role in promoting chromosome biorientation (6, 41). Ipl1 kinase also is required for cells to activate the spindle checkpoint in the absence of tension on kinetochore-microtubule attachments, and hence ipl1 mutant cells fail to delay anaphase despite their many mono-oriented chromosomes (2). Depending on the circumstances, the checkpoint role of Ipl1 involves either the generation of unattached kinetochores (26) or the phosphorylation of the checkpoint protein Mad3 (19). Ipl1 also is required in the absence of the BimC family kinesin Cin8p, probably reflecting a role in spindle assembly (9, 21), and is involved in regulating spindle disassembly following anaphase (5).
Ipl1 kinase is highly conserved, and its metazoan ortholog (Aurora B) is involved in both chromosome biorientation and the spindle assembly checkpoint, forming part of the chromosomal passenger complex that also contains INCENP, Survivin, and Borealin (27, 40). The chromosomal passenger complex is so called because although these proteins colocalize throughout the cell cycle, their location changes dynamically from the chromosome arms in G1 to centromeres in prometaphase and finally to the central spindle in anaphase. Such coordinated behavior is consistent with the recent crystal structure of the complex between INCENP, Survivin, and Borealin, in which they interact via tightly entwined helical domains (16).
In budding yeast, Ipl1 interacts with Sli15, Bir1, and Nbl1, which have been proposed to be orthologs of INCENP, Survivin, and Borealin, respectively (6, 18). All three proteins are the products of essential genes. Like INCENP, Sli15 has a conserved C-terminal domain (the IN-box) that is required for Ipl1 kinase activation, and sli15 mutants have a phenotype that is very similar to that of ipl1 mutants (17, 18). Although yeast cells with reduced Bir1 function show chromosome instability, the first-described bir1 mutants failed to reveal a chromosome biorientation defect but instead conferred defects in septin dynamics during anaphase (38). Bir1 interacts with Ndc10 and is responsible for taking Ndc10 to the anaphase spindle (38, 42, 43), a role that may be linked to this septin defect (4). Yeast Bir1 is much larger than its metazoan counterpart (Survivin) and shows little sequence conservation outside the conserved BIR domain, yet this region is nonessential in yeast (42) and therefore unlikely to be involved in chromosome biorientation. Conversely, metazoan Borealin proteins are much larger than yeast Nbl1, which consists of little more than the helical region proposed to form the tight interaction with INCENP/Sli15 and Survivin/Bir1 complexes. Furthermore, a significant fraction of both Sli15 and Bir1 are present in a complex that lacks Ipl1 (29, 38) and that recent work has shown to contain Nbl1 (25), bringing into question the importance of Bir1 for chromosome biorientation. The extent to which Bir1 and Survivin function in conserved or analogous ways within the chromosomal passenger complexes of yeast and metazoans therefore was unclear at the start of our work.
The Sli15-Bir1 complex has been proposed to interact both with microtubules (via the central domain of Sli15) and with kinetochores (through the Bir1-Ndc10 interaction) and through these interactions to function as a tension sensor, relaying information concerning the state of microtubule-kinetochore connections to Ipl1 kinase. Thus, when chromosomes are mono-oriented, the Bir1-Sli15-Nbl1 complex might activate Ipl1 in the absence of tension so as to promote chromosome biorientation by detaching incorrect microtubule attachments (29). This model predicts an essential role for Bir1 in promoting chromosome biorientation, but such evidence has been lacking. By generating a temperature-sensitive bir1 allele (bir1-17) and showing that it confers a profound defect in chromosome biorientation, we demonstrate that Bir1 does play a key role in the correction process needed to ensure that all yeast chromosomes become correctly aligned on the mitotic spindle. Furthermore, since the bir1-17 mutant fails to activate the spindle assembly checkpoint properly in response to reduced sister kinetochore tension, like Ipl1 it forms part of the tension checkpoint mechanism. Our data therefore are consistent with a role for Bir1 in conferring tension responsiveness on Ipl1 function.
MATERIALS AND METHODS
Yeast strains and general methods.
All yeast strains used in this study (Table 1) are derivatives of W303-1a (37) and have the following markers unless indicated to the contrary: ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d2 Gal+. Basic yeast methods, growth media, and routine recombinant DNA methodology were performed as previously described (1, 10, 28). For creating temperature-sensitive bir1 mutants, error-prone PCR, gapped plasmid repair, and plasmid shuffling were used as previously described (32). The region selected for mutagenesis encompassed amino acids 489 to the end of the protein (954) and was defined by the two PCR primers used (5′-GGCATAAATACAGACACAAAAGGAGC-3′ and 5′-AAGCATCAGGAACGCCCG-3′). For the gap repair step, YCplac22-BIR1 was digested with PpuMI and NsiI to generate a suitable fragment missing the C-terminal BIR1 coding region. To make YCplac22-BIR1, a 3.7-kb genomic PvuII fragment starting 484 bp upstream of BIR1 first was inserted into pSP72 (Promega) and then moved as an XhoI-HindIII fragment into YCplac22 (11) cut with SalI and HindIII. The shuffle strain (AKY4) was generated by the sporulation and tetrad dissection of AKYD3, a strain heterozygous for a bir1::KANMX6 knockout and carrying YCplac33-BIR1, a plasmid identical to YCplac22-BIR1 except for its YCplac33 backbone (11). Plasmids carrying potential temperature-sensitive bir1 alleles were verified by plasmid rescue and retesting by plasmid shuffling, and then the sites of mutations were located by DNA sequencing. To integrate the bir1-17 allele using the pop in-pop out method (32), the bir1-17 fragment from YCplac22-bir1-17 was excised and cloned into the URA3 plasmid YIplac204 (11). This construct was targeted to the BIR1 locus in AY925 by linearizing it at its unique SwaI site prior to transformation. Following selection on 5-fluoroorotic acid for recombination between the repeated BIR1 sequences thus generated, colonies were screened for temperature sensitivity, and a strain in which the bir1-17 allele had replaced the wild-type sequence was identified by the DNA sequencing of a suitable genomic PCR fragment encompassing the mutated region. The temperature sensitivity of strains was assessed by diluting mid-exponential-phase cultures of cells in yeast extract-peptone-dextrose (YPD) to an optical density at 600 nm (OD600) of 0.1 and by spotting 5 μl of this and of five 10-fold serial dilutions. Plates were photographed after growth for 2 days at the indicated temperature.
TABLE 1.
