Abstract
The expression of oncogenic ras in normal human cells quickly induces an aberrant proliferation response that later is curtailed by a cell cycle arrest known as cellular senescence. Here, we show that cells expressing oncogenic ras display an increase in the mitochondrial mass, the mitochondrial DNA, and the mitochondrial production of reactive oxygen species (ROS) prior to the senescent cell cycle arrest. By the time the cells entered senescence, dysfunctional mitochondria accumulated around the nucleus. The mitochondrial dysfunction was accompanied by oxidative DNA damage, a drop in ATP levels, and the activation of AMPK. The increase in mitochondrial mass and ROS in response to oncogenic ras depended on intact p53 and Rb tumor suppression pathways. In addition, direct interference with mitochondrial functions by inhibiting the expression of the Rieske iron sulfur protein of complex III or the use of pharmacological inhibitors of the electron transport chain and oxidative phosphorylation was sufficient to trigger senescence. Taking these results together, this work suggests that mitochondrial dysfunction is an effector pathway of oncogene-induced senescence.
Mitochondria are central to cell metabolism and energy production. High-energy electrons coming from the oxidation of different carbon sources such as glucose and fatty acids enter the mitochondrial electron transport chain as reduced equivalents, and their energy gradually is converted into a proton gradient. Mitochondria use this gradient to synthesize ATP that later is used for biosynthetic reactions (9, 30). Mitochondria also control decisions for life and death. Changes in mitochondrial membrane permeability lead to the release of proapoptotic mediators that can kill cells with DNA damage or activated oncogenes (16). In this way, mitochondria control one of the major tumor suppressor responses: apoptosis (27). Some oncogenes, such as RasV12, STAT5, and Bcl2, have antiapoptotic activity, and some cell types have a high apoptosis threshold. Another tumor suppressor response, called cellular senescence, serves as a fail-safe mechanism against the transforming activity of antiapoptotic oncogenes (29, 40, 43). However, currently it is unknown whether mitochondria also can play a role in oncogene-induced senescence (OIS).
OIS is phenotypically similar to the senescence response triggered by short telomeres, also known as replicative senescence (6). Replicative senescence is, in essence, the consequence of a DNA damage response triggered by short telomeres (11). OIS also involves the DNA damage response (2, 15, 28), but the mechanism of DNA damage and the contribution of mitochondria to it are unclear. It has been demonstrated that mitochondria play a critical role in replicative senescence, and several mitochondrial changes, including an increase in the production of reactive oxygen species (ROS), were reported in cells with short telomeres (34, 35). Mitochondrion-derived ROS contribute to the senescent phenotype by damaging the DNA (35) and therefore amplifying the DNA damage signals originally caused by short telomeres. We reasoned that a similar amplifying mechanism involving the mitochondria could operate in cells expressing oncogenes.
Here, we use Ha-RasV12, an oncogenic allele of Ha-Ras, to study the role of mitochondria in OIS. RasV12 is a very important human oncogene and was the first linked to the senescence program (43). We report that oncogenic ras induces an increase in mitochondrial mass, mitochondrial DNA, and mitochondrial superoxide production before any sign of senescent cell cycle arrest. With time, these mitochondrial changes evolved into a severe mitochondrial dysfunction characterized by a further increase in ROS production, the accumulation of depolarized mitochondria around the cell nucleus, a decrease in ATP, and the activation of AMPK. The mechanism of the increase in mitochondrial mass and ROS in response to oncogenic ras was found to be dependent on either p53 or Rb. In addition, direct interference with mitochondrial functions by downregulating the mitochondrial Rieske iron sulfur protein (RISP) or by using pharmacological inhibitors of oxidative phosphorylation induced senescence. We suggest that the senescence effector mechanism acting downstream of p53 and Rb involves mitochondrial dysfunction.
MATERIALS AND METHODS
Cells, chemicals, and retroviruses.
Normal human diploid IMR90 fibroblasts (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Logan, UT) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin G-streptomycin sulfate (Invitrogen). Rotenone and oligomycin were purchased from Sigma. Retroviruses pBabe, pBabeRasV12, pWZL, pWZLRasV12, pLPC, and pLPCE1A were described by Serrano et al. (43), pRetroSuper-shp53 and pRetroSuper-shGFP were described by Voorhoeve and Agami (47), and pLXSNE7 was a gift from D. Galloway. pSIRENshRISP was a gift from N. Chandel (5). MSCVshp16 was a gift from S. W. Lowe (32).
mtDNA quantitation.
Total DNA was extracted as described previously (7). The real-time PCR mixture contained 30 ng of total DNA, forward and reverse primers (250 nM), deoxynucleoside triphosphates (0.2 mM), SYBR green (0.33×), buffer for Jump Start Taq supplemented with 2.5 mM MgCl2, and Jump Start Taq (0.5 U; Sigma) in a final volume of 20 μl. After denaturation at 95°C for 10 min, samples went through 40 cycles of amplification (20 s at 95°C, 20 s at 58°C, and 30 s at 72°C). A dissociation protocol followed the amplification program to characterize the amplified product(s). Quantitative PCR (qPCR) was performed using a LightCycler 480 (Roche Applied Science, Canada), and relative quantification of mitochondrial DNA (mtDNA) over nuclear DNA (nuDNA) levels was determined using the ΔΔCp method, where Cp is the crossing point. Experiment was performed three times in triplicate. The primers used were the following: mtDNA PCR (within D-loop), forward, GATTTGGGTACCACCCAAGTATTG; reverse, GTACAATATTCATGGTGGCTGGCA; and nuDNA PCR (within the TBP nuclear region on chromosome 6), forward TTCACTTCCCCTTGGCCACAACAT; reverse, TGTTCCATGCAGGGGAAAACAAGC.
Cell proliferation and senescence determination.
To determine cell proliferation rates, we estimated cell counts at different times after plating using a crystal violet retention assay (14). Bromodeoxyuridine-propidium iodide (BrdU/PI) staining and fluorescence-activated cell sorting (FACS) was done according to Ferbeyre et al. (14). Data were analyzed by the software CellQuest (version 3.3). Senescence-associated β-galactosidase (SA-β-gal) activity was assayed as described previously (14). Data were quantified from 200 cells counts in three independent experiments (n = 3).
Protein expression.
To prepare total cell protein, cells were collected by trypsinization, washed with phosphate-buffered saline (PBS), lysed in sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 100 mM dithiothreitol), and boiled for 5 min. For Western blotting, 25 μg of total protein was separated on SDS-polyacrylamide gel electrophoresis gels and transferred to Immobilon-P membranes (Millipore, Bedford, MA). We used the following primary antibodies: anti-α1-α2-AMPK (2532; 1:1,000; Cell Signaling), anti-phospho-acetyl coenzyme A carboxylase (AAC) (3661; 1:1,000; Cell Signaling), anti-p53 (9282; 1:1,000; Cell Signaling), anti-p21 (Sc-397, 1:200, Santa Cruz Biotechnology), anti-HDM2 (4B11; 1:250; A. Levine), anti-H-Ras (F235) (sc-29; 1:200; Santa Cruz Biotechnology), anti-Rb (G3-245; 1:500; BD), anti-RISP (A21346; 1:1,000; Molecular Probes), antitubulin (T5168; 1:5,000; Sigma), and anti-phospho-Thr172-AMPK (40H9; 1:1,000; Cell Signaling). Signals were revealed after incubation with anti-mouse or anti-rabbit secondary antibodies coupled to peroxidase (Amersham Pharmacia, Amersham, United Kingdom) by using enhanced chemiluminescence (Amersham Pharmacia).
