Abstract
CtBPs (CtBP1 and CtBP2) act in the nucleus as transcriptional corepressors and in the cytoplasm as regulators of Golgi apparatus fission. Studies in which the expression or function of CtBPs has been inhibited have independently identified roles for CtBPs in both suppressing apoptosis and promoting cell cycle progression. Here, we have analyzed the consequences of ablating CtBP expression in breast cancer-derived cell lines. We found that loss of CtBP expression suppresses cell proliferation through a combination of apoptosis, reduction in cell cycle progression, and aberrations in transit through mitosis. The third phenotype includes errors in mitotic chromosome segregation that are associated with decreased association of the chromosome passenger protein aurora B with mitotic chromatin and that are likely to be a primary cause of the proapoptotic and antiproliferative effects of CtBP loss. We also show that loss of CtBP expression results in the activation of the transcription factor p53 and that loss of p53 function renders cells more susceptible to CtBP small interfering RNA-induced apoptosis.
Chromosome segregation during mitosis involves the bipolar (amphitelic) attachment of paired sister chromatids to microtubules of the mitotic spindle, equal distribution of the replicated chromosomes, and finally cytokinesis. The spindle assembly checkpoint (SAC) is a key determinant of the fidelity of chromosome segregation; it can be viewed as an “anaphase wait signal,” holding cells in metaphase until all criteria for equal chromosome segregation are met (25, 30, 38). SAC proteins accumulate at kinetochores of unattached chromosomes, providing a signal that maintains inhibition of the anaphase-promoting complex until bipolar attachments have been made and the SAC proteins are displaced from the kinetochores. Many aspects of mitosis are regulated by a complex of chromosomal passenger proteins, notably aurora B kinase and its regulators survivin, borealin, and INCENP (38, 43, 55). Inhibiting the expression and/or activity of chromosomal passenger proteins results in severe mitotic defects (17, 20, 32). Cell death may occur as a consequence of such defects, and thus, proteins such as survivin and aurora B kinase are currently under intense investigation as targets for anticancer therapeutics (13, 61).
CtBPs (C-terminal binding proteins) were originally identified as cellular binding partners of type 2/5 adenovirus 243R E1A proteins (4). What are now known to be consensus CtBP-binding motifs, PXDLS, were subsequently identified within the C termini of E1A and EBNA3C (45, 52, 54). Deletion of these regions from either of these two proteins markedly alters their ability to transform cells in cooperation with mutant RAS, providing compelling evidence for a key role of CtBPs in cellular transformation (4, 45). Recent studies have provided more direct evidence; CtBPs form functional interactions with multiple cellular proteins that have diverse roles in intracellular signaling and transcriptional control (1, 5, 6, 54), experimental suppression of CtBP expression renders cancer cells hypersensitive to apoptosis (19, 37, 63), and the cytotoxic effects of certain genotoxic cancer chemotherapeutics can be, in part, due to the activation of signaling pathways that promote CtBP degradation (58).
In mammals, CtBPs are expressed from two genes, CTBP1 and CTBP2. Human and rodent CTBP1 genes are expressed as two main splice forms, CtBP1-L and CtBP1-S, which differ by 13 amino acids at their N termini (5). Murine Ctbp2 undergoes similar alternative splicing (56). Human CtBP1-L and CtBP2-L are highly similar proteins of 440 and 445 amino acids, respectively. They are widely expressed in normal tissues. CtBPs are characterized by a conserved central NADH-dependent dimerization domain. Their N- and C-terminal regions form a single globular domain, which binds PXDLS-containing proteins (35). CtBPs can function interchangeably in many assays; there are, however, differences in their regulation, particularly at the level of their subcellular distribution; notably, only CtBP2-L contains a nuclear localization/retention sequence, located at its unique N terminus (2, 56, 64).
CtBPs have been ascribed two distinct functions; in the nucleus, they act as transcriptional corepressors (1, 5, 6, 54), whereas in the cytoplasm, they are involved in the fission of Golgi and endocytic membranes (3, 5, 11). Mechanistically, as corepressors, CtBPs primarily function as a scaffold to recruit chromatin-modifying enzymes, including histone deacetylases (HDACs), histone methyltransferases, and polycomb group proteins, to DNA-binding transcription factors (26, 47). Of the many CtBP-recruiting transcription factors, the best characterized include SLUG and ZEB/δEF1, which repress the expression of epithelium-specific genes (15, 19, 41, 47, 53). In the Golgi apparatus, membrane fission has been reported to be dependent upon a CtBP1-S-associated lipid-specific acyl-transferase activity (11), though the nature of this activity is unclear (16).
The diverse functions of CtBP proteins in the nucleus and cytoplasm are reflected in studies in which the phenotypic consequences of genetic or functional ablation of CtBPs have been investigated. Disruption of dCtBP in Drosophila results in embryonic segmentation defects consistent with aberrant transcriptional control (36, 40). Similarly, Ctbp1−/−/Ctbp2−/− mice die early in development, with their embryonic fibroblasts displaying abnormal transcription and hypersensitivity to apoptosis signals but a normal Golgi apparatus architecture (19, 23). The hypersensitivity to apoptosis is associated with an upregulation in the expression of multiple proapoptotic genes through an as yet undetermined p53-independent pathway (19). RNA interference (RNAi)-mediated depletion of CtBPs in various cancer cell lines is also, on its own, sufficient to induce p53-independent apoptosis (37, 63). CtBP-dependent transcriptional regulation also plays an important role in the control of tumor cell migration (62). In apparent contrast, studies focusing upon the role of CtBPs in Golgi membrane fission have demonstrated that the ablation of CtBP function or expression can result in a “Golgi checkpoint” in G2, preventing entry into mitosis (10, 22). To clarify our understanding of the role of CtBPs in the proliferation and survival of cancer cells, we performed a detailed analysis of the phenotypic consequences of inhibiting CtBP expression in breast cancer-derived cell lines. These studies have revealed a previously unidentified role of CtBPs in the regulation of mitotic fidelity and the SAC that is an important component of the prosurvival activity of these proteins.
MATERIALS AND METHODS
Statistical analysis.
Unless otherwise stated, error bars on graphs represent the standard error of the mean of three independent experiments. The statistical significance of differences was determined using paired two-tailed Student's t tests (Microsoft Excel) comparing CtBP small interfering RNA (siRNA)- to control siRNA-transfected cells, unless otherwise indicated. P values of less than 0.05 are marked with one asterisk, and those less than 0.02 are marked with two asterisks.
Cell culture and RNAi.