Yeast strains
| Straina | Genotype | Source or reference |
|---|---|---|
| AKY4 | MATabir1::HIS3MX6 [YCplac33-BIR1] | A. Kozak |
| AKYD3 | MATa /MATα BIR1/bir1::HIS3MX6 [YCplac33-BIR1] | A. Kozak |
| AY925 | MATa | K. Arndt |
| BP635 | MATadam1-S265A,S292A,S257A | T. Tanaka |
| K5043 | MATaCFIII (CEN3.L.YPH278) URA3 SUP11 | 24 |
| PKY63 | MATaipl1-321-myc13::KANMX6 | P. Keating |
| T2608 | MATaIPL1-myc12::URA3 | T. Tanaka |
| T2997 | MATacen3::pGAL-CEN3-tCYC1-tetOs::ura3 leu2::tetR-GFP::LEU2 trp1::YFP-TUB1::TRP1 cdc20::pMET3-CDC20::TRP1 ask1-3 | T. Tanaka |
| T5241 | MATaIPL1-GFP::KANMX6 CTF19-3ECFP-SkHIS3MX6 NDC80-3ECFP-SkHIS3MX6 | T. Tanaka |
| VMY1 | MATabir1-17 | This study |
| VMY5 | MATaIPL1-myc12::URA3 bir1-17 | This study |
| VMY7 | MATabir1-36 | This study |
| VMY14 | MATaIPL1-GFP::KANMX6 ctf19::CTF19-3ECFP-SkHIS3MX6 ndc80::NDC80-3ECFP-SkHIS3MX6 bir1-17 | This study |
| VMY33 | MATaCEN3-tetO336::URA3 leu2::tetR-GFP::LEU2 trp1::YFP-TUB1::TRP1 cdc20::pMET3-CDC20::TRP1 | This study |
| VMY34 | MATaCEN3-tetO336::URA3 leu2::tetR-GFP::LEU2 trp1::YFP-TUB1::TRP1 cdc20::pMET3-CDC20::TRP1 bir1-17 | This study |
| VMY36 | MATascc1-73 leu2::tetR-GFP::LEU2 pds1::PDS1-myc18::LEU2 ura3::tetOs::URA3 | This study |
| VMY48 | MATapds1::PDS1-myc18::LEU2 ura3-1::tetOs::URA3 leu2-3::tetR-GFP::LEU2 bir1-17::NATRMX | This study |
| VMY56 | MATaPds1::PDS1-myc18::LEU2 ura3-1::tetOs::URA3 leu2-3::tetR-GFP::LEU2 | This study |
| VMY113 | MATabir1-17::NATRMX CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| VMY137 | MATapds1::PDS1-myc18::LEU2 ura3::pGAL-CDC6::URA3 | This study |
| VMY132 | MATapds1::PDS1-myc18::LEU2 bir1-17::NATRMX ssc1Δ::TRP1 leu2::LEU2::pGAL-SCC1 | This study |
| VMY134 | MATapds1::PDS1-myc18::LEU2 ssc1Δ::TRP1 leu2::LEU2::pGAL-SCC1 | This study |
| VMY139 | MATapds1::PDS1-myc18::LEU2 ura3::pGAL-CDC6::URA3 bir1-17::NATRMX | This study |
All strains are in the W303 background: ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d2 Gal+.
Biochemical and immunological techniques.
For the synchronization of cells in G1, 1.25 μg/ml α-factor was used. Standard assays were performed as previously described (26). This involved α-factor treatment for 2 h, followed by the release of the α-factor arrest by filtration and a wash with prewarmed medium, followed by being shaken in fresh YPD medium at 26 or 37°C as indicated. To prevent cells from entering the next cell cycle, α-factor was added back (7.5 μg/ml) when small buds appeared in the majority (>80%) of cells. This procedure was modified for the depletion of cohesin (using pGAL-SCC1) or CDC6 (using pGAL-CDC6) as described elsewhere (19). Briefly, cells were grown in YP medium containing 2% raffinose and 2% galactose at 26°C, treated with α-factor for 50 min, filtered as described above and transferred to fresh YPD medium containing α-factor for a further 2 h, and finally released into fresh YPD at 37°C as described above. Where cells were released into YPD containing nocodazole, the drug was used at a concentration of 30 μg/ml. Cell cycle progression was studied by monitoring budding and Pds1 levels in strains containing PDS1-myc18. Samples were taken during a 180-min time course for immunoblotting, lysing cells either by using glass beads as described in King et al. (19) or by using sequential NaOH and trichloroacetic acid treatment (23). Pds1-myc18 was detected with an anti-myc antibody (c-myc A-14; sc-789; Santa Cruz Biotechnology). Histone H3 phosphorylated on serine-10 was detected with an anti-phospho-histone H3 (Ser10) from Upstate Biotechnology (no. 06-570), total histone H3 was detected using ab-1791 (Abcam), and Cdc28 was detected with an anti-Cdc28 antibody from Santa Cruz Biotechnology (sc-6709).
Immunoprecipitation and protein kinase assay.
Cultures of mid-log-phase cells grown at 37°C were collected and lysates were prepared and immunoprecipitated, and where indicated, in vitro kinase assays were carried out as previously described (5). Ipl1-myc was immunoprecipitated using an anti-myc antibody (sc-789; Santa Cruz Biotechnology), and recombinant glutathione S-transferase (GST)-Dam1 was used as a kinase substrate at 5 μg/reaction. Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to Immobilon-P membranes (Millipore), and then treated with the indicated antibodies (typically at a 1:1,000 dilution), followed by an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. The detection of HRP-labeled secondary antibodies utilized enhanced chemiluminescence that was detected by exposing the membrane to X-ray film. Kinase assays were processed similarly, and the incorporated 32P was detected by exposure to X-ray film. Films from either Western blotting or kinase assays were scanned using an Epson V750 scanner in transmissive mode, and band intensities were quantified using Image J (http://rsb.info.nih.gov/ij/; National Institutes of Health, Bethesda, MD).
Microscopy.
The time-lapse fluorescence microscopy of live cells for monitoring chromosome biorientation was performed as previously described (34). Time-lapse images of cells released from G1 (α-factor) to metaphase arrest (Cdc20 depletion) at the restrictive temperature for the bir1-17 mutant (37°C) were collected for 4 min using a DeltaVision RT microscope (Applied Precision), a UplanSApo ×100 objective lens (numeric aperture, 1.40; Olympus), SoftWoRx software (Applied Precision), and a CoolSnap HQ (Photometrics) charge-coupled device camera. Seven Z sections (0.3 μm apart) were acquired and subsequently deconvoluted, projected as two-dimensional images, and analyzed with SoftWoRx software. Yellow fluorescent protein (YFP) and green fluorescent protein (GFP) signals were visualized with a JP3 filter set, and cyan fluorescent protein (CFP) and GFP signals were visualized with an ET filter set. Sister CEN3 centromeres that remained unseparated at one end of the metaphase spindle were scored as mono-oriented, whereas sister CEN3 centromeres that showed dynamic separation and reassociation or that remained separated for the duration of the time-lapse sequence were scored as bioriented. When fixed cells were imaged to monitor biorientation, samples of cells were fixed in 2% paraformaldehyde for 2 min at room temperature and then resuspended in 0.1 M potassium phosphate buffer (pH 6.6). Images were collected as described above, except that cells were immobilized on an agarose pad and a single exposure was taken from each field. Sister CEN3 centromeres on a metaphase spindle were scored as either unseparated or separated. For imaging live cells without time lapse, a setup identical to that described above was used. Statistical analysis of microscopy data was carried out using Fisher's exact test, and all P values are two tailed.