Fluorescence microscopy.
For fluorescence microscopy, IMR90 (5 × 104) cells were plated on coverslips placed on 6-well plates (Costar, Corning, NY). Twenty-four hours after plating, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Subsequently, the cells were washed in PBS and permeabilized using ice-cold 0.2% Triton X-100 in PBS-3% bovine serum albumin (BSA) solution for 5 min. The cells then were washed three times with PBS-3% BSA and incubated for 1 h at room temperature with the following primary antibodies: anticatalase (ab1877-10; 1:200; Abcam), anti-γH2AX (05-636; 1:200; Upstate), and anti-PML (sc-966; 1:200; Santa Cruz Biotechnology).
For 8-hydroxy-2′-deoxyguanosine (8-OHdG) detection, cells were incubated with 2 M HCl for 20 min at room temperature after fixation. After the removal of HCl, 0.1 M sodium borate, pH 8.5, was added for 2 min. Cells were washed with PBS and permeabilized with 0.2% Triton X-100 in PBS-3% BSA solution for 5 min at room temperature. Cells then were washed three times with PBS-3% BSA. Incubation with primary antibody against 8-OHdG (ab26842; 1:50; Abcam) was performed overnight at 4°C in a humidification chamber. We tested this protocol with cells treated with 100 μM hydrogen peroxide (see Fig. S6 in the supplemental material).
After primary antibodies cells were washed three times with PBS-3% BSA solution, they were incubated with the appropriate secondary antibody (1:5,000) (Alexa Fluor 488-goat anti-mouse, Alexa Fluor 488-goat anti-rabbit, Alexa Fluor 568-goat anti-mouse, or Alexa Fluor 568-goat anti-rabbit antibody) for 1 h at room temperature. Cells then were washed three times with PBS alone and once with PBS containing 300 nM 4′,6′-diamidino-2-phenylindole (DAPI) for 10 min. Images from independent fields were captured and processed with the software Metamorph. Each experiment was repeated three times, and representative fields are presented in each figure.
Determination of mitochondrial mass, mitochondrial membrane potential, and ROS.
To measure mitochondrial mass, we used MitoFluor Green (M-7502; Molecular Probes, Eugene, OR). Cells were incubated for 15 min with 100 nM MitoFluor Green and washed with Hanks' balanced salt solution (HBSS; Gibco), and the intensity of labeling was measured by FACS or immunofluorescence.
To measure mitochondrial membrane potential, we used the cationic dye JC-1 (Molecular Probes). Cells were incubated with 0.77 μM JC-1 for 20 min and washed with HBSS (Gibco).
To measure hydrogen peroxide cells, we used 2 μM dichlorodihydrofluorescein diacetate (DCFDA; Molecular Probes) using conditions indicated by the manufacturer. To measure superoxide, we used 1 μM dihydroethidium (DHE) or 5 μM MitoSox from Molecular Probes. Fluorescence probes were detected by immunofluorescence microscopy or FACS. Nuclei were visualized by 4 μg/ml Hoechst stain (Molecular Probes).
Determination of ATP concentration and glucose consumption.
The ATP concentration was determined using an ATP determination kit, and the data were normalized for total cell protein (Invitrogen). For glucose consumption, 5 × 104 cells 6 days postselection were plated into 6-well plates, and media were taken 24, 48, and 72 h later. The glucose concentration in a medium was measured using the QuantiChrom glucose assay kit (BioAssay Systems). The rate of consumption was calculated by dividing the concentration of glucose measured 72 h after plating by the concentration measured 24 h after plating. Each concentration was normalized to cell numbers measured by a Casy cell counter.
qPCR.
Total RNA was extracted from IMR90 cells expressing a vector control or RasV12 at day 8 after selection using Tryzol (Invitrogen, Burlington, Ontario, Canada). Reverse transcription was performed on 2 μg of total RNA using the RevertAid H first minus-strand cDNA synthesis kit (MBI Fermentas, Burlington, Ontario, Canada) and oligo(dT) as recommended by the manufacturer. The resulting cDNA was diluted 10 times to a final volume of 200 μl prior to real-time PCR. Each real-time PCR amplification reaction mixture contained the reverse transcription dilution (2 μl), forward and reverse primers (250 nM), MgCl2 (4 mM), deoxynucleoside triphosphates (0.2 mM), SYBR green (0.33×), buffer for Jump Start Taq, and Jump Start Taq (0.5 U; Sigma) in a final volume of 20 μl. After denaturation at 95°C for 10 min, samples went through 40 cycles of amplification (30 s at 95°C, 30 s at 58°C, and 30 s at 72°C). A dissociation protocol followed the amplification program to characterize the amplified product(s). qPCR was performed using a LightCycler 480 (Roche Applied Science, Canada), and the relative quantification of gene expression levels above those of the levels of the housekeeping gene β-actin was determined using the ΔΔCp method. The experiment was performed three times in triplicate, and a typical result is shown. The primers used were the following: for SOD2, forward, CAGGATCCACTGCAAGGAACAA; reverse, GTAAGCGTGCTCCCACACATCA; for β-actin, forward, AGAGCTACGAGCTGCCTGAC; reverse, AAAGCCATGCCAATCTCATC; for NRF1, forward, GCTACAGTCACTATGGCGCTTAACA; reverse, ACGATCTGTCCCCCACCTTGTA; for NRF2a, forward, TGAAATACTCCAGCCATGACTAAAAGA; reverse, CGCGTAGGTTTGTTCTACAATGC; for TFAM, forward, CCCATATTTAAAGCTCAGAACCCAGA; reverse, TTGAATCAGGAAGTTCCCTCCA; for PGC1β, forward, GGCAGGCCTCAGATCTAAAAGTCA; reverse, GCATCATGGGAGCCTTCTTGTC; and for PGC1α, forward, GCTACGAGGAATATCAGCACGA; reverse, AATCACACGGCGCTCTTCAA.
RESULTS
Oncogenic ras induces an increase of mitochondrial mass and the production of ROS in normal human fibroblasts.
Ras-induced senescence can be inhibited by using antioxidants or growing cells in low-oxygen conditions (8, 25, 33, 51). These treatments act in part by preventing the accumulation of ROS. The source of ROS and the mechanism linking oncogenic activity to their production presently are not clear. To investigate these issues, we introduced oncogenic ras into normal human IMR90 fibroblasts by retroviral gene transfer using vectors that confer puromycin resistance. We then selected a population of Ras-expressing cells after treatment with puromycin for 2 days. We analyzed these cell populations at two time points. The first one chosen was 2 days after selection (4 days after infection), a time at which Ras-expressing cells were not senescent but showed signs of hyperproliferation, as reported before (14, 26). The second one was 8 days after selection, when all cells are arrested and senescent (14, 26). First, we stained the cells with the mitochondrial superoxide indicator MitoSox. We found that Ras-expressing cells exhibited a dramatic increase in superoxide production according to this marker at day 2 after selection (Fig. 1A). This superoxide signal in mitochondria was even higher in fully senescent cells (day 8 after selection) (Fig. 1A). Taken together, these data suggest that mitochondria in cells expressing oncogenic ras are an important source of superoxide.