All cells were maintained in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Autogen Bioclear) and penicillin (100 U/ml), streptomycin (100 μg/ml), and l-glutamine (292 μg/ml) (Invitrogen). RNAi-mediated CtBP knockdown was performed using the siRNA 5′-GGGAGGACCUGGAGAAGUUdTdT-3′/3′-dTdTCCCUCCUGGACCUCUUCAA-5′, obtained from Ambion. CtBP1 siRNA (5′-ACGACUUCACCGUCAAGCAdTdT-3′/3′-dTdTUGCUGAAGUGGCAGUUCGU-5′), CtBP 2 siRNA (5′-GCGCCUUGGUCAGUAAUAGdTdT-3′/3′-dTdTCGCGGAACCAGUCAUUAUC-5′), p53 siRNA (Hs_TP53_9 HP Validated; catalog no.S102655170) and aurora B siRNA (target, AAGGTGATGGAGAATAGCAGT) were obtained from Qiagen. Negative control siRNA no. 1 (Ambion) was used for experimental controls. siRNA was transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The growth medium was replaced 4 h after transfection and, unless otherwise stated, was not replaced before the assay. To make RNAi-insensitive plasmids for rescue experiments, plasmids pcDNA3.1CtBP2(1-445)mh (34) and pcDNA3.1CtBP1(1-440)mh (2) were modified by site-directed mutagenesis (Quickchange; Stratagene) to make synonymous changes in the RNAi target sequence (mh stands for myc-His tag). The sequence of the forward mutagenesis primer for CtBP1 was 5′-CACCATCACTCTCACGCGTGAAGATTTAGAAAAATTCAAAGCCCTCCGC-3′, and for CtBP2 it was 5′-ACCATCACCCTCACGCGTGAAGATTTAGAAAAATTCAAGGCCCTGAGA-3′. Caspase inhibitor Q-VD-OPH (QVD) was from Calbiochem.
Cell proliferation and viability assays and live-cell imaging.
For cell proliferation assays, adherent cells in wells of 24-well plates were detached using trypsin and counted using a hemocytometer. For colony-forming assays, cells were detached with trypsin 2 days posttransfection in 96-well plates and replated into 6-well plates. The cells were grown for a further 10 to 12 days with a change of growth medium every 3 to 4 days. Colonies were stained with Giemsa (Sigma) and counted manually. Fluorescence time-lapse microscopy on H2B-GFP (green fluorescent protein) HeLa cells was carried out using a Zeiss Live Cell Observer with a 37°C incubator and a 5% CO2 hood. Images were captured every 15 min using a 20× objective on a Zeiss AxioCam MRm camera with AxioVision software. Time lapse microscopy of MCF-7 cells was performed using an Olympus IX81 microscope with a CO2- and temperature-controlled environmental chamber controlled by SIS Cell P software. Image J software was used for analysis.
Flow cytometry.
For determination of DNA content, all floating and attached cells were collected and combined for analysis. Ethanol-fixed cell samples were washed with phosphate-buffered saline (PBS) and stained in freshly prepared propidium iodide buffer (20 μg/ml propidium iodide, 0.2 mg/ml RNase A, 0.1% [vol/vol] Triton X-100 in PBS) for 30 min at room temperature. Flow cytometry was performed using a FACSCalibur (Becton Dickinson) equipped with CellQuest software (Becton Dickinson).
Antibodies and immunoblotting.
The following antibodies were used: mouse monoclonal antibodies against CtBP1 (E12; Santa Cruz Biotechnologies), CtBP2 (E16 [Santa Cruz Biotechnologies] or clone 16 [Becton Dickinson]), anti-p21WAF1 (OP64; Calbiochem), CENP-A and six-His (ab13939, and ab9108; AbCam), AIM-1 (aurora B) (611082; BD Biosciences), p53 (clone DO-1; Serotec), phosphohistone H3 (Ser10) (clone 3H10; Upstate), and rabbit polyclonal against actin (20-33; Sigma). Antibody E12, which was raised against human CtBP1, is marketed as recognizing both CtBP1 and CtBP2; however, it does not recognize recombinant human CtBP2 (see Fig. S6 in the supplemental material; data not shown) and can be used to selectively detect CtBP1 in humans. For immunoblotting, adherent cells were washed with PBS, scraped into PBS, and pelleted by centrifugation. Cell lysis was performed for 15 min at 4°C in 2 volumes of urea lysis buffer (7 M urea, 0.05% Triton X-100, 25 mM NaCl, 20 mM HEPES, pH 7.6, 100 mM dithiothreitol). Lysates were clarified by centrifugation at 13,000 × g for 10 min. Protein was quantified using Bio-Rad protein assay reagent. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Hybond-ECL membranes (GE Healthcare). Blocking and antibody incubations were done in 3% low-fat milk in PBS-0.025% Tween 20, and washes were in PBS-0.1% Tween 20. Detection of horseradish peroxidase-labeled secondary antibody was done with Supersignal (Pierce), and images were collected and analyzed on a Fluor-S MultiImager (Bio-Rad) equipped with Quantity One software (Bio-Rad).
Immunofluorescence analysis, mitotic index, and micronucleus counts.
Cells growing on 13-mm glass coverslips were fixed with 4% paraformaldehyde in PBS for 10 min. For immunofluorescence assays, cells were permeabilized using 0.1% Triton X-100/PBS for 20 min, blocked for 30 min in 3% bovine serum albumin/PBS, and incubated with primary antibody for 1 h in 0.6% bovine serum albumin/PBS, followed by species-specific secondary antibody. The secondary antibodies were as follows: Alexa Fluor 594-conjugated anti-mouse immunoglobulin G (Molecular Probes), fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit immunoglobulins, and tetramethyl rhodamine isothiocyanate-conjugated anti-mouse immunoglobulins (DakoCytomation). Cells were counterstained with 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) during the secondary-antibody incubation. Coverslips were mounted on slides with fluorescent mounting medium (DakoCytomation). All cells were visualized using a Zeiss Axiovert 200 fluorescence microscope with a 40× or 100× objective, and images were collected using an Orca-ER digital camera (Hamamatsu) and processed using Openlab 3.5.1 Software (Improvision). Identical exposure times were applied for different images within the same experiment. Images were cropped using Corel Photo-Paint 12. Aurora B fluorescence was quantified using Openlab 3.5.1 software and was normalized to the DNA content as assessed by DAPI staining. The mitotic index was determined by counting ≥500 cells per slide and scoring the cells as either mitotic or interphase according to the state of chromosome condensation. For micronucleus analysis, ≥200 cells per slide were analyzed.
RESULTS
Inhibition of CtBP synthesis inhibits breast cancer cell proliferation.
To investigate the role of CtBPs in the proliferation of breast cancer cells, we used an siRNA to target a sequence conserved in known human CtBP1- and CtBP2-encoding mRNAs (63). In referring to experiments using this reagent, we have therefore used the terms “CtBP” and “CtBPs” to refer to both family members and denote individual proteins when appropriate. We first transfected MCF-7 cells with CtBP-specific or control siRNA, harvested cells at 24-h intervals posttransfection, and determined the abundances of CtBP1 and CtBP2 proteins (Fig. 1A [days 1 to 4 shown]). CtBP siRNA downregulated CtBP expression within 24 h, with maximum effect at days 2 to 5. CtBP1 was depleted by up to 85% and CtBP2 to below the limits of detection. By day 7, CtBP protein abundance had begun to recover (not shown). In MDA-MB-231 and ZR75.1 cells, CtBP siRNA similarly reduced CtBP abundance (Fig. 1A). Expression of both CtBP1 and CtBP2 was reduced in up to 95% of the cells (see Fig. S1 in the supplemental material).
FIG. 1.