Chromosome loss assay.
The chromosome loss assay was performed by monitoring the loss of a previously described chromosome III fragment (CFIII) carrying URA3 and SUP11 (31) in a colony-sectoring assay, where the loss of CFIII (and, hence, SUP11) leads to a red colony color due to the failure to suppress the ade2-1 mutation. K5043 (BIR1 wild-type strain) and its bir1-17 derivative (VMY113) were grown in minimal medium lacking uracil to mid-log phase at 26°C, and then cells were washed and resuspended in YPD medium lacking additional adenine. After suitable dilutions were plated on unsupplemented YPD agar, CFIII loss was monitored visually after growth at 26°C for 6 days by the production of red sectors. The CFIII loss rate per cell division was calculated by dividing the number of half-sectored red colonies (i.e., at least 50% red) by the total number of colonies (excluding any completely red colonies derived from cells that already had lost CFIII before plating).
RESULTS
A conditional bir1 allele conferring chromosome instability and showing genetic interactions with ipl1 and sli15 mutants.
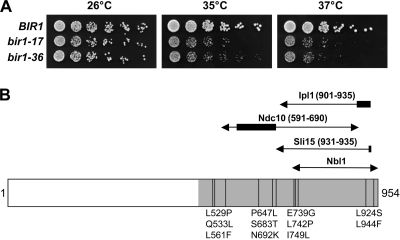
To address whether Bir1 is required for efficient chromosome biorientation, we generated temperature-sensitive bir1 mutants by error-prone PCR, focusing mutagenesis on the C-terminal domain that had been shown previously to be sufficient for providing the essential function of Bir1 (42). Two alleles (bir1-17 and bir1-36) were obtained that supported normal proliferation at 26°C but showed a reproducible temperature-sensitive lethal phenotype at 37°C on centromeric plasmids, which was retained when these alleles were used to replace the genomic copy of BIR1 (Fig. 1). However, bir1-17 was chosen for study because of its stronger temperature-sensitive phenotype. Of the amino acid substitutions encoded by bir1-17 (Fig. 1), two (L924S and L944F) fall within the minimal region required to provide the essential function of Bir1 (residues 876 to 954 [42]). Both of these substitutions affect conserved positions, and one of them (L924S), which also was identified in a bir1 allele recently described by Shimogawa et al. (30), changes a residue predicted to contribute directly to the triple-helical interaction between Nbl1, Sli15, and Bir1 (25). To examine chromosome stability in a bir1-17 strain, we made use of a colony-sectoring assay in which the loss of a supernumerary chromosome III fragment could be quantitated. Table 2 shows that chromosome loss at 26°C in the bir1-17 strain was elevated 18.5-fold relative to that of the control, which is consistent with a role for Bir1 in promoting efficient chromosome segregation.
FIG. 1.
Characterization of the bir1-17 allele. (A) Temperature sensitivity of the bir1-17 and bir1-36 alleles. Equivalent 10-fold dilutions of wild-type BIR1 (AY925), bir1-17 (VMY1), and bir1-36 (VMY7) strains were spotted onto YPD agar and grown for 2 days at 26, 35, or 37°C. (B) Location of the amino acid substitutions in bir1-17. The region of the 954-amino-acid Bir1 open reading frame that was mutagenized is shaded, and the 11 amino acid substitutions are indicated. The limits to the regions proposed to interact with Ndc10, Sli15, Nbl1, and Ipl1 from the two-hybrid analysis of Thomas and Kaplan (38) and Nakajima et al. (25) are indicated (the regions denoted by thicker lines abolish the indicated interactions when deleted or mutated). Only residues 876 to 954 are essential, provided a nuclear localization signal is included (42). Unlike the other substitutions, L924S and L944F affect residues that are highly conserved in Bir1 orthologs from other fungi, such as Ashbya gosypii, Saccharomyces bayanus, and Kluyveromyces lactis (not shown).
TABLE 2.
Chromosome loss rates
| Strain | No. of red colonies | No. of half-sectored colonies | Total no. of colonies | Chromosome loss rate per cell divisiona (103) | Fold increase |
|---|---|---|---|---|---|
| Wild type (K5043) | 3 | 2 | 2,380 | 0.84 | |
| bir1-17 (VMY113) | 35 | 39 | 2,540 | 15.6 | 18.5 |
Calculated as half-sectored/(total − red).
The bir1-17 allele next was crossed with several strains carrying mutations in genes encoding cell cycle regulators, and then tetrad analysis was performed to test for synthetic lethal interactions. Double bir1-17 mad2Δ and bir1-17 mad3Δ mutants were viable, indicating that bir1-17 strains do not require a functional spindle assembly checkpoint for survival. However, bir1-17 showed synthetic lethal interactions with both sli15-3 and ipl1-321, which is consistent with a close functional relationship between Bir1 and these other components of the yeast chromosomal passenger complex. SLI15 originally was isolated through its synthetic lethality with ipl1 mutants (18), so mutations affecting all three pairwise combinations of SLI15, BIR1, and IPL1 are inviable. Like ipl1 mutants (9), bir1-17 was inviable in the absence of functional Cin8, a BimC kinesin involved in yeast mitotic spindle assembly and spindle pole separation (12).
bir1-17 cells are defective in the establishment but not the maintenance of bioriented chromosomes.