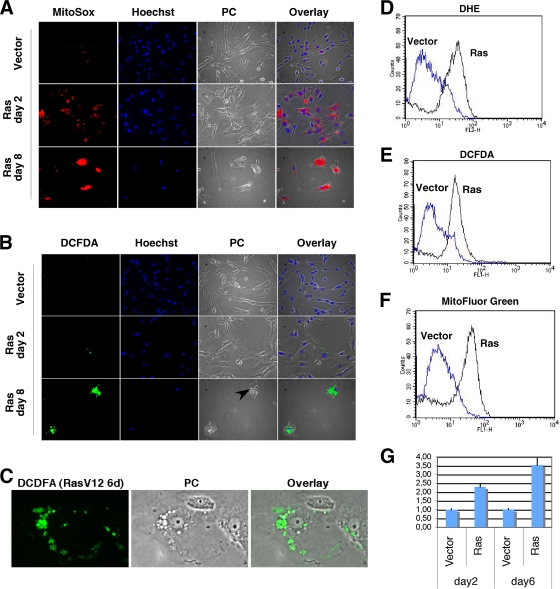
FIG. 1.
Mitochondrial changes in Ras-expressing cells. (A) MitoSox fluorescence images of Ras-expressing cells and cells with a control vector obtained from cells 2 and 8 days after selection. (B) DCFDA fluorescence images of cells as described for panel A. Nuclei were counterstained with Hoechst 33342. PC means phase-contrast images, and the arrows point to vacuoles that characterize the cytoplasm of Ras-senescent cells. (C) Magnified immunofluorescence image to visualize the vacuoles in the cytoplasm of Ras-senescent cells stained with DFCDA. (D to F) Fluorescence intensity of cells analyzed by flow cytometry and stained with DHE, DCFDA, or Mitofluor Green as indicated. (G) Real-time PCR measuring the relative levels of mtDNA versus nuDNA in cells with control vector or its derivative expressing oncogenic ras.
The superoxide anion in the mitochondria can be converted into hydrogen peroxide (H2O2) by manganese superoxide dismutase (MnSOD) (49). We studied the accumulation of H2O2 using the fluorescent probe DCFDA. In cells expressing oncogenic ras, we did not detect a large accumulation of H2O2 with DCFDA early after the introduction of oncogenic ras (2 days after selection) (Fig. 1B). However, 8 days after selection, senescent cells accumulated H2O2 around the nucleus in vacuolated compartments (Fig. 1B). These vacuoles are visible in the light microscope in Ras-senescent cells (Fig. 1B and C), and their identity and role remain to be investigated. Labeling mitochondria with green fluorescent protein-MnSOD did not show a predominant localization of mitochondria into these vacuoles (see Fig. S1 in the supplemental material). We confirmed and quantified the accumulation of ROS seen by immunofluorescence using flow cytometry with cells obtained 8 days after selection. In this case, superoxide was detected using DHE, another superoxide indicator (Fig. 1D), and H2O2 was measured with DCFDA (Fig. 1E). Taken together, the data show that cells expressing oncogenic ras have an increased generation of both anion superoxide and hydrogen peroxide.
Since the staining of Ras-expressing cells with MitoSox suggests that mitochondria are the source of this ROS, we used MitoFluor Green to study how oncogenic ras affects mitochondria. This compound preferentially accumulates in mitochondria, becoming fluorescent in their lipid environment, and thus it gives an estimate of the mitochondrial mass in cells. We observed green fluorescence all over the cytoplasm, but it was predominantly perinuclear in both control cells and cells expressing oncogenic ras 2 days after selection (see Fig. S2 in the supplemental material). In cells obtained 8 days after selection, at a time when the entire cell population is senescent, we found a dramatic increase in perinuclear MitoFluor Green fluorescence (see Fig. S2 in the supplemental material). These immunofluorescence data were confirmed by flow cytometry, showing again an increase in MitoFluor Green labeling in Ras senescence 8 days after selection (Fig. 1F). To have an independent confirmation of an increase in mitochondria induced by oncogenic ras, we measured the levels of mtDNA by real-time PCR. We found that Ras-expressing cells also have an increase in mtDNA after day 2, when cells are not yet senescent (Fig. 1G).
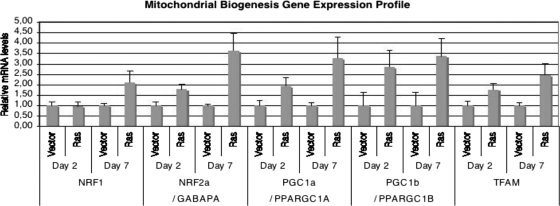
The increase in mitochondrial mass and DNA in cells expressing oncogenic ras may result from the induction of a mitochondrial biogenesis program. This program is known to be controlled by several transcription factors and coactivators (21). In agreement with this idea, oncogenic ras induced the expression of NRF2a, PGC1α, PGC1β, and TFAM but not NRF1 before the cells were senescent, 2 days after the introduction of oncogenic ras (Fig. 2). The expression of these genes was even higher when the cells already were senescent (Fig. 2). Moreover, that the expression of NRF1 also was increased 8 days after selection is consistent with the fact that NRF1 is an NRF2a target gene (37). Taken together, the results indicate that the expression of oncogenic ras led to both mitochondrial ROS production and mitochondrial biogenesis. We view the latter as a programmed response to mitochondrial damage that could help to replace injured mitochondria. However, in cells expressing oncogenic ras, these newly formed mitochondria also would be exposed to the oncogenic stress and also eventually will be damaged. Therefore, we next evaluated mitochondrial functionality after several days of ras expression, when cells already are senescent.
FIG. 2.
Oncogenic ras induces multiple regulators of mitochondrial biogenesis. Results are shown for real-time PCR measuring mRNA levels of the indicated genes at days 2 and 7 after selection with a retroviral vector control or its derivative expressing oncogenic ras.
Collapse of mitochondrial membrane potential in cells expressing oncogenic ras.
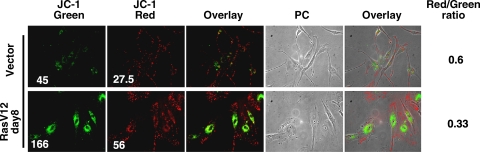
Functional mitochondria maintain a gradient of protons and ions across the inner mitochondrial membrane. High levels of ROS may damage mitochondria, leading to the opening of mitochondrial pores and allowing the influx of protons and ions, which leads to a loss of membrane polarization (13, 52). Moreover, superoxide in the mitochondria can cause a loss of membrane potential by stimulating uncoupling proteins (13). Therefore, we next studied mitochondrial polarization in ras-expressing cells to evaluate overall mitochondrial health. For this, we used the cationic dye JC-1, which localizes to normal mitochondrial membranes as red aggregates and turns into a green monomer upon membrane depolarization.