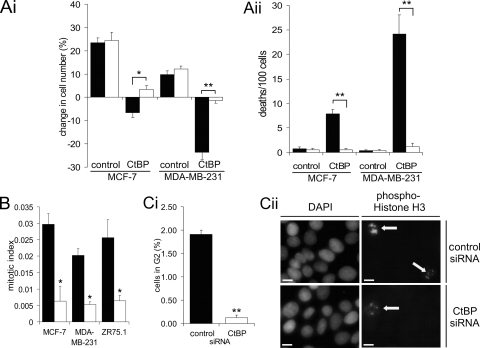
Downregulation of CtBP protein expression in breast cancer cell lines by RNAi. (A) Immunoblots showing CtBP1 and CtBP2 proteins compared to actin loading control (42 kDa) at the indicated times following transfection. Lanes: a, nontransfected; b, control siRNA; c, CtBP siRNA. (B) Colony-forming assays 12 to 14 days posttransfection. Solid bars, control siRNA; open bars, CtBP siRNA (MCF-7, n = 9; MDA-MB-231, n = 9; ZR75.1, n = 6). (C) Cells were plated at 5 × 104 (MCF-7) or 7 × 104 (MDA-MB-231) cells per well 1 day prior to transfection (note that plating efficiency was <100%). The numbers of adherent cells at the indicated time points following transfection with control (solid symbols) or CtBP (open symbols) siRNA are shown. Triangles, MCF-7 cells; diamonds, MDA-MB-231 cells. The data represent means of two duplicate dishes. (D) Time lapse microscopy of MCF-7 and MDA-MB-231 cells following siRNA-mediated inhibition of CtBP expression 2.5 to 3.5 days posttransfection (see Videos S1 to S4 in the supplemental material). (Di) Change in total cell numbers in the imaged fields over 24 h. (Dii) Number of mitotic events observed. (Dii) Number of cell deaths observed. Solid bars, control siRNA; open bars, CtBP siRNA (MCF-7, n = 5 fields; MDA-MB-231, n = 5 fields.) The data are representative of two independent experiments for each cell line.
We next established the effects of inhibiting CtBP synthesis on the ability of single breast cancer cells to establish colonies when plated 2 days posttransfection with siRNA (Fig. 1B). CtBP siRNA resulted in significant decreases in the number of colonies relative to control siRNA-transfected cells (2-fold, 6-fold, and 36-fold for ZR75.1, MCF-7, and MDA-MB-231 cells, respectively). We also determined the total numbers of adherent MCF-7 and MDA-MB-231 cells following their transfection with siRNA (Fig. 1C). The numbers of CtBP-depleted cells were lower than those of control cells by 2 days posttransfection and were at their lowest (and more than fourfold less than control transfectants) at day 4, after which cell numbers began to increase. Similar results were obtained with ZR75.1 cells (data not shown). To determine whether this was due to effects on cell division or cell death, we performed live-cell imaging analysis. Cells were imaged for 24 h over the period from 2.5 to 3.5 days posttransfection (see Videos S1 to S4 in the supplemental material). Whereas the numbers of control siRNA-transfected MCF-7 and MDA-MB-231 cells increased during this period by 49% and 29%, respectively, following CtBP siRNA there was a significant decrease in both MCF-7 cells (9%) and MDA-MB-231 cells (37%) (Fig. 1Di). These data are consistent with those obtained by cell counting (Fig. 1C). CtBP siRNA caused a significant (eight- to ninefold) decrease in the number of mitotic events in this period (Fig. 1Dii). The rate of cell death was also increased, from 1.8 to 9.7 deaths per 100 cells in MCF-7 and from 1.7 to 35.7 deaths per 100 cells in MDA-MB-231 (Fig. 1Diii). The greater effect of CtBP siRNA on cell death in MDA-MB-231 cells would account for the much larger decrease in cell population over that seen with MCF-7 cells during this period. Together, these data clearly establish that loss of CtBP expression results in a marked reduction in the proliferative capacity of breast cancer cells.
Loss of CtBP results in increased apoptosis and reduced cell division.
To determine whether the cell death induced by CtBP siRNA is apoptosis, we determined whether this death was inhibited by the caspase inhibitor QVD (Fig. 2A; see Videos S5 and S6 in the supplemental material). As previously shown, CtBP depletion induces cell death in both MCF-7 and MDA-MB-231 cells. However, in the presence of QVD, deaths over the 10-h period of analysis were significantly reduced, from 7.8/100 to 0.5/100 in MCF-7 cells and from 24.2/100 to 1.2/100 in MDA-MB-231 cells (Fig. 2Aii). This effect was reflected in the total change in cell numbers during the imaging period (Fig. 2Ai) (QVD treatment had no effect on the number of mitoses [not shown]). Therefore, inhibition of CtBP expression results in a marked induction of apoptosis in MDA-MB-231 cells, with a more modest increase occurring in MCF-7 cells.
FIG. 2.
Analysis of apoptosis and cell proliferation in CtBP-depleted cells. (A) Time lapse microscopy of MCF-7 and MDA-MB-231 from 2.5 days posttransfection with the indicated siRNA. Where indicated, 25 μM QVD was added 4 h prior to the start of time lapse analysis (see Videos S5 and S6 in the supplemental material). (Ai) Change in total cell numbers over 10 h. (Aii) Number of cell deaths observed. Solid bars, no QVD; open bars, 25 μM QVD (MCF-7, n = 5; MDA-MB-231, n = 5). (B) Cells were fixed for analysis of the mitotic index 4 days posttransfection with control (solid bars) or CtBP (open bars) siRNA. (Ci) Analysis of MCF-7 cells in late G2, as indicated by speckled phosphohistone H3 staining. More than 500 cells were scored per slide. (Cii) Immunofluorescence staining of phosphohistone H3 expression in MCF-7 cells 4 days posttransfection. The arrows indicate cells in late G2. Bar, 10 μm. Color images, including staining of a mitotic nucleus for comparison, are shown in Fig. S2 in the supplemental material.
Live-cell imaging also demonstrated that CtBP ablation resulted in reduced mitosis of breast cancer cells. We therefore counted the mitotic index of cells 4 days posttransfection with siRNA (Fig. 2B). Compared to control siRNA transfection, CtBP siRNA resulted in a significant (P < 0.05) four- to sixfold reduction in the mitotic index in all three cell lines examined. We therefore examined whether this was due to arrest of CtBP-depleted cells in G2 by examining histone H3 phosphorylation (Fig. 2C). During late G2, histone H3 is phosphorylated primarily in pericentromeric heterochromatin, giving a very distinct speckled staining pattern, which allows the identification of cells in late G2 (21). When a “Golgi checkpoint” is activated by inhibition of CtBP expression or function, cells accumulate in late G2 and can be detected by this assay (22). There was a significant (P < 0.02) decrease in the number of MCF-7 cells compared to control cells in late G2 after CtBP siRNA. Therefore, the reduced entry into mitosis seen in CtBP-depleted breast cancer cells is not due to a late G2 arrest, but rather, must be due to fewer cells entering G2, or possibly successfully traversing this cell cycle phase.
Loss of CtBP leads to multiple mitotic defects.