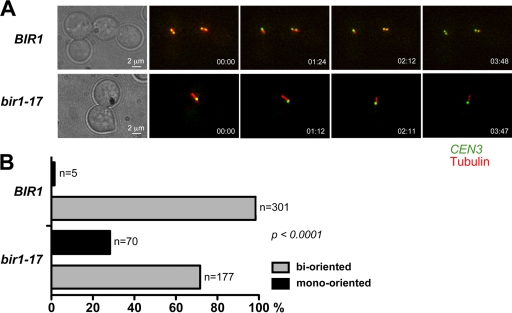
To examine the potential role of Bir1 in chromosome biorientation, we generated control and bir1-17 strains in which we could visualize both microtubules (YFP-TUB1) and sister centromeres on chromosome III by virtue of tetR-GFP binding to an array of tet operators inserted at CEN3 (designated CEN-dot). In such strains, bioriented chromosomes can be scored by the dynamic splitting and reassociation of the two sister CEN-dots in preanaphase cells, whereas mono-oriented chromosomes appear as a single, unresolved CEN-dot that localizes toward one of the two spindle poles (35). These strains also were dependent on the expression of CDC20 from the methionine-repressible MET3 promoter, enabling cells to be arrested in metaphase by Cdc20 depletion in the presence of methionine (39). This experimental design monitors biorientation directly in real time in metaphase rather than relying on failed sister chromatid segregation following anaphase as a readout, which also could result from other types of defect. Cells of the control and bir1-17 strains were arrested in G1 with α-factor, shifted to 37°C, and then released from the α-factor arrest into methionine-containing medium. Once the majority of cells had reached metaphase, chromosome biorientation was monitored by the time-lapse imaging of CEN-dot behavior in the metaphase cells. Figure 2 shows representative images for the control and mutant cells, and representative time-lapse movies are included in the supplemental material. Our data show that bir1-17 cells have a marked, statistically significant (P < 0.0001) defect in chromosome biorientation such that sister CEN3-dots remained mono-oriented in almost 30% of metaphase-arrested cells. Thus, Bir1 clearly is required for efficient chromosome biorientation. Given that haploid yeast cells contain 16 chromosomes, the magnitude of the bir1-17 defect implies that under restrictive conditions virtually no cells in the population will have a complete set of bioriented chromosomes when they undergo anaphase.
FIG. 2.
bir1-17 mutant cells show mono-oriented attachment of sister centromeres to microtubules during metaphase. Wild-type BIR1 (VMY33) and bir1-17 (VMY34) cells containing CEN3-(tetO)336, tetR-GFP, YFP-TUB1, and pMET3-CDC20 were arrested in G1 with α-factor at 26°C and then released to a metaphase block in rich medium (containing 2 mM methionine to deplete Cdc20) at 37°C for 2.5 h. (A) Representative time-lapse images taken from live cells during 4 min. Bioriented chromosomes show the dynamic splitting and reassociation of sister CEN3 centromeres, whereas mono-oriented chromosomes show continuously unresolved sister CEN3 centromeres localized to one end of the spindle. Green, CEN3 labeled with tetR-GFP; red, YFP-tubulin. (B) Quantification of chromosome biorientation in metaphase-arrested cells from multiple time-lapse fields (n is the number of cells scored in each category). Examples of wild-type and bir1-17 time-lapse movies can be found in the supplemental material.
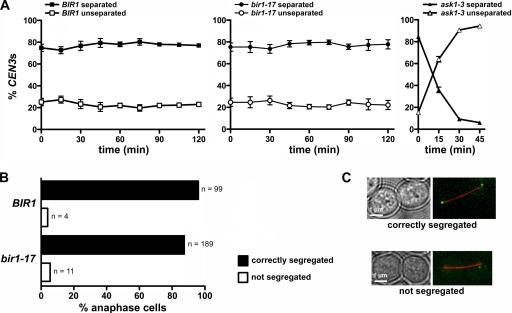
Although required for promoting chromosome biorientation, Ipl1 function is not needed to maintain chromosomes in the amphitelic state once it has been attained (35). We therefore tested whether Bir1 similarly is dispensable for the maintenance of chromosome biorientation by shifting metaphase-arrested control and bir1-17 cells to 37°C and monitoring the behavior of sister CEN3 centromeres during the following 2 h in samples of fixed cells. Neither control nor mutant cells showed any significant decline in the proportion of cells with two clearly separated CEN-dots, which is indicative of bioriented sister CEN3 centromeres (Fig. 3A, left and center). In contrast, an ask1-3 kinetochore mutant showed rapid loss of chromosome biorientation in this assay (Fig. 3A, right). Furthermore, when these metaphase-arrested cells subsequently were released into anaphase at the restrictive temperature, chromosomes segregated with normal efficiency in the bir1-17 mutant compared to the rate of segregation of the control strain (Fig. 3B, C). Thus, Bir1 is required for the establishment but not the maintenance of bioriented chromosomes, which segregate normally during anaphase in the absence of BIR1 function.
FIG. 3.
Bir1 is not required for maintenance of biorientation. (A) Wild-type BIR1 (VMY33), bir1-17 (VMY34), and ask1-3 (T2997) cells with CEN3-(tetO)336, tetR-GFP, YFP-TUB1, and pMET3-CDC20 were arrested in G1 with α-factor, incubated for 2.5 h at 26°C in rich medium containing 2 mM methionine (to deplete Cdc20), and then shifted to 37°C in the same medium (defined as time zero). Samples were fixed with paraformaldehyde at the indicated time points. The graphs show the percentage of cells with separated and unseparated sister CEN3 centromeres on the bipolar spindle, plotting the mean values from three separate experiments (error bars indicate standard errors of the means). Sister CEN3 centromere separation was not scored after 45 min in ask1-3 cells, as they exhibited a collapsed spindle at later times. Note that while essentially all separated sister CEN3 centromeres should represent instances where chromosome III is bioriented, the unseparated sister CEN3 category will contain both mono-oriented chromosomes and bioriented chromosomes where the sister CEN centromeres had transiently reassociated at the point of fixation. (B) Metaphase-arrested cells at 37°C from panel A were released at 37°C in methionine-free medium, and the distribution of sister CEN3 centromeres was scored on elongated, anaphase spindles observed within the first 30 min. Cumulative data from two experiments are shown. (C) Representative images of cells to indicate the phenotypes scored in panel B. Red, tubulin (YFP-TUB1); green, CEN3 (tetR-GFP).
bir1-17 cells respond normally to nocodazole but show a tension checkpoint defect.