We found, using immunofluorescence, that mitochondria in Ras-senescent cells were mostly green (depolarized) and perinuclear (Fig. 3). This indicates a massive mitochondrial collapse around the nucleus in senescent cells. The ratio of red/green mitochondria was calculated by FACS, confirming that Ras-expressing cells have an increase in the proportion of depolarized mitochondria (Fig. 3). Since these cells keep generating large quantities of ROS (Fig. 1), we think that the loss of membrane potential is not due to a lack of electrons entering the electron transport chain but to a loss of the proton gradient due to increased mitochondrial membrane permeability. In turn, the loss of membrane potential may increase ROS production (4). For this reason, it remains challenging to determine what comes first in cells expressing oncogenic ras, superoxide production or the loss of membrane potential. Whatever the mechanism, the data indicate that mitochondria in Ras-senescent cells are dysfunctional. They produce more ROS and should be compromised in ATP generation due to the loss of the proton gradient used by the F1ATPase for ATP synthesis. Also, mitochondrial biogenesis, which normally would compensate for a mitochondrial dysfunction, actually increases the pool of dysfunctional mitochondria due to the continuous nature of the oncogenic stress.
FIG. 3.
Mitochondrial dysfunction in RasV12-expressing cells: JC-1 fluorescence. Red fluorescence indicates a normal membrane potential and green fluorescence indicates mitochondrial membrane depolarization. A decrease in the ratio of red/green fluorescence measured by FACS is a measure of the extent of mitochondrial membrane depolarization. PC, phase-contrast images.
Oxidative DNA damage and metabolic checkpoint in OIS.
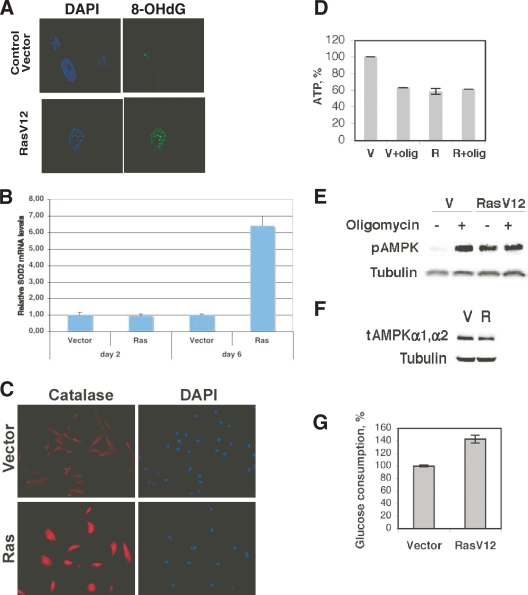
The location of altered mitochondria around the cell nucleus suggests that they release ROS in the proximity of the nuclear envelope, where they can gain access to the DNA. DNA damage in the nucleus of normal cell-expressing oncogenes already has been characterized using techniques that detect DNA breaks, and they could arise by either replication stress or oxidative damage (2, 15, 28). To investigate a more direct link between ROS and DNA damage, we used an antibody against 8-OHdG, an oxidation derivative of deoxyguanosine (10). To facilitate the access of the antibody to the modified guanine in the DNA, we adjusted our immunofluorescence protocol by including a DNA denaturation step that potentially could expose 8-OHdG for antibody binding. Consistently with the accumulation of ROS in Ras-expressing cells, their nuclei were strongly stained by this antibody compared to the staining of control cells (Fig. 4A).
FIG. 4.
Oxidative DNA damage during Ras-induced senescence. (A) 8-OHdG detection in Ras-expressing cells. Cells were fixed 8 days after selection with a control vector or its derivative expressing RasV12 and were left untreated or were treated with HCl. 8-OHdG was visualized by indirect immunofluorescence using an antibody against 8-OHdG. Representative images of cells in the population were obtained by immunofluorescence microscopy. Nuclei were visualized by DAPI. (B) Real-time qPCR indicating relative levels of SOD2 mRNA at the designated times. (C) Immunofluorescence for catalase in cells fixed at day 8 post selection. (D) ATP content of control growing cells with an empty vector (V) (set to 100%) or cells expressing oncogenic ras (R). Cells also were treated with oligomycin (olig) at 3.5 μg/ml for 24 h or with vehicle. Data are means ± standard deviations. (E) Activation of AMPK as measured using an antibody against phospho-AMPK (pAMPK). (F) Total levels of the α1 and α2 subunits of AMPK (tAMPK) as measured by immunoblots with a specific antibody. (G) Glucose consumption in cells expressing oncogenic ras or a control vector. Cells were taken at day 6 after selection. The value for control cells was taken as a reference. Data represent the means and standard deviations from four measurements.
To further characterize the degree of oxidative stress in Ras-senescent cells, we measured the expression of antioxidant enzymes. Two days after the introduction of oncogenic ras, the levels of SOD2 were not significantly elevated compared to those of control cells, but they dramatically increased by day 6 after the introduction of oncogenic ras (Fig. 4B). This explains why superoxide levels but not hydrogen peroxide were found to be elevated early in the process of Ras-induced senescence (Fig. 1A). We also measured catalase levels by immunofluorescence and found that it was elevated by day 6 after the introduction of oncogenic ras (Fig. 4C). Since SOD2 and catalase both are known for being induced by oxidative stress (24, 44), these data support the conclusion that normal cells expressing oncogenic ras are under oxidative stress.
Although the role of oxidative damage has been considered very important for the induction of senescence in response to oncogenic ras (8, 25, 33, 51), mitochondrial dysfunction may contribute to senescence through additional mechanisms. In fact, in our hands the use of antioxidants retarded the establishment of OIS but did not prevent it completely (data not shown). The loss of mitochondrial membrane potential we have observed during OIS suggests a defect in oxidative phosphorylation. To investigate this possibility, we measured ATP levels in cells expressing oncogenic ras and control cells 6 days after the introduction of oncogenic ras. Cells expressing oncogenic ras displayed a decrease in ATP levels, which is consistent with the hypothesis that the loss of membrane potential compromised ATP synthesis (Fig. 4D). This decrease in ATP was equivalent to that observed in control cells treated with oligomycin, an inhibitor of mitochondrial ATP synthesis. In addition, oligomycin did not further decrease ATP levels in cells expressing oncogenic ras, confirming once again a state of mitochondrial dysfunction. The oligomycin-resistant ATP levels in Ras-expressing cells presumably are produced by substrate-level phosphorylation.
A decrease in ATP increases the AMP/ATP ratio, which triggers a metabolic checkpoint via the enzyme AMPK (19). In agreement, we found that AMPK was highly active in Ras-senescent cells compared to its activity in control cells, according to a phosphospecific antibody against the active form of α-AMPK (Fig. 4E). Also, the levels of total α-AMPK in Ras-expressing cells were not increased in oncogenic ras (Fig. 4F), which is consistent with the idea that the activation of this enzyme responds to a decrease in ATP levels. It has been reported that AMPK activation mediates a metabolic checkpoint and senescence in cells cultured with a low concentration of glucose (19). Ras-induced senescence occurs in medium having high-glucose content, but it still is possible that a defect in glucose consumption rather than a mitochondrial dysfunction is responsible for the activation of this checkpoint. To eliminate this possibility, we measured glucose consumption in cells expressing oncogenic ras or a vector control. We found that Ras-induced senescence was associated with higher glucose consumption (Fig. 4G) compared to that of control growing cells. Taken together, the data are in agreement with a model proposing that a mitochondrial dysfunction and not a defect in glucose consumption is the underlying cause of the bioenergetic defects of Ras-senescent cells.
p53 and Rb are required for mitochondrial changes and ROS production during Ras-induced senescence.