To investigate whether breast cancer cells arrest at a specific point of the cell cycle after CtBP siRNA, we performed flow cytometry analysis of DNA content (Fig. 3A). This analysis did not reveal any striking single effect of the CtBP siRNA that alone might account for its effect on cell proliferation. Consistent with induction of apoptosis, there was a modest increase in the number of events with a sub-G1 DNA content in CtBP-depleted MCF-7 and MDA-MB-231 cells at both 2 and 4 days posttransfection (not shown). There was also a modest but consistent increase in the proportion of cells with 4N DNA content in both cell lines at 2 and 4 days posttransfection. This is generally indicative of accumulation in G2 or M phase. However, we have already shown that there is a decrease in cells in M and late G2 phases after CtBP depletion. An alternative cause of an increase of 4N cells is failures in mitosis that result in the generation of binucleate cells. At day 4 posttransfection, MDA-MB-231 cells also consistently showed a small increase in the 2N to 4N (usually S phase) fraction; this was not observed in MCF-7 cells.
FIG. 3.
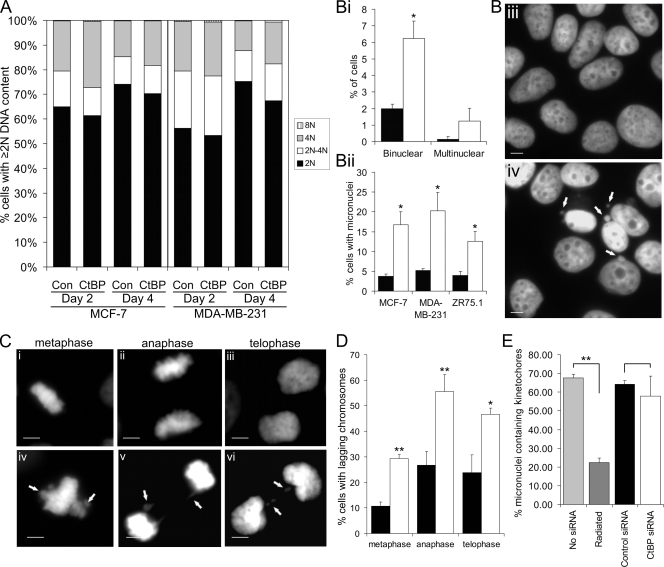
Evidence of chromosome segregation defects following loss of CtBP. (A) Graph showing DNA content as assessed by flow cytometry analysis of the indicated cell lines transfected with the indicated siRNA and analyzed 2 to 4 days later. The data are representative of at least three separate experiments. Fragments with sub-2N DNA content were excluded from the analysis. Con, control. (B) Mitotic phenotypes 4 days posttransfection with control (solid bars) or CtBP (open bars) siRNA. (Bi) Percentages of MCF-7 cells with binuclear and multinuclear phenotypes 4 days after transfection with control or CtBP siRNA. (Bii) Quantification of micronucleus occurrence. (Biii and Biv) DAPI-stained MCF-7 nuclei of control-siRNA (Biii)- and CtBP siRNA (Biv)-transfected cells 4 days posttransfection. Micronuclei are indicated by arrows. Bar, 10 μm. Of the three cell lines included in this study, MCF-7 cells possess the best combination of morphology and transfection efficiency for detailed cellular analyses. (C) MCF-7 cells at different stages of mitosis 3 days posttransfection with control (i, ii, and iii) or CtBP (iv, v, and vi) siRNA. Lagging chromosomes and chromatin bridges are indicated by arrows. Bar, 10 μm. (D) Quantification of mitotic defects in MCF-7 cells 3 days posttransfection with control (solid bars) or CtBP (open bars) siRNA. More than 25 events were scored per stage of mitosis per slide. (E) Quantification of immunofluorescence analysis of CENP-A in MCF-7 cells 4 days posttransfection with no siRNA, control siRNA, or CtBP siRNA or 2 days postexposure to 5 Gy ionizing radiation. More than 30 micronuclei were scored per slide as staining either positive or negative for CENP-A.
We then further analyzed the phenotype of CtBP-depleted cells by detailed morphological analysis of DAPI-stained cell nuclei. In addition to the previously described reduction in the mitotic index (Fig. 2B), we identified multiple morphological defects consistent with aberrant transit through mitosis. First, there was a significant increase in the occurrence of binucleate cells, from 2.0% of the total population in controls to 6.2%, following CtBP siRNA transfection. This suggests that a proportion of the cells undergo cell division without segregating their DNA and is consistent with the modest increase in cells with 4N DNA content seen with flow cytometry. Furthermore, in all three cell lines examined, CtBP siRNA caused a three- to fourfold increase in the proportion of the cells containing micronuclei (Fig. 3Bii to iv) (a similar result was obtained in nontransformed human fibroblasts [see Fig. S7 in the supplemental material], indicating that this response is not restricted to cancer cell lines). Analysis of the alignment and segregation of condensed chromosomes in mitosis also revealed that CtBP siRNA resulted in increased numbers of cells containing lagging chromosomes and chromatin bridges (Fig. 3C and D). For example, 26.7% ± 5.3% of control siRNA-transfected MCF-7 cells undergoing anaphase exhibited lagging chromosomes compared to 55.5% ± 6.7% following CtBP siRNA (P < 0.02).
Micronuclei can arise from whole chromosomes that lag at mitosis due to, for example, a damaged kinetochore or faulty spindle apparatus or from acentric chromosome fragments created by DNA breaks. It was thus important to determine whether the micronuclei found in CtBP-depleted cells were derived from whole chromosomes or chromosome fragments. To do this, we assessed the presence of the kinetochore protein CENP-A in the micronuclei (see Fig. S3 in the supplemental material). CENP-A was detected in 60 to 70% of micronuclei in both control and CtBP siRNA-transfected cells (Fig. 3E). As a control, we used ionizing radiation to cause DNA breaks, as ∼85% of radiation-induced micronuclei arise from acentric chromosome fragments (57). Less than 20% of the micronuclei formed in irradiated MCF-7 cells stained positive for CENP-A. Therefore micronuclei seen in CtBP-depleted cells contain whole chromosomes as opposed to chromosome fragments, and these are probably formed from the lagging chromosomes observed during mitosis.
Known functions of CtBPs provide little clue as to why their depletion might result in these abnormalities in mitosis; one possibility, given the role of CtBPs in Golgi apparatus fission during G2 phase, is that it is a result of cells entering mitosis with an incorrectly organized Golgi apparatus. Normal rat kidney cells injected with a CtBP-inhibiting antibody or transfected with antisense CtBP siRNA primarily arrest in late G2; however, the cells that do enter mitosis following CtBP antibody injection are clearly characterized by the presence of aggregated Golgi complexes (22). We therefore determined the structure of the Golgi apparatus during mitosis in siRNA-transfected MCF-7 cells by immunolocalization of golgin 97. Particular attention was paid to cells in metaphase, as this is the stage of mitosis in which fragmentation of the Golgi apparatus is most apparent. Compared to control siRNA-transfected cells, there was no observable difference in the phenotype of the Golgi apparatus during mitosis in CtBP siRNA-transfected cells (see Fig. S4 in the supplemental material). It is possible that these cells had only escaped a Golgi-dependent G2 checkpoint and entered mitosis as a consequence of insufficient knockdown of the CtBP. However, chromosome segregation defects were clearly observable in CtBP siRNA-transfected cells that had normal mitotic Golgi apparatus fragmentation (see Fig. S4 in the supplemental material), and we therefore consider it highly unlikely that these defects were a consequence of a loss of CtBP-dependent Golgi apparatus fission.