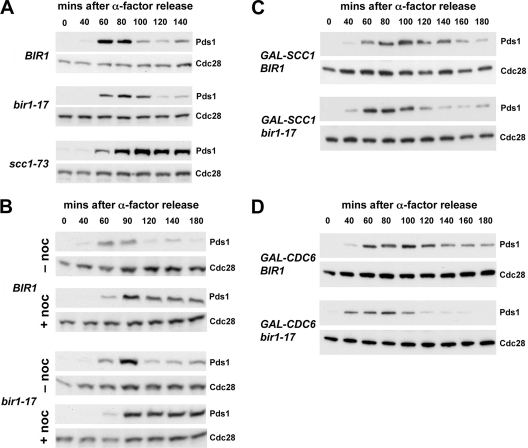
In normal cells, the spindle assembly checkpoint can respond to both unattached kinetochores and incorrect microtubule-kinetochore attachments by delaying anaphase, allowing time for all chromosomes to become correctly bioriented on the mitotic spindle (22). To determine whether the defect in bir1-17 cells results in spindle assembly checkpoint activation, we monitored the kinetics of Pds1 degradation, a marker for anaphase entry (8), in cells synchronized in G1 using α-factor and then released into a new cell cycle at 37°C. Control and bir1-17 cells budded with similar kinetics at 37°C following release from α-factor (not shown) and initiated anaphase after 100 to 120 min, with little or no delay in the mutant cells compared to data for the wild type (Fig. 4A). In contrast, Pds1 was stabilized in the scc1-73 cohesin mutant in which the lack of sister chromatid cohesion relaxes the tension applied to sister kinetochores through their microtubule attachments, indicating that anaphase was delayed as expected. Thus, bir1-17 does not lead to significant checkpoint activation, consistently with the lack of synthetic lethality between bir1-17 and either mad2Δ or mad3Δ (as described above). We next tested whether the bir1-17 mutation affected the normal checkpoint-dependent arrest seen in response to microtubule depolymerization using nocodazole. Figure 4B shows that at 37°C, control and bir1-17 cells both showed a robust and prolonged stabilization of Pds1 in response to nocodazole treatment compared to results for untreated cells.
FIG. 4.
Bir1 is required for the checkpoint response to reduced cohesion. (A) Wild-type BIR1 (VMY56), bir1-17 (VMY48), and scc1-73 (VMY36) strains expressing Pds1-myc18 were arrested in G1 with α-factor and synchronously released into YPD medium at 37°C (the restrictive temperature for bir1-17), and we collected samples at the indicated times. Levels of Pds1-myc18 (Pds1) and Cdc28 (loading control) were monitored by immunoblotting using anti-myc and anti-Cdc28 antibodies, respectively. (B) Wild-type BIR1 (VMY56) and bir1-17 (VMY48) mutant cells were synchronized as described for panel A, except that cells were released in the presence (+ noc) or absence (− noc) of 30 μg/ml nocodazole at 37°C. (C) Wild-type pGAL-SCC1 BIR1 (VMY134) and pGAL-SCC1 bir1-17 (VMY132) cells expressing Pds1-myc18 were arrested with α-factor for 2 h in medium containing galactose and then released at 37°C in medium containing glucose to repress pGAL-SCC1. Pds1 and Cdc28 (loading control) were monitored as described for panel A. Pds1 levels declined faster in pGAL-SCC1 bir1-17 cells than in pGAL-SCC1 BIR1 cells, indicating that Bir1 is required for the checkpoint arrest following pGAL-SCC1 shutoff. (D) Wild-type pGAL-CDC6 BIR1 (VMY137) and pGAL-CDC6 bir1-17 (VMY139) cells were arrested with α-factor for 2 h in medium containing galactose and then released at 37°C in medium containing glucose to repress pGAL-CDC6. Pds1 and Cdc28 (loading control) were monitored as described for panel A. Pds1 levels declined faster in pGAL-CDC6 bir1-17 cells than in pGAL-CDC6 BIR1 cells, indicating that Bir1 is required for the checkpoint arrest in response to unreplicated sister chromatids.
However, since ipl1 mutants specifically are defective in the tension checkpoint (i.e., the activation of the spindle assembly checkpoint mechanism in response to reduced sister kinetochore tension), we next examined whether the bir1-17 mutant affected the tension checkpoint by examining the response to cohesin depletion using strains dependent on pGAL-SCC1 for Scc1 expression. Synchronous cultures of such strains show a profound anaphase delay when grown on glucose to repress SCC1 expression due to the activation of the spindle checkpoint (15), and this appears to be mediated through the phosphorylation of Mad3 by Ipl1 kinase (19). Figure 4C shows that following cohesin depletion, bir1-17 cells failed to stabilize Pds1 to anywhere near the same extent as control cells with normal BIR1, indicating that they are defective in spindle checkpoint activation under these conditions. Similar results were obtained using the scc1-73 mutant as an alternative to cohesin depletion using pGAL-SCC1 (data not shown). We also used strains dependent on pGAL-CDC6 to reduce tension on microtubule-kinetochore attachments by a different means. When such strains are released from α-factor arrest into medium containing glucose, the failure to express Cdc6 blocks DNA replication initiation and causes cells to undergo a spindle checkpoint-dependent arrest in metaphase with unreplicated chromosomes (33). Figure 4D shows that while Pds1 is stabilized in control Cdc6-depleted cells, indicating that the spindle checkpoint has been activated, bir1-17 cells enter anaphase and degrade Pds1. Thus, bir1-17 cells are defective in restraining anaphase through the activation of the spindle assembly checkpoint when tension on microtubule-kinetochore attachments is reduced, either by interfering with sister chromatid cohesion or by blocking DNA replication. This defect is specific for tension checkpoint activation, because bir1-17 cells arrest normally in response to unattached kinetochores generated by treatment with the microtubule-depolymerizing drug nocodazole (Fig. 4B).
Ipl1 localization in the bir1-17 mutant.
Since Ipl1 kinase is thought to promote chromosome biorientation by phosphorylating proteins at the kinetochore-microtubule interface, either the failure to localize Ipl1 kinase correctly or a reduced ability to activate Ipl1 kinase could explain the chromosome biorientation defect in the bir1-17 mutant. We therefore examined the effect of the bir1-17 mutation on Ipl1 localization by generating wild-type and mutant bir1-17 cells in which Ipl1 was tagged with GFP (Ipl1-GFP) and that expressed CFP-tagged Ndc80 and Ctf19 as kinetochore markers.
In both wild-type and bir1-17 cells after 1 h at 37°C, Ipl1-GFP fluorescence was closely associated with the single kinetochore cluster in unbudded cells, while in anaphase cells Ipl1 was localized along the anaphase spindle (data not shown) as previously described (5, 38). In metaphase cells (defined by two kinetochore clusters showing up to ∼1.6 μm separation), most wild-type and the majority of bir1-17 cells showed kinetochore-associated Ipl1-GFP (Fig. 5A). However, in contrast to wild-type cells, no Ipl1-GFP was evident at either of the kinetochore clusters in around one-third of bir1-17 cells (Fig. 5A). This difference between wild-type and bir1-17 populations was statistically significant (P < 0.0001) (Fig. 5B), but since a similar difference was apparent in bir1-17 cells at 26°C (data not shown), it seems unlikely that it is solely responsible for the chromosome biorientation defect seen specifically at the higher temperature. We did not observe any bright foci of Ipl1-GFP at the nuclear periphery seen in a different bir1 mutant (30), possibly due to allele-specific differences or to the use of different fluorescent protein tags in the two studies.