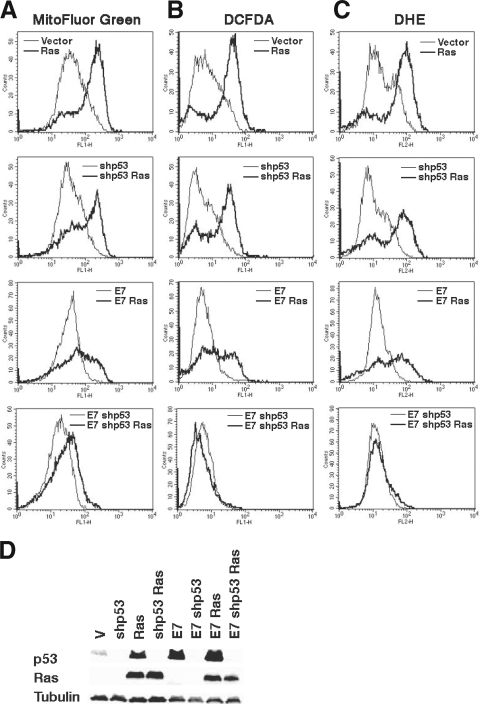
Ras-induced senescence in human cells can be mediated by either the p53 or the Rb family of tumor suppressors (28, 29, 43, 48). This means that oncogenic ras can trigger senescence in human cells where p53 or Rb is disabled but cannot do it if both pathways are inhibited. This well-known fact (28, 29, 43, 48) indicates that the pathway activated by p53 or Rb converges into a common senescence effector mechanism that has yet to be identified. For this reason, we wanted to investigate whether p53 and/or Rb also could mediate the mitochondrial changes and ROS production induced by oncogenic ras. To study the role of p53, we used a small hairpin RNA (shRNA) previously validated to specifically inactivate p53 (47). To study the role of the Rb family, we used the oncoprotein E7. We found that the shRNA against p53 did not prevent the increase in mitochondria, as measured by MitoFluor Green fluorescence, or the ROS accumulation induced by oncogenic ras (Fig. 5A to C, top). On the other hand, E7 was able to partially reduce MitoFluor Green fluorescence and ROS induced by Ras (Fig. 5A to C, middle). Of note, as reported before (28, 29, 43, 48), the shRNA against neither p53 nor E7 rescued the senescence phenotype (see Fig. S3 in the supplemental material), although the biochemical effects of these reagents on the Rb and the p53 pathways indicate that they efficiently inhibited their targets (Fig. 5D). If changes in mitochondria leading to the generation of ROS are an essential part of the mechanism that mediates cell senescence, then it is logical to propose that both p53 and Rb individually are sufficient to promote these changes. In agreement with this idea, when we combined the shRNA against p53 with E7, we totally prevented the accumulation of ROS, the increase in mitochondrial mass (Fig. 5A to C, bottom), and senescence (see Fig. S3A and B in the supplemental material) that oncogenic ras normally induces in normal cells.
FIG. 5.
p53 and Rb pathways can independently connect oncogenic ras signals to the mitochondria. Fluorescence intensity of cells taken 8 days after selection for the indicated vectors and analyzed by flow cytometry after staining with MitoFluor Green (A), DCFDA (B), and DHE (C). (D) Immunoblots for p53, Ras, and tubulin for the cells used for panels A to C. V, empty vector.
E7 has multiple cellular targets, and it also could stabilize p53. Thus, we used an shRNA against p16INK4 (32) alone or in combination with shp53 to evaluate the roles of the p16/Rb and p53 pathways in the mitochondrial changes induced by oncogenic ras. As we found with E7, shp16 was not sufficient to prevent the accumulation of ROS and the increase in mitochondrial mass induced by RasV12, but it did so in cooperation with shp53 (see Fig. S4 in the supplemental material).
The adenoviral oncoprotein E1A is known for blocking Ras senescence (43), facilitating Ras-induced transformation (42). It is known that E1A binds and inhibits Rb (50) and blocks the senescence functions of p53 (43). In particular, E1A inhibits p53 phosphorylation at serine 15 (14), a hallmark of the activation of the DNA damage response during senescence (3, 12). To investigate the effects of E1A on mitochondria and ROS production, we coinfected normal human fibroblasts with E1A and oncogenic ras. We found that E1A inhibited the Ras-dependent induction of mitochondrial mass and the accumulation of ROS (see Fig. S5A to C in the supplemental material). Taken together, our data are consistent with the idea of a senescent mitochondrial effector pathway that can be activated independently by the p53 or the Rb tumor suppressor pathway.
Mitochondrial dysfunction triggers cellular senescence.
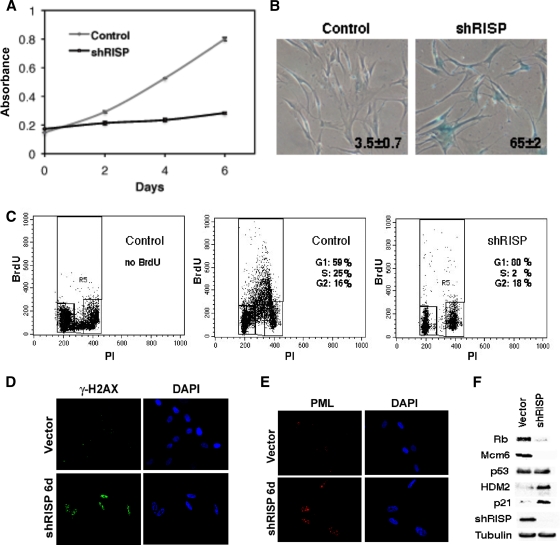
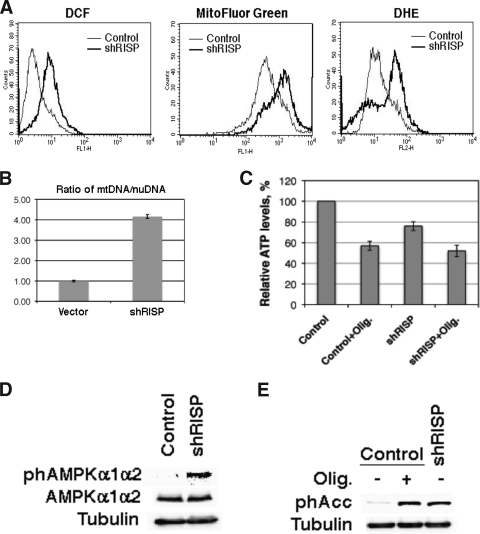
We next asked whether a mitochondrial dysfunction could be sufficient to trigger senescence. First, we knocked down the mitochondrial RISP, which normally transfers electrons from ubiquinol to cytochrome c1 in complex III (18). Shortly after the introduction of the anti-RISP shRNA, cells slowed their proliferation without a noticeable hyperproliferative phase, as is observed with oncogenic ras (Fig. 6A). Six days after the introduction of the shRNA, the cells were arrested with a flat morphology and several senescent markers, such as SA-β-Gal (Fig. 6B), the DNA content of G1-arrested cells (Fig. 6C), low BrdU incorporation (Fig. 6C), DNA damage foci (Fig. 6D), and promyelocytic leukemia (PML) bodies (Fig. 6E). The characterization of the Rb and the p53 pathways also was typical of senescent cells, displaying the presence of hypophosphorylated Rb, low E2F target expression (Mcm6), and the induction of p53 targets such as p21 and HDM2 (Fig. 6F). The mitochondrial profile after RISP knockdown was similar to that of OIS, including increased staining for the ROS markers DCFDA and DHE and for the mitochondrial mass marker MitoFluor Green (Fig. 7A) and higher levels of mtDNA (Fig. 7B). Also, as in cells expressing oncogenic ras, knocking down RISP decreased ATP production (Fig. 7C) and activated the AMPK pathway, as measured with a phosphospecific antibody against AMPK (Fig. 7D) or the AMPK target ACC (Fig. 7E). Interestingly, RISP knockdown blocks the arrival of electrons into complex III and inhibits ROS production by this complex during hypoxia (5). Therefore, the observed increase in ROS after RISP knockdown should be the consequence of electron transfer to molecular oxygen from components of the electron transport chain upstream of complex III.