Reduction in mitotic fidelity by CtBP siRNA is due to on-target effects.
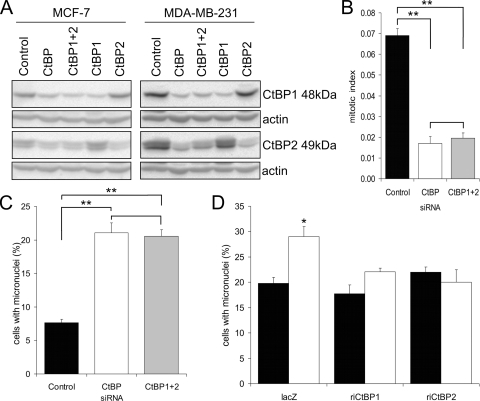
As loss of mitotic fidelity has not been reported following other methods of CtBP ablation, we next used the two independent approaches of rescue and redundancy (14) to establish that this was not due to off-target effects of the CtBP siRNA. In each approach, we scored the occurrence of micronuclei as a marker of mitotic abnormalities. Figure 4A, B, and C show that the effects of the CtBP siRNA on micronucleus formation were replicated when a combination of two independent siRNAs to CtBP1 and CtBP2 were cotransfected (Fig. 4C). For a rescue approach, we transfected CtBP1 or CtBP2 protein expression plasmids containing synonymous mutations in the CtBP siRNA target sequence together with the siRNA (Fig. 4D). While this experimental approach resulted in an overall increase in the numbers of cells containing micronuclei in control siRNA-transfected cells, an effect which we believe to be either a consequence of cellular stress following plasmid DNA transfection or formation of micronuclei directly from cytoplasmic plasmid DNA (31), when cotransfected with the control LacZ plasmid, CtBP siRNA still caused a significant increase in micronucleus-containing cells. In contrast, when cotransfected with either siRNA-insensitive CtBP1 or CtBP2 expression vectors, CtBP siRNA had no effect on micronucleus formation. Together, these two independent approaches confirm that CtBP siRNA induces the formation of micronuclei through specific on-target effects.
FIG. 4.
CtBP siRNA induces chromosome segregation defects through on-target effects. (A) Immunoblots showing CtBP1 and CtBP2 depletion in MCF-7 and MDA-MB-231 cells using either CtBP siRNA or CtBP1 and CtBP2 siRNAs individually or in combination (total siRNA concentration, 100 nM) 3 days posttransfection. Actin (42 kDa) was included as a loading control. (B) MCF-7 cells were fixed for analysis of the mitotic index 3 days posttransfection with control, CtBP, or CtBP1 plus CtBP2 siRNA. Analysis of variance (ANOVA) was used for statistical analysis. (C) Quantification of micronuclei in MCF-7 cells 3 days posttransfection with control, CtBP, or CtBP1 plus CtBP2 siRNA. ANOVA was used for statistical analysis. (D) MCF-7 cells were cotransfected with control (solid bars) or CtBP (open bars) siRNA and plasmids encoding His-tagged RNAi-insensitive CtBP proteins or His-tagged β-galactosidase control (lacZ). The cells were immunostained with anti-six-His antibody, and the percentage of His fusion protein-expressing cells that contained micronuclei was determined at day 4 posttransfection.
Evidence that CtBPs promote mitotic fidelity through effects on the cellular activity of chromosome passenger proteins.
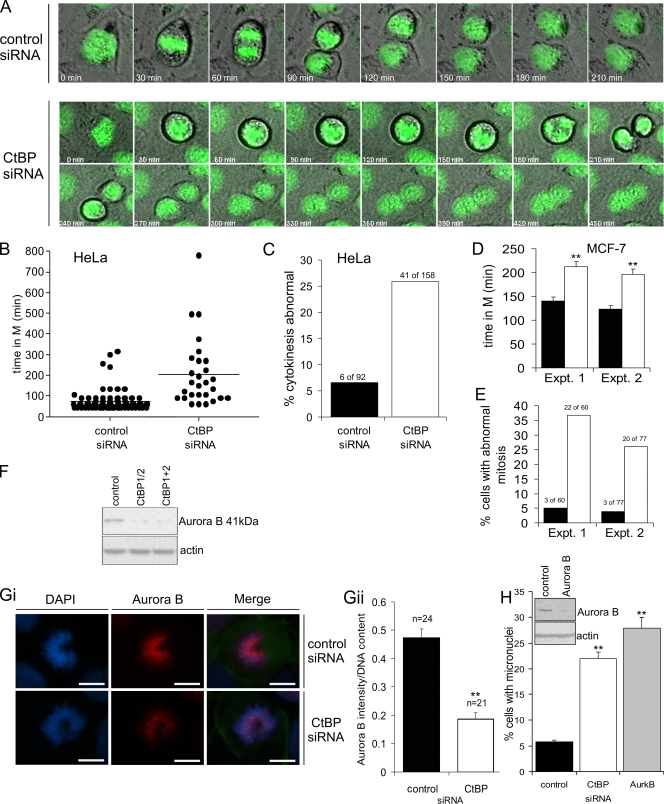
To further define the nature of the mitotic abnormalities that follow inhibition of CtBP synthesis, we used live-cell imaging to assess transit through mitosis. We initially analyzed HeLa cells stably expressing GFP-histone H2B, in order to permit visualization of chromatin during mitosis. CtBP siRNA resulted in a striking increase in the mean length of time a cell spent in mitosis (Fig. 5A and B; see Videos S7 and S8 in the supplemental material). This delay was primarily a consequence of cells spending a longer time in prometaphase, which can indicate prolonged activation of the SAC. Additionally, >25% of the cells that did enter mitosis ultimately failed to complete cytokinesis, resulting in the generation of a binucleate cell (Fig. 5A and C). Specifically, these cells commence cytokinesis and successfully form a cleavage furrow; however, this subsequently regresses, and no abscission occurs; the cells rejoin shortly afterward (Fig. 5A). CtBP siRNA had very similar effects on MCF-7 cells, with a significant extension of the time spent in mitosis, and an increase in the proportion of mitoses that ended in failed cytokinesis (Fig. 5D and E). Together with the analysis of fixed cells, these results indicate that loss of CtBPs results in chromosome alignment defects and activation of the SAC. We have confirmed that the SAC is indeed activated in these cells by determining that the SAC protein BubR1 is recruited to kinetochores (see Fig. S8 in the supplemental material). However, this checkpoint is ultimately not sustained, and cells initiate anaphase with errors in chromosome segregation and either fail in cytokinesis or divide to form aberrant daughter cells.
FIG. 5.