FIG. 5.
Effect of bir1-17 on the cellular localization of Ipl1 kinase in cells in metaphase. Asynchronous, exponentially growing cultures of wild-type BIR1 (T5241) and bir1-17 (VMY14) mutant cells expressing Ipl1-GFP and both CFP-tagged Ctf19 and Ndc80 as kinetochore markers were shifted to 37°C for 1 h, and then images were acquired. (A) Representative differential interference contrast (DIC), Ipl1-GFP (green), Ctf19/Ndc80 (red), and merged live cell images from two wild-type cells showing kinetochore-associated Ipl1-GFP (a and b) and two bir1-17 cells where no kinetochore-associated Ipl1-GFP was evident (c and d). Note, however, that both populations contained examples of both types of localization. (B) Quantitation of kinetochore-associated Ipl1-GFP in metaphase-arrested cells. n is the number of cells scored in each category.
While carrying out this analysis, we noticed that in metaphase cells of both wild-type and mutant strains, although Ipl1-GFP was often in close proximity to one or both kinetochore clusters defined by Ndc80 and Ctf19 fluorescence, it was seldom coincident with the kinetochore clusters. The Ipl1-GFP signal frequently localized to one side of each cluster, often with significant fluorescence surrounding the kinetochore clusters or located between them. When examined by time-lapse microscopy, Ipl1 localization was observed to change considerably from frame to frame and showed larger frame-to-frame differences than the two kinetochore clusters themselves; a representative time-lapse movie is included in the supplemental material. Thus, while Ipl1 clearly is associated with the kinetochore clusters, it behaves differently from a bone fide kinetochore protein.
Ipl1 kinase activity is reduced in the bir1-17 mutant.
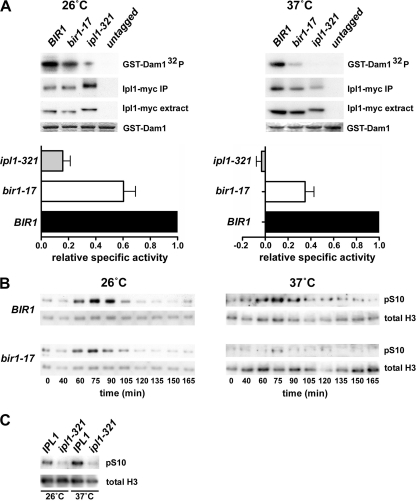
We next adopted two different but complementary approaches to determine whether Ipl1 protein kinase activity shows dependence on Bir1p. First, we generated Ipl1 immune precipitates from both wild-type and bir1-17 cell extracts prepared from cells grown at either 26 or 37°C, and we measured their ability to phosphorylate recombinant GST-Dam1 in an in vitro protein kinase assay, taking care to normalize the activity measured to the Ipl1-myc input. Figure 6A shows that the specific activity of Ipl1 kinase recovered from the bir1-17 strain was reduced when cells were grown at either temperature. However, the defect was more severe when Ipl1 was precipitated from cells grown at 37°C, with a specific protein kinase activity under these conditions of only around one-third of that of Ipl1 from control cells. Second, we examined the phosphorylation of ser-10 on histone H3 in synchronous cultures of control and bir1-17 cells. In yeast, histone H3 phosphorylation on this residue is Ipl1 dependent (13) (Fig. 6C) and represents an in vivo readout of Ipl1 kinase activity. Compared to that of control cells, bir1-17 cells showed greatly reduced histone H3 phosphorylation at 37°C, with a much smaller, but noticeable, reduction in cells grown at 26°C (Fig. 6B). Thus, two different approaches both indicate much lower levels of Ipl1 protein kinase activity in bir1-17 cells at the restrictive temperature.
FIG. 6.
Ipl1 kinase activity is significantly reduced in bir1-17 mutant cells. (A) IPL1 BIR1 (AY925; untagged), IPL1-myc12 BIR1 (T2608), IPL1-myc12 bir1-17 (VMY5), and ipl1-321-myc13 BIR1 (PKY63) cells were cultured at 26 or 37°C for 2 h as indicated, and the myc-tagged Ipl1 was recovered by immune precipitation. The level of tagged Ipl1 in each extract (Ipl1-myc extract) and recovery of Ipl1 (Ipl1-myc IP) was quantified by Western blotting. The in vitro protein kinase activity of the immunoprecipitates was measured by the incorporation of radiolabel from [γ-32P]ATP into recombinant GST-Dam1 as a substrate. Kinase assays were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (GST-Dam1-32P). GST-Dam1 input was visualized by Coomassie staining (GST-Dam1). Histograms show the relative specific activity of Ipl1 kinase in the different immune precipitates. 32P incorporation was normalized to the level of Ipl1-myc recovered in each immunoprecipitate, and the values for the bir1-17 and ipl1-321 strains are expressed as a fraction of the value obtained for the wild-type strain (set to 1.00). Values for the two mutant strains represent the means ± standard errors of the means for four (37°C) or six (26°C) experiments. (B) In vivo phosphorylation of histone H3 is severely reduced in the bir1-17 mutant at 37°C. Wild-type BIR1 (VMY56) and bir1-17 (VMY48) cells were synchronized in G1 using α-factor and then released at either 26 or 37°C as indicated. Samples were analyzed across the first cell cycle by Western blotting to detect the phosphorylation of histone H3 serine 10 and total histone H3 levels. (C) Serine 10 phosphorylation of histone H3 is an Ipl1-dependent phosphorylation event, as confirmed by the use of an ipl1-321 mutant strain.
The bir1-17 biorientation defect increases in severity on recovery from nocodazole treatment.
Despite showing a normal metaphase arrest in response to microtubule depolymerization (Fig. 4B), we noticed that bir1-17 cells nonetheless were hypersensitive to benomyl (Fig. 7A). We therefore examined how proficiently bir1-17 cells could generate bioriented chromosomes when released from nocodazole arrest at 37°C rather than during a normal cell cycle. This experiment was performed using pMET3-CDC20-dependent strains in the presence of methionine such that cells remained arrested in metaphase following nocodazole removal. Surprisingly, we found that under these conditions the biorientation defect was far more severe, with around 80% of mutant cells persistently failing to achieve biorientation during a 120-min time course after nocodazole removal (Fig. 7B, C). In the majority of control BIR1 cells, the marked chromosome became bioriented within 30 to 60 min (Fig. 7B, C). Thus, cells have a greater need for Bir1 when they are forced to attach their sister kinetochores to microtubules during metaphase rather than immediately following CEN replication (20), as in a normal cell cycle.