FIG. 6.
Senescence after knocking down mitochondrial RISP. (A) Growth curves. Cells were plated in triplicate 2 days after infection with a retroviral vector expressing shRNA control or shRNA against RISP. Cell counts were estimated every 2 days using a crystal violet staining assay. (B) SA-β-Gal assay of cells described for panel A and plated 8 days after infection with the indicated vectors. (C) BrdU/PI profiles of cells described for panel A and plated 8 days after infection. (D and E) Immunofluorescence for γH2AX (D) or PML bodies (E) of cells described for panel A plated 6 days after infection. (F) Immunoblots for RISP and members of the p53 and Rb pathways in cells described for panel A 8 days after infection.
FIG. 7.
Mitochondrial dysfunction after knocking down mitochondrial RISP. (A) Fluorescence intensity of cells taken 6 days after infection with shRNA control of shRNA against RISP and analyzed by flow cytometry after being stained with the indicated fluorescent probes. (B) mtDNA levels in cells described for panel A. (C) Relative ATP levels of cells expressing shRNA control of shRNA against RISP and treated with oligomycin (olig.; 7 μg/ml for 24 h) or vehicle. (D and E) Activation of AMPK as measured using an antibody against phospho-AMPK (phAMPK) (D) or phospho-ACC (phACC) (E).
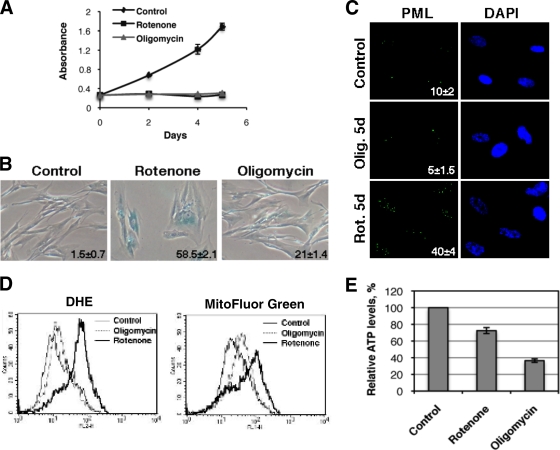
To further characterize the role of mitochondrial dysfunction in senescence, we took advantage of the specific effects that rotenone and oligomycin have on mitochondrial functions. Both inhibitors immediately inhibited cell growth when used at concentrations that do not induce cell killing (Fig. 8A), but rotenone was more efficient at inducing the senescence marker SA-β-Gal (Fig. 8B) and PML bodies (Fig. 8C), the production of superoxide (Fig. 8D), and the increase in mitochondrial mass (Fig. 8D). Rotenone inhibits complex I of the electron transport chain, leading to an increase in the production of superoxide by this complex (36, 38). Since cells still can use complex II to channel electrons into the electron transport chain, rotenone does not compromise ATP production as does oligomycin, an inhibitor of the mitochondrial ATP synthase (Fig. 8E). Thus, blocking mitochondrial functions at the levels of complex I, which leads to ROS production, or at the level of the F1ATPase, which leads to a decrease in ATP synthesis, can induce a partial senescence phenotype. On the other hand, interfering electron transport at the level of complex III leads to ROS production, a decrease in ATP synthesis, and a robust senescence phenotype. Taken together, these results suggest that ROS and ATP shortage contribute distinctly to the senescence phenotype, and these two defects cooperate in the context of Ras-induced senescence.
FIG. 8.
Rotenone and oligomycin induce a cell cycle arrest with some characteristics of senescence. (A) For growth curves, 5 × 104 cells were plated in 12-well plates. Twenty-four hours later, rotenone (1.5 μM) or oligomycin (7 μg/ml) was added (day 0). Drugs were added with fresh medium every 2 days, and cells were counted at the indicated time points. (B) SA-β-Gal was added 5 days after treatment. (C) PML bodies were detected by immunofluorescence. We quantified the number of cells having 5 or more PML bodies, and the numbers are indicated at the bottom right of each panel. Rot. 5d, rotenone administration for 5 days; Olig. 5d, oligomycin administration for 5 days. (D) Fluorescence intensity of cells analyzed by flow cytometry after being stained with the indicated fluorescent probes. (E) Relative ATP levels of cells treated with rotenone or oligomycin.
DISCUSSION
We show here that the expression of oncogenic ras in normal human cells leads to notable changes in mitochondria, characterized by the production of ROS and a loss in mitochondrial membrane potential and mitochondrial biogenesis. These changes appeared before the senescence cell cycle arrest and were totally prevented by disabling the p53 and Rb tumor suppressors, which are known to regulate independently the senescence effector mechanism (28, 29, 43, 48). These mitochondrial alterations may contribute to Ras-induced senescence, because reproducing its effects by RNA interference against a component of the electron transport chain or the use of drugs that interfere with electron transport and oxidative phosphorylation was sufficient to trigger a senescent cell cycle arrest.
Normally, mitochondria use electrons from different carbon sources to generate a proton gradient across the inner mitochondrial membrane. The energy of this gradient is used to generate ATP (9, 30). In Ras-senescent cells, mitochondria are dysfunctional because they allow electrons to partially reduce molecular oxygen, generating high levels of superoxide. Also, most mitochondria in Ras-expressing cells have lost their membrane potential and thus should be compromised for ATP generation. Consistently with this hypothesis, Ras-senescent cells have lower ATP levels than control cells, but their levels are similar to those attained after the inhibition of oxidative phosphorylation with oligomycin. In addition, ATP levels in Ras-senescent cells could not be reduced further by oligomycin. Taken together, the data suggest that the observed drop in ATP levels in Ras-expressing cells is the consequence of mitochondrial dysfunction. Nevertheless, we cannot eliminate the possible contribution of an increase in cellular ATPase activities or metabolic futile cycles to this drop in ATP levels.
We do not think that mitochondrial dysfunction is a direct consequence of Ras signaling. Our results clearly show that in the absence of p53 and Rb, oncogenic ras does not induce mitochondrial dysfunction. A critical question is how p53 and Rb pathways affect mitochondria. Data from the literature suggest several mechanisms. First, p53 can be exported from the nucleus and localize to mitochondria, where it may affect electron transport, permeability, and ROS production (31). Second, several p53 target genes may contribute to mitochondrial changes and/or ROS production (39). Rb also could contribute to mitochondrial accumulation, since it has been reported that Rb was required for mitochondrial biogenesis during erythropoiesis (41). More studies will be required to elucidate the molecular pathways linking p53 and Rb to mitochondria during OIS.