Analysis of the effects of CtBP siRNA on progression through mitosis. (A, B, and C) HeLa cells stably expressing GFP-histone H2B were transfected with either control or CtBP siRNA and analyzed by time lapse microscopy 24 h later (see Videos S7 and S8 in the supplemental material). (A) Representative time lapse images of individual cells undergoing mitosis. (B) Time taken for cells to complete mitosis. The bars through clusters of time points represent the mean times taken to complete mitosis. Similar results were obtained in an independent experiment. (C) Percentage of cells that displayed defects in cytokinesis. Similar results were obtained in an independent experiment. (D) Time taken for MCF-7 cells to complete mitosis 3 days posttransfection with either control (solid bars [see Video S1 in the supplemental material]) or CtBP (open bars [see Video S2 in the supplemental material]) siRNA. The bars indicate means plus standard errors of the mean for ≥60 mitoses. Results from two independent experiments are shown. (E) Percentage of the mitoses analyzed in panel D that exhibited abnormal cytokinesis. (F) Immunoblot showing aurora B in MCF-7 cells 3 days posttransfection with either control siRNA, CtBP siRNA (CtBP1/2), or both CtBP1 and CtBP2 siRNAs (CtBP1+2). (Gi) Immunofluorescence staining of aurora B (red) in prometaphase MCF-7 cells 3 days posttransfection with either control or CtBP siRNA. Bar, 10 μm. (Gii) Quantification of aurora B immunofluorescence intensity (normalized to the DNA content as assessed by DAPI staining) in prophase and prometaphase MCF-7 cells. The data are from a representative of three separate experiments in which >20 cells were scored per slide. (H) MCF-7 cells were transfected with the indicated siRNA and assessed for the presence of micronuclei 3 days later.
Causes of the abnormalities in chromosome alignment in metaphase could include either gross abnormalities in assembly of the spindle or failure of the normal mechanism of spindle connection to kinetochores and chromosome orientation. We have excluded gross microtubule spindle abnormalities, as immunodetection of α- and γ-tubulin in CtBP-depleted MCF-7 cells revealed no observable defects in the morphology of the microtubule spindle or centrosome numbers (see Fig. S5 in the supplemental material; data not shown). The defects in chromosome segregation and cytokinesis following CtBP silencing are similar to those seen following inhibition of the activity of aurora B kinase (20, 46, 49) or expression of its regulator, survivin (29). We therefore examined whether aurora B expression and/or localization were abnormal in CtBP-depleted cells. Western blot analysis (Fig. 5F) showed that aurora B is reduced to an undetectable level in MCF-7 cells transfected with either CtBP-specific siRNA or the combination of CtBP1 and CtBP2-specific siRNAs. As aurora B expression is cell cycle regulated (24), this reduction could be due to the reduced number of cells in mitosis. We therefore used immunofluorescence to visualize and quantify aurora B in mitotic cells. During mitosis, aurora B first localizes to centromeres in prophase and remains concentrated at centromeres until it relocalizes to the central spindle at the start of anaphase, where it subsequently concentrates at the midbody (51). Compared to control cells, CtBP-siRNA-treated cells showed a >2-fold reduction (P < 0.01) in the accumulation of aurora B on chromatin in prophase and prometaphase cells (Fig. 5Gi and Gii). Aurora B subsequently localized to the midbody in anaphase in both control and CtBP siRNA-treated cells; we were unable to reliably quantify its abundance within these structures. We have discovered, therefore, that CtBPs have a novel function in regulating the association of the chromosome passenger protein aurora B with chromatin in early mitosis. Based on what is known of the functions of aurora B, compromising this association would be expected to result in phenotypic abnormalities in mitosis of the type that occurs in CtBP-depleted cells. To confirm this, we used siRNA to reduce aurora B expression in MCF-7 cells and found the expected increase in the frequency of cells containing micronuclei (Fig. 5H).
p53 protects breast cancer cells from CtBP siRNA-induced apoptosis.
It was important to determine whether this novel role of CtBPs in the regulation of mitotic fidelity plays a role in its prosurvival activity. We therefore examined whether cells that had exited mitosis with incorrectly segregated chromosomes showed an increased propensity to undergo apoptosis. Figure 6A shows that when cells are prevented from undergoing apoptosis with the caspase inhibitor QVD, the proportion of cells in the population that contain micronuclei increases. This accumulation was seen in both MCF-7 and MDA-MB-231 cells with or without the transfection of CtBP siRNA. Interestingly, whereas in CtBP siRNA-transfected MDA-MB-231 cells QVD markedly increased the proportion of the population containing micronuclei from 22.5% ± 0.5% to 32.4% ± 0.7%, this accumulation was much less in CtBP siRNA-transfected MCF-7 cells. Thus, the increased rates of apoptosis in response to CtBP siRNA in MDA-MB-231 compared to MCF-7 cells can be accounted for, at least in part, by apoptosis in cells that have failed to accurately segregate their chromosomes in mitosis.
FIG. 6.
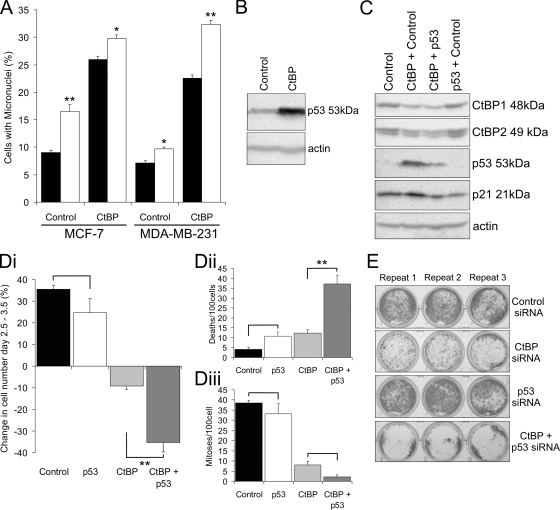
p53 protects breast cancer cells from CtBP siRNA-induced apoptosis. (A) Quantification of micronuclei in MCF-7 and MDA-MB-231 cells 3 days posttransfection with control or CtBP siRNA. Solid bars, no QVD; open bars, 25 μM QVD added 24 h prior to fixing the cells. (B) Immunoblot showing p53 protein abundance in MCF-7 cells 3 days posttransfection with CtBP siRNA compared to control siRNA. (C) Immunoblot confirming effects of siRNAs on the abundance of their targets in MCF-7 cells. The abundance of the p53-induced protein p21WAF1 was also examined. (D) Time lapse microscopy analysis of MCF-7 cells 2.5 to 3.5 days posttransfection with either control, p53, CtBP, or CtBP plus p53 siRNA (n = 5). The transfection conditions were the same as for panel C. (See Videos S9 and S10 in the supplemental material.) (Di) Change in total cell numbers over 24 h. (Dii) Number of cell deaths observed. (Diii) Number of mitotic events observed. Analysis of variance was used for statistical analysis. (E) Giemsa staining of MCF-7 cell colonies 10 days posttransfection with either control, CtBP, p53, or CtBP plus p53 siRNA.