FIG. 7.
Bir1 inactivation causes a severe biorientation defect following nocodazole (NOC) treatment. (A) Equivalent 10-fold dilutions of wild-type (AY925), bir1-17 (VMY1), and dam1-S257A,S265A,S292A (BP635; nocodazole-hypersensitive control) cells were plated on YPD medium containing 8 μg/ml benomyl. (B) BIR1 (VMY33) and bir1-17 (VMY34) cells containing CEN3-(tetO)336, tetR-GFP, YFP-TUB1, and pMET3-CDC20 were arrested in G1 with α-factor at 26°C and then released for 2.5 h to a metaphase block in rich medium containing 2 mM methionine (to deplete Cdc20) and nocodazole (30 μg/ml; to depolymerize microtubules). Subsequently, nocodazole was removed and the temperature concurrently shifted to 37°C (to inactivate Bir1). Cells showing a clear bipolar spindle were scored for mono-oriented or bioriented sister CEN3 genes based on 2-min time-lapse movies taken during a 30- to 120-min time course (n is the number of cells scored). (C) Examples of cells from panel B at the point of nocodazole release (0 min) showing the absence of spindles and at 90 min showing bipolar spindles. Red, YFP-Tub1; green, sister CEN3 centromeres (tetR-GFP).
DISCUSSION
Despite its known interactions with Sli15 and Ipl1, both of which are required for effective yeast chromosome biorientation, evidence to support a role for Bir1 in promoting chromosome biorientation has been lacking. However, our analysis of the bir1-17 mutant provides strong support for such a role and is consistent with a need for Bir1 to support Ipl1 function. Thus, despite their lack of obvious conservation outside the BIR domains, which are not required for chromosome biorientation in yeast, Bir1 and its metazoan counterpart Survivin may fulfill similar roles. Our findings therefore concur with those reported recently in a concurrent study (30). Although we find that functional Bir1 is required for the efficient establishment of chromosome biorientation, it clearly is dispensable for maintaining the bioriented state either during metaphase or anaphase and so, like Ipl1, its role appears specifically to be in promoting the reorientation of chromosomes that have acquired monopolar (syntelic) attachment.
Although the bir1-17 mutation confers a significant biorientation defect, the phenotype is less severe than that of the ipl1-321 mutant analyzed in the same manner, which shows 60 to 70% mono-orientation (35 and data not shown). This may reflect fortuitous differences in allele strength between ipl1-321 and bir1-17 but also may have deeper significance. Both the bir1-107 allele described by Shimogawa et al. (30) and the nbl1-6 allele (25) have relatively mild phenotypes compared to those of ipl1 mutants, whereas sli15-3 (affecting the IN-box required for Ipl1 kinase activation) has a stronger biorientation phenotype that is comparable to that of ipl1-321 (35). Furthermore, it is possible to isolate very-slow-growing, polyploid Bir1- and Nbl1-deficient cells (25, 29), whereas such survivors are not observed following the deletion of IPL1 or SLI15 (our unpublished data). Perhaps this reflects the difference between mutations that largely abolish Ipl1 catalytic activity (ipl1-321 and sli15-3) and mutations that reduce Ipl1 activity or interfere with its correct targeting (bir1 and nbl1 mutations).
Sandall et al. (29) have proposed that a Bir1-Sli15 complex forms a tension-sensitive linkage at the kinetochore-microtubule interface such that when tension is reduced, the Sli15 IN-box is able to bind to and activate Ipl1 kinase. This complex presumably contains Nbl1, because the Bir1-Sli15 interaction is Nbl1 dependent (25). Our data showing the Bir1 dependence of Ipl1 kinase activity and a requirement for Bir1 to activate the tension checkpoint are consistent with its involvement in a tension-responsive mechanism. However, if Bir1 has a role in switching off Ipl1 activity once a chromosome has bioriented and its sister kinetochores are under tension, bir1 loss-of-function mutants might fail to inactivate Ipl1, leading to the destabilization of kinetochore-microtubule attachments. However, since bir1-17 cells can maintain their chromosomes in the bioriented state when shifted to 37°C, it seems unlikely that Bir1 functions to restrain Ipl1 kinase activity once chromosomes are bioriented. Our finding that Bir1 functions positively for Ipl1 kinase activity also is inconsistent with such a role. Thus, while the tension sensor model is attractive, it may be an oversimplification.
How might Bir1 affect the reorientation of syntelically attached sister chromatids? Our finding that the protein kinase activity of Ipl1 clearly is reduced by the bir1-17 mutation both in vitro and in vivo provides a possible explanation. Bir1 might activate Ipl1 kinase through a direct protein-protein interaction within the chromosomal passenger complex or may support Ipl1 kinase activity through stabilizing the chromosomal passenger complex as proposed for Nbl1 (25), but in either case the reduced kinase activity would compromise the cell's ability to detach incorrectly attached microtubules from kinetochores. Alternatively, Bir1 may be required to localize Ipl1 to tensionless kinetochore-microtubule connections so as to promote their reorientation, and Shimogawa et al. (30) have provided evidence that Bir1 localization responds to the tension status of such connections. Although we find a significant defect in the localization of Ipl1 to the vicinity of clustered kinetochores in the bir1-17 mutant, because this defect is observed at both the restrictive and permissive temperatures, it is not easy to see how it could account for a chromosome biorientation defect that becomes significant only at the restrictive temperature. If the Bir1-dependent localization of Ipl1 is important, then perhaps the need for the Ipl1-dependent reorientation of mono-oriented chromosomes is greater at 37°C than at 26°C. The reduced kinetochore association of Ipl1 in bir1-17 therefore might be tolerable at 26°C but become limiting for efficient biorientation at higher temperatures. However, since the bir1-17 mutation adversely affects Ipl1 kinase activity more severely at 37°C, particularly in vivo, it seems likely that the effect of the mutation on Ipl1 kinase activity forms an important part of the biorientation defect. Such a role for Bir1 in Ipl1 kinase activation is consistent with previous work showing that Survivin is needed to promote the full activation of Aurora B in metazoans, at least in vitro (3, 7).