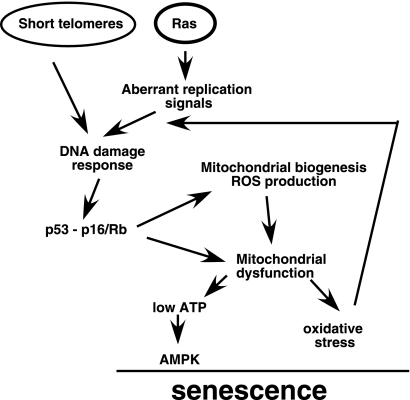
Mitochondrial dysfunctions give rise to cellular stresses that could further activate p53 and Rb. For example, the oxidative stress observed in Ras-senescent cells is a well-known stimulus for p53 activation (8, 46, 51). The DNA damage resulting from the interaction of ROS with nuclear or mtDNA also could activate p53 (1) and Rb (17). Finally, the drop in ATP levels with the consequent activation of AMPK could stimulate p53 as well (19). The result of this interplay is the generation of positive feedback loops that could sustain p53 and Rb activation during senescence. This situation is reminiscent of cells in replicative senescence, where mitochondrial dysfunction was found to play an important role (34). We propose that during replicative senescence, short telomeres constitute the first signal leading to the activation of p53 and Rb. p53 and Rb then affect mitochondria, leading to the generation of ROS and mitochondrial dysfunction, as we have shown here during OIS (Fig. 9).
FIG. 9.
Mitochondrial dysfunction as an effector mechanism of OIS and replicative senescence (short telomeres). The oncogenic activity of the Ras/mitogen-activated protein kinase pathway induces a replication stress (12) and mitochondrial biogenesis leading to DNA damage and the activation of the p53 and Rb pathways. The activity of these tumor suppressors converge in the mitochondria to induce a senescence effector pathway characterized here as a metabolic checkpoint and oxidative stress. This is not a linear pathway, since the effects of mitochondrial dysfunction can activate the p53 and Rb pathways, generating positive feedback mechanisms that we propose contribute to keeping cells senescent.
If OIS involves mitochondrial dysfunction leading to DNA damage and bioenergetic checkpoints, how are these processes circumvented during tumorigenesis? Clearly, any process that helps to restore ATP production and prevent oxidative damage may help to bypass the senescence checkpoint. It is known that cancer cells preferentially use glycolysis instead of respiration to obtain energy (Warburg effect) (20). This metabolic rearrangement would achieve precisely what is needed to prevent senescence, avoiding the entry of pyruvate (and, indirectly, electrons) into the mitochondria and generating ATP by substrate-level phosphorylation. Consistently with this idea, it has been reported that the overexpression of phosphoglyceromutase or glucose phosphate isomerase increases the glycolytic flux and the resistance to oxidative stress, rendering mouse fibroblasts refractory to Ras-induced senescence (22). A high glycolytic flux also correlated with the unlimited proliferative potential that characterizes embryonic stem cells (23). These cells are resistant to a variety of stresses that induce senescence in primary cells (23), suggesting that some tissue stem cells do not use the senescence anti-tumor barrier in response to oncogenic insults.
One important part of our study indicates that interfering with mitochondrial functions using RNA interference against RISP is sufficient to trigger a senescence phenotype in normal human fibroblasts. This is consistent with other studies that found that it was possible to induce senescence in normal fibroblasts by using antimycin A or oligomycin (45). Since oncogenic ras triggers mitochondrial changes similar to those obtained by knocking down RISP, it is possible to conclude that mitochondrial dysfunctions contribute to Ras-induced senescence. The mitochondrial dysfunction we observe in cells expressing oncogenic ras depends on an intact p53 or Rb pathway. Therefore, mitochondria are not the initial targets of oncogenic activity but play a role as an effector mechanism where the activity of the p53 and Rb tumor suppression pathways converge to regulate senescence (Fig. 9). Cancer cells are sensitive to mitochondrial poisons such as rotenone (36), suggesting that mitochondria can be targeted for cancer therapeutics. However, rotenone can induce growth arrest and senescence in normal cells. Further studies are required to find strategies that can engage the mitochondrial senescence effector pathway in tumor cells but spare normal cells.
Supplementary Material
Acknowledgments
We thank N. Chandel, C. Fernandez, and D. Galloway and S. Lowe for reagents and comments and Serge Sénéchal for assistance with FACS.
This research was supported by NCIC grant 18131. G.F. is chercheur senior FRSQ.
Footnotes
Published ahead of print on 15 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Achanta, G., R. Sasaki, L. Feng, J. S. Carew, W. Lu, H. Pelicano, M. J. Keating, and P. Huang. 2005. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 243482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434864-870. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova, J., N. Rezaei, M. Liontos, P. Karakaidos, D. Kletsas, N. Issaeva, L. V. Vassiliou, E. Kolettas, K. Niforou, V. C. Zoumpourlis, M. Takaoka, H. Nakagawa, F. Tort, K. Fugger, F. Johansson, M. Sehested, C. L. Andersen, L. Dyrskjot, T. Orntoft, J. Lukas, C. Kittas, T. Helleday, T. D. Halazonetis, J. Bartek, and V. G. Gorgoulis. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444633-637. [DOI] [PubMed] [Google Scholar]
- 4.Batandier, C., X. Leverve, and E. Fontaine. 2004. Opening of the mitochondrial permeability transition pore induces reactive oxygen species production at the level of the respiratory chain complex I. J. Biol. Chem. 27917197-17204. [DOI] [PubMed] [Google Scholar]
- 5.Brunelle, J. K., E. L. Bell, N. M. Quesada, K. Vercauteren, V. Tiranti, M. Zeviani, R. C. Scarpulla, and N. S. Chandel. 2005. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 1409-414. [DOI] [PubMed] [Google Scholar]
- 6.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120513-522. [DOI] [PubMed] [Google Scholar]
- 7.Carew, J. S., Y. Zhou, M. Albitar, J. D. Carew, M. J. Keating, and P. Huang. 2003. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia 171437-1447. [DOI] [PubMed] [Google Scholar]
- 8.Catalano, A., S. Rodilossi, P. Caprari, V. Coppola, and A. Procopio. 2005. 5-Lipoxygenase regulates senescence-like growth arrest by promoting ROS-dependent p53 activation. EMBO J. 24170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, D. C. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 1251241-1252. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Q., A. Fischer, J. D. Reagan, L. J. Yan, and B. N. Ames. 1995. Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc. Natl. Acad. Sci. USA 924337-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426194-198. [DOI] [PubMed] [Google Scholar]
- 12.Di Micco, R., M. Fumagalli, A. Cicalese, S. Piccinin, P. Gasparini, C. Luise, C. Schurra, M. Garre, P. G. Nuciforo, A. Bensimon, R. Maestro, P. G. Pelicci, and F. d'Adda di Fagagna. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444638-642. [DOI] [PubMed] [Google Scholar]
- 13.Echtay, K. S., D. Roussel, J. St-Pierre, M. B. Jekabsons, S. Cadenas, J. A. Stuart, J. A. Harper, S. J. Roebuck, A. Morrison, S. Pickering, J. C. Clapham, and M. D. Brand. 2002. Superoxide activates mitochondrial uncoupling proteins. Nature 41596-99. [DOI] [PubMed] [Google Scholar]