Cells that exit mitosis with incorrectly segregated DNA can undergo a postmitotic G1 arrest that prevents replication of aberrant cells (27). This checkpoint can be an important determinant of cellular viability in such cells, notably when chromosome segregation defects have been induced by the inhibition of aurora kinases (18). Given that p53 mutation status is one of the known molecular differences between MCF-7 (wild-type) and MDA-MB-231 (R280K-inactivating mutant) cells, we assessed the role of p53 in the cellular response to CtBP siRNA. In MCF-7 cells, CtBP siRNA resulted in a >10-fold upregulation of the p53 protein 3 days posttransfection (Fig. 6B), consistent with p53 activation. A similar p53 response to CtBP knockdown was found in a nontransformed human fibroblast cell line (see Fig. S7 in the supplemental material). We therefore cotransfected cells with siRNAs for both CtBP and p53 (Fig. 6E); again, CtBP siRNA induced an increase in p53 protein abundance, as well as upregulation of p21WAF1, the transcription of which is induced by p53. p53 siRNA suppressed the CtBP siRNA-induced increase in the abundance of both p53 and p21WAF1 proteins. Twenty-four-hour live-cell imaging analysis (Fig. 6D; see Videos S9 and S10 in the supplemental material) showed that, whereas p53 siRNA alone had no significant effect on MCF-7 cell numbers and CtBP depletion resulted in a 10% decrease as shown previously, when CtBP and p53 were depleted simultaneously, cell numbers were reduced by 35% (Fig. 6Di), a figure that is similar to that seen after CtBP depletion in MDA-MB-231 cells (Fig. 1Di). This large reduction in cell numbers after combined CtBP-p53 ablation was primarily a consequence of a large increase in cell death to 40/100 cells (Fig. 6Dii). There was also a further reduction in the mitotic index (Fig. 6Diii). These effects of p53 on cell survival were also seen in long-term clonogenic assays (Fig. 6E) and were confirmed using a second siRNA to p53 (see Fig. S9 in the supplemental material). Together, these data demonstrate that loss of CtBP expression in wild-type p53-expressing cells results in the activation of a p53 response. In the absence of this response, cells depleted of CtBPs are prone to high rates of apoptotic cell death.
DISCUSSION
The aim of this study was to better define the mechanistic basis of the prosurvival and proproliferative activities of CtBPs (reviewed in references 1 and 6). We discovered that CtBPs are required for the maintenance of mitotic fidelity. In addition to being a previously unidentified cellular role for these proteins, this is an important component of their prosurvival function. We also found, consistent with previous studies, that the cell death that is induced by CtBP knockdown does not require p53 function. However, in cells with functional p53, we found that loss of CtBP does induce a cellular p53 response that includes upregulation of the cyclin-dependent kinase inhibitor p21WAF1. Furthermore, by suppressing the p53-independent apoptotic response, p53 has a protective effect in CtBP-depleted cells.
CtBP siRNA-treated cells display a marked reduction in the number of cells that enter mitosis. Based on previous studies, the failure of cells to enter mitosis was expected, due to a demonstrated requirement for CtBPs for fissioning of the Golgi apparatus prior to mitotic entry (10, 22). However in our experiments, rather than activation of a “Golgi checkpoint” in late G2, CtBP siRNA resulted in a reduction in the number of cells in late G2 sufficient to account for the reduced mitotic entry. This appears to be due to a cell line- and/or p53-dependent combination of death and arrest, most likely predominantly in G1 and/or S phase. Presumably, the lack of a Golgi apparatus phenotype in our experiments indicates that these breast cancer cells express endophilin B or other such factors that are now known to be interchangeable with CtBPs in Golgi apparatus fission (60).
We found three distinct, though potentially interrelated, mitotic phenotypes in CtBP siRNA-treated cells: (i) lagging chromosomes during mitosis and, in interphase cells, micronuclei that contained primarily centromere-containing chromatin, together indicative of entry into anaphase prior to completion of amphitelic attachment of paired sister chromatids to microtubules; (ii) an extended average time spent in mitosis, with mitosis being delayed predominantly at the prometaphase stage; and (iii) failure of a proportion of cells to complete the final, abscission, phase of cytokinesis, resulting in the generation of binucleate cell progeny. Given that all known delays in mitosis are mediated by the SAC (42), it can be concluded from the second phenotype that this checkpoint is activated for longer in CtBP-compromised cells than in controls. Many conditions are able to induce a delay in mitosis, including defects in centrosome separation, anchorage of microtubules to kinetochores, formation of stable microtubule-kinetochore attachments, prevention of cyclin B/CDK1 inactivation, or global DNA damage (42). The combined mitotic phenotype of CtBP-compromised cells most closely resembles a modest form of that obtained when both the formation of stable microtubule attachments and cytokinesis are disrupted through the experimental targeting of chromosome passenger proteins, notably aurora B kinase (12, 20, 46) and survivin (29). We therefore examined the effects of CtBP knockdown on accumulation of aurora B on mitotic chromosomes and found the accumulation to be reduced.
Early in mitosis, SAC proteins localize to outer kinetochores lacking attached microtubules and maintain the anaphase wait signal until microtubule attachment leads to their release from the kinetochore (30, 33). Kinetochore-microtubule attachments that result in both sister chromatids being connected to the same spindle pole (merotelic attachments) are thought to be severed through a chromosome passenger protein-dependent pathway, resulting in rerecruitment of SAC proteins and ensuring the anaphase wait signal is maintained until all pairs of sister chromatids have correct bipolar attachments (7, 38, 43). Reduced efficacy of this aurora B kinase-dependent process would provide an explanation for the mitotic phenotypes i and ii we observed in CtBP-depleted cells, i.e., the overall process would take longer, resulting in a prolonged SAC and extended time in mitosis (phenotype ii). However, when only a small number of inappropriate connections are left, some may theoretically remain undetected by chromosomal passenger proteins for long enough for anaphase-promoting complex activity to be derepressed and anaphase initiated. Such a mechanism would result in lagging chromosomes and the subsequent micronuclei that we observed (phenotype i). A recent study in which cells were exposed to a low concentration of the kinase inhibitor ZM447439 clearly demonstrated that the frequency of formation of lagging chromosomes is very sensitive to partial inhibition of aurora enzyme activity (9). Subsequent to anaphase, chromosome passenger proteins migrate to the central spindle and are required for cytokinesis, which fails if their expression is compromised, producing binucleate cells (12, 20, 29, 46, 55). Critically, aurora B activity is required for the activation of an abscission checkpoint in response to chromatin being trapped within the cleavage furrow due to chromatin bridges. Loss of this activity results in a failure to resolve the bridges, and the cleavage furrow regresses, resulting in the formation of a tetraploid cell (49). Even partial inhibition of aurora activity can cause a modest increase in the frequency of failed cytokinesis (9), again comparable to the effects of CtBP siRNA (phenotype iii).