We also have found that Bir1 is a key element in the tension checkpoint, in which the spindle assembly checkpoint mechanism is activated in response to a lack of sister kinetochore tension. Ipl1 may activate the tension checkpoint either by the generation of unattached kinetochores (26) or by the phosphorylation of Mad3 (19), and our observation that Ipl1 is associated with, but not stably and precisely colocalized at, the two clusters of bioriented sister kinetochores is entirely consistent with the recent findings of Shimogawa et al. (30) that the chromosomal passenger complex is delocalized from sister kinetochores under tension. Both the Bir1 requirement for full Ipl1 activity and the failure of Ipl1 to associate with clustered kinetochores in a significant fraction of bir1-17 mutant cells may contribute to the tension checkpoint defect.
The chromosome biorientation defect in bir1-17 cells is enhanced following release from nocodazole arrest, demonstrating a greater dependence on Bir1 under these conditions. This phenotype is highly reminiscent of that shown by the sgo1-100 mutant. Yeast Sgo1 has been proposed to act in the pathway used to monitor sister chromatid tension (15), yet it is a nonessential gene, which is inconsistent with an obligatory role for Sgo1 in normal chromosome biorientation during an unperturbed S phase and contrasts with Bir1, Sli15, Nbl1, and Ipl1, all of which are essential. However, Sgo1 becomes essential for chromosome biorientation following release from nocodazole arrest (14). Chromosomes normally reattach to spindle microtubules very soon after the replication of the centromeric DNA (20). Since Bir1 is more critical for chromosome biorientation upon release from nocodazole, this may indicate that cells have a greater requirement for the Ipl1-dependent correction of mono-oriented chromosomes when spindle formation is delayed relative to replication.
Supplementary Material
Acknowledgments
This research was supported by grants BB/C007077/1 and BB/G003440/1 from the Biotechnology and Biological Sciences Research Council to M.S.
Thanks are due to Tomoyuki Tanaka and members of his laboratory for yeast strains and helpful comments, Etsushi Kitamura and Jean-François Maure for advice on microscopy, Agnieszka Kozak for yeast strains, and Patrick Keating for yeast strains and useful advice.
Footnotes
Published ahead of print on 15 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics. A Cold Spring Harbor Laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Biggins, S., and A. W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 153118-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton, M. A., W. Lan, S. E. Powers, M. L. McCleland, J. Kuang, and P. T. Stukenberg. 2002. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 133064-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouck, D. C., and K. S. Bloom. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA 1025408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buvelot, S., S. Y. Tatsutani, D. Vermaak, and S. Biggins. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheeseman, I. M., S. Anderson, M. Jwa, E. M. Green, J. Kang, J. R. Yates III, C. S. Chan, D. G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111163-172. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., S. Jin, S. K. Tahir, H. Zhang, X. Liu, A. V. Sarthy, T. P. McGonigal, Z. Liu, S. H. Rosenberg, and S. C. Ng. 2003. Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells. J. Biol. Chem. 278486-490. [DOI] [PubMed] [Google Scholar]
- 8.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 931067-1076. [DOI] [PubMed] [Google Scholar]
- 9.Geiser, J. R., E. J. Schott, T. J. Kingsbury, N. B. Cole, L. J. Totis, G. Bhattacharyya, L. He, and M. A. Hoyt. 1997. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell 81035-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz, R. D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74527-534. [DOI] [PubMed] [Google Scholar]
- 12.Hoyt, M. A., L. He, K. K. Loo, and W. S. Saunders. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102279-291. [DOI] [PubMed] [Google Scholar]
- 14.Indjeian, V. B., and A. W. Murray. 2007. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 171837-1846. [DOI] [PubMed] [Google Scholar]
- 15.Indjeian, V. B., B. M. Stern, and A. W. Murray. 2005. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307130-133. [DOI] [PubMed] [Google Scholar]
- 16.Jeyaprakash, A. A., U. R. Klein, D. Lindner, J. Ebert, E. A. Nigg, and E. Conti. 2007. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131271-285. [DOI] [PubMed] [Google Scholar]
- 17.Kang, J., I. M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C. S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. H., J. S. Kang, and C. S. Chan. 1999. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1451381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, E. M., N. Rachidi, N. Morrice, K. G. Hardwick, and M. J. Stark. 2007. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 211163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura, E., K. Tanaka, Y. Kitamura, and T. U. Tanaka. 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 213319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotwaliwale, C. V., S. B. Frei, B. M. Stern, and S. Biggins. 2007. A pathway containing the Ipl1/Aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell 13433-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May, K. M., and K. G. Hardwick. 2006. The spindle checkpoint. J. Cell Sci. 1194139-4142. [DOI] [PubMed] [Google Scholar]
- 23.Mekhail, K., J. Seebacher, S. P. Gygi, and D. Moazed. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 9135-45. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima, Y., R. G. Tyers, C. C. Wong, J. R. Yates III, D. G. Drubin, and G. Barnes. 2009. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 201772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinsky, B. A., C. Kung, K. M. Shokat, and S. Biggins. 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 878-83. [DOI] [PubMed] [Google Scholar]
- 27.Ruchaud, S., M. Carmena, and W. C. Earnshaw. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8798-812. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Sandall, S., F. Severin, I. X. McLeod, J. R. Yates III, K. Oegema, A. Hyman, and A. Desai. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 1271179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimogawa, M. M., P. O. Widlund, M. Riffle, M. Ess, and T. N. Davis. 2009. Bir1 is required for the tension checkpoint. Mol. Biol. Cell 20915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spencer, F., and P. Hieter. 1992. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 898908-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark, M. J. R. 2007. Studying essential genes: generating and using promoter fusions and conditional alleles, p. 79-102. In I. Stansfield and M. J. R. Stark (ed.), Methods in microbiology, 2nd ed., vol. 36. Elsevier, New York, NY. [Google Scholar]
- 33.Stern, B. M., and A. W. Murray. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 111462-1467. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, K., E. Kitamura, Y. Kitamura, and T. U. Tanaka. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka, T. U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M. J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108317-329. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, T. U., M. J. Stark, and K. Tanaka. 2005. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 6929-942. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, S., and K. B. Kaplan. 2007. A Bir1p-Sli15p kinetochore passenger complex regulates septin organization during anaphase. Mol. Biol. Cell 183820-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlmann, F., D. Wernic, M. A. Poupart, E. V. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103375-386. [DOI] [PubMed] [Google Scholar]
- 40.Vader, G., A. F. Maia, and S. M. Lens. 2008. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Div. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, R. R., J. Al-Bassam, and S. C. Harrison. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 1454-59. [DOI] [PubMed] [Google Scholar]
- 42.Widlund, P. O., J. S. Lyssand, S. Anderson, S. Niessen, J. R. Yates III, and T. N. Davis. 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell 171065-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon, H. J., and J. Carbon. 1999. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA 9613208-13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.