- 14.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 142015-2027. [PMC free article] [PubMed] [Google Scholar]
- 15.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434907-913. [DOI] [PubMed] [Google Scholar]
- 16.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407770-776. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, Y., M. Kitagawa, and Y. Taya. 2007. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 262083-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwata, S., J. W. Lee, K. Okada, J. K. Lee, M. Iwata, B. Rasmussen, T. A. Link, S. Ramaswamy, and B. K. Jap. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 28164-71. [DOI] [PubMed] [Google Scholar]
- 19.Jones, R. G., D. R. Plas, S. Kubek, M. Buzzai, J. Mu, Y. Xu, M. J. Birnbaum, and C. B. Thompson. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18283-293. [DOI] [PubMed] [Google Scholar]
- 20.Jones, R. G., and C. B. Thompson. 2009. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18357-368. [DOI] [PubMed] [Google Scholar]
- 22.Kondoh, H., M. E. Lleonart, J. Gil, J. Wang, P. Degan, G. Peters, D. Martinez, A. Carnero, and D. Beach. 2005. Glycolytic enzymes can modulate cellular life span. Cancer Res. 65177-185. [PubMed] [Google Scholar]
- 23.Kondoh, H., M. E. Lleonart, Y. Nakashima, M. Yokode, M. Tanaka, D. Bernard, J. Gil, and D. Beach. 2007. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid. Redox Signal. 9293-299. [DOI] [PubMed] [Google Scholar]
- 24.Kops, G. J., T. B. Dansen, P. E. Polderman, I. Saarloos, K. W. Wirtz, P. J. Coffer, T. T. Huang, J. L. Bos, R. H. Medema, and B. M. Burgering. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419316-321. [DOI] [PubMed] [Google Scholar]
- 25.Lee, A. C., B. E. Fenster, H. Ito, K. Takeda, N. S. Bae, T. Hirai, Z. X. Yu, V. J. Ferrans, B. H. Howard, and T. Finkel. 1999. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J. Biol. Chem. 2747936-7940. [DOI] [PubMed] [Google Scholar]
- 26.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 123008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe, S. W., E. Cepero, and G. Evan. 2004. Intrinsic tumour suppression. Nature 432307-315. [DOI] [PubMed] [Google Scholar]
- 28.Mallette, F. A., M. F. Gaumont-Leclerc, and G. Ferbeyre. 2007. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2143-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallette, F. A., M. F. Gaumont-Leclerc, G. Huot, and G. Ferbeyre. 2007. Myc down-regulation as a mechanism to activate the Rb Pathway in STAT5A-induced senescence. J. Biol. Chem. 28234938-34944. [DOI] [PubMed] [Google Scholar]
- 30.McBride, H. M., M. Neuspiel, and S. Wasiak. 2006. Mitochondria: more than just a powerhouse. Curr. Biol. 16R551-R560. [DOI] [PubMed] [Google Scholar]
- 31.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11577-590. [DOI] [PubMed] [Google Scholar]
- 32.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113703-716. [DOI] [PubMed] [Google Scholar]
- 33.Nicke, B., J. Bastien, S. J. Khanna, P. H. Warne, V. Cowling, S. J. Cook, G. Peters, O. Delpuech, A. Schulze, K. Berns, J. Mullenders, R. L. Beijersbergen, R. Bernards, T. S. Ganesan, J. Downward, and D. C. Hancock. 2005. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol. Cell 20673-685. [DOI] [PubMed] [Google Scholar]
- 34.Passos, J. F., G. Saretzki, S. Ahmed, G. Nelson, T. Richter, H. Peters, I. Wappler, M. J. Birket, G. Harold, K. Schaeuble, M. A. Birch-Machin, T. B. Kirkwood, and T. von Zglinicki. 2007. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 5:e110. [DOI] [PMC free article] [PubMed]
- 35.Passos, J. F., and T. Von Zglinicki. 2006. Oxygen free radicals in cell senescence: are they signal transducers? Free Radic. Res. 401277-1283. [DOI] [PubMed] [Google Scholar]
- 36.Pelicano, H., L. Feng, Y. Zhou, J. S. Carew, E. O. Hileman, W. Plunkett, M. J. Keating, and P. Huang. 2003. Inhibition of mitochondrial respiration: a novel strategy to enhance drug-induced apoptosis in human leukemia cells by a reactive oxygen species-mediated mechanism. J. Biol. Chem. 27837832-37839. [DOI] [PubMed] [Google Scholar]
- 37.Piantadosi, C. A., M. S. Carraway, A. Babiker, and H. B. Suliman. 2008. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 1031232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitkanen, S., and B. H. Robinson. 1996. Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J. Clin. Investig. 98345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polyak, K., Y. Xia, J. L. Zweier, K. W. Kinzler, and B. Vogelstein. 1997. A model for p53-induced apoptosis. Nature 389300-305. [DOI] [PubMed] [Google Scholar]
- 40.Rincheval, V., F. Renaud, C. Lemaire, N. Godefroy, P. Trotot, V. Boulo, B. Mignotte, and J. L. Vayssiere. 2002. Bcl-2 can promote p53-dependent senescence versus apoptosis without affecting the G1/S transition. Biochem. Biophys. Res. Commun. 298282-288. [DOI] [PubMed] [Google Scholar]
- 41.Sankaran, V. G., S. H. Orkin, and C. R. Walkley. 2008. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 22463-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seger, Y. R., M. Garcia-Cao, S. Piccinin, C. L. Cunsolo, C. Doglioni, M. A. Blasco, G. J. Hannon, and R. Maestro. 2002. Transformation of normal human cells in the absence of telomerase activation. Cancer Cell 2401-413. [DOI] [PubMed] [Google Scholar]
- 43.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88593-602. [DOI] [PubMed] [Google Scholar]
- 44.Shull, S., N. H. Heintz, M. Periasamy, M. Manohar, Y. M. Janssen, J. P. Marsh, and B. T. Mossman. 1991. Differential regulation of antioxidant enzymes in response to oxidants. J. Biol. Chem. 26624398-24403. [PubMed] [Google Scholar]
- 45.Stöckl, P., E. Hutter, W. Zwerschke, and P. Jansen-Durr. 2006. Sustained inhibition of oxidative phosphorylation impairs cell proliferation and induces premature senescence in human fibroblasts. Exp. Gerontol. 41674-682. [DOI] [PubMed] [Google Scholar]
- 46.Toussaint, O., E. E. Medrano, and T. von Zglinicki. 2000. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp. Gerontol. 35927-945. [DOI] [PubMed] [Google Scholar]
- 47.Voorhoeve, P. M., and R. Agami. 2003. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4311-319. [DOI] [PubMed] [Google Scholar]
- 48.Wei, W., W. A. Jobling, W. Chen, W. C. Hahn, and J. M. Sedivy. 2003. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol. Cell. Biol. 232859-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisiger, R. A., and I. Fridovich. 1973. Mitochondrial superoxide simutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem. 2484793-4796. [PubMed] [Google Scholar]
- 50.Whyte, P., N. M. Williamson, and E. Harlow. 1989. Cellular targets for transformation by the adenovirus E1A proteins. Cell 5667-75. [DOI] [PubMed] [Google Scholar]
- 51.Wu, C., I. Miloslavskaya, S. Demontis, R. Maestro, and K. Galaktionov. 2004. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature 432640-645. [DOI] [PubMed] [Google Scholar]
- 52.Zorov, D. B., C. R. Filburn, L. O. Klotz, J. L. Zweier, and S. J. Sollott. 2000. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med. 1921001-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.