At present, based on known functions of CtBPs, it is not clear how they may exert their effects on aurora B, mitotic progression, and mitotic fidelity. There are, however, a number of important observations that provide avenues for investigation. Might CtBPs play a direct role in mitosis, or is the effect indirect, for example, through regulating the transcription of other genes with roles in mitosis? The latter is certainly possible, as FoxM1, a gene with a critical role in many aspects of cell division, including mitotic entry, centrosome amplification, and the spindle checkpoint (28), is upregulated in Ctbp1−/−Ctbp2−/− murine embryo fibroblasts (19). The reduced accumulation of aurora B on prophase and prometaphase chromatin in CtBP-depleted cells could be due to either effects on the overall abundance of aurora B in mitotic cells or its recruitment to centromeres as a component of the chromosome passenger complex. With respect to the latter, it is striking that treatment of cells in S phase with pharmacological inhibitors of HDAC activity cause delay in the subsequent mitosis, as well as defects in chromosome alignment and increased frequency of lagging chromosomes, and failed cytokinesis (8, 50, 59). A reduction in the concentration of chromosomal passenger proteins, including aurora B, in prophase and prometaphase centromeres, with no effect on the overall abundance of these proteins in mitotic cells, appears to underlie this phenotype (50), though further details of the mechanism remain to be elucidated. Given that HDACs are a core component of CtBP complexes in cells (26, 47), the mitotic phenotypes of CtBP siRNA-treated cells could be a consequence of hyperacetylation of histones in S phase. Finally, in contrast, CtBPs may have a direct role in mitosis; it has long been recognized that CtBP1 is differentially phosphorylated through the cell cycle, with overall phosphorylation reaching a peak during mitosis (4). In addition CtBPs localize to centrosomes throughout mitosis in a complex including γ-tubulin and Plk1 (48).
Several reports have clearly demonstrated that CtBPs play a prosurvival role, with loss of their expression resulting in either apoptosis or hypersensitivity to apoptotic stimuli through a p53-independent mechanism (19, 37, 63; reviewed in references 1 and 6). This is an important observation, because CtBPs are also targeted for degradation in response to cancer chemotherapeutic agents, such as cisplatin, a response that is part of the cytotoxic mechanism of such agents (58). However, the involvement of CtBPs in such a wide range of key intracellular signaling pathways (1, 6) has made it difficult to define the mechanistic basis of this prosurvival activity. Indeed, it seems probable that CtBPs impinge upon more than one antiapoptotic pathway; suppression of anoikis through the inhibition of epithelial gene expression in cells with a mesenchymal phenotype appears to be one such pathway (19); here, we show the maintenance of mitotic fidelity to be another. Studies with aurora kinase inhibitors (12, 18) have found that, in the absence of a functional p53 stress response pathway, cells that exit mitosis following failed cytokinesis enter subsequent rounds of S phase, in which they show a high propensity to undergo cell death. We found that the inhibition of apoptosis in the breast cancer cell lines led to an increase in the proportion of cells that contained micronuclei. This effect was very apparent in MDA-MB-231 cells in which CtBP expression had been inhibited. Thus, in this mutant p53-expressing cell line, cells that have undergone chromosome segregation defects in response to CtBP siRNA show an increased propensity to undergo subsequent apoptosis compared to cells without such defects.
Inhibition of CtBP expression in the MCF-7 breast cancer cell line caused a clear increase in p53 protein abundance and upregulation of its transcriptional target, p21WAF1. By extrapolation from previous studies in which the effects of chromosome segregation defects in wild-type p53-expressing cells were examined (12, 18, 44), it seems likely that this p53 activation occurs in G1 cells in response to the consequences of chromosome segregation defects in the prior mitosis. It is quite possible that loss of CtBP expression also leads to p53 activation through additional pathways in these cells, particularly given the previously described physical and functional interaction between CtBPs and the p53 negative-regulatory partner HDM2 (34). However, despite the mechanism of p53 induction, it is clear from our data that p53 protects MCF-7 cells from loss of CtBP expression in short-term apoptosis assays and also in longer-term tests of clonogenic survival. While p53 is generally considered to be a proapoptotic molecule, previous studies have shown that p53-dependent expression of p21WAF1, which induces cell cycle arrest primarily in G1 phase, can protect cells from p53-dependent apoptosis (39). This p21WAF1 induction can also protect from apoptosis in cells that contain chromosome segregation defects as a consequence of their exposure to aurora kinase inhibitors (18). It seems probable that a similar response protects CtBP-depleted cells. The ability to induce cell death with selectivity for cells with a mutation in p53 would be a useful property in any cancer chemotherapeutic drug. Our study therefore supports previous assertions that CtBPs warrant further investigation for their potential as targets for cancer therapeutics (58). It would also be important to determine the extent to which the loss of CtBP-dependent maintenance of mitotic fidelity plays a role in the cellular response to other agents with anticaner activity, notably genotoxics, such as cisplatin, and HDAC inhibitors.
Supplementary Material
Acknowledgments
This work is supported by grants (no. 2003:713 and 2007MayPR15) from the U.K. Breast Cancer Campaign.
We are grateful to Peter Lackie, University of Southampton, for assistance with the live-cell imaging of breast cancer cells.
Footnotes
Published ahead of print on 8 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bergman, L. M., and J. P. Blaydes. 2006. C-terminal binding proteins: emerging roles in cell survival and tumorigenesis. Apoptosis 11879-888. [DOI] [PubMed] [Google Scholar]
- 2.Bergman, L. M., L. Morris, M. Darley, A. H. Mirnezami, S. C. Gunatilake, and J. P. Blaydes. 2006. Role of the unique N-terminal domain of CtBP2 in determining the subcellular localisation of CtBP family proteins. BMC Cell Biol. 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonazzi, M., S. Spano, G. Turacchio, C. Cericola, C. Valente, A. Colanzi, H. S. Kweon, V. W. Hsu, E. V. Polishchuck, R. S. Polishchuck, M. Sallese, T. Pulvirenti, D. Corda, and A. Luini. 2005. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat. Cell Biol. 7570-580. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnadurai, G. 2007. CtBP family proteins: unique transcriptional regulators in the nucleus with diverse cytosolic functions, p. 1-17. In G. Chinnadurai (ed.), CtBP family proteins. Springer, New York, NY.
- 6.Chinnadurai, G. 2009. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 69731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimini, D. 2007. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle 61558-1564. [DOI] [PubMed] [Google Scholar]
- 8.Cimini, D., M. Mattiuzzo, L. Torosantucci, and F. Degrassi. 2003. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell 143821-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimini, D., X. Wan, C. B. Hirel, and E. D. Salmon. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 161711-1718. [DOI] [PubMed] [Google Scholar]
- 10.Colanzi, A., C. H. Carcedo, A. Persico, C. Cericola, G. Turacchio, M. Bonazzi, A. Luini, and D. Corda. 2007. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 262465-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corda, D., A. Colanzi, and A. Luini. 2006. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 16167-173. [DOI] [PubMed] [Google Scholar]
- 12.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditchfield, C., N. Keen, and S. S. Taylor. 2005. The Ipl1/Aurora kinase family: methods of inhibition and functional analysis in mammalian cells. Methods Mol. Biol. 296371-381. [DOI] [PubMed] [Google Scholar]
- 14.Echeverri, C. J., P. A. Beachy, B. Baum, M. Boutros, F. Buchholz, S. K. Chanda, J. Downward, J. Ellenberg, A. G. Fraser, N. Hacohen, W. C. Hahn, A. L. Jackson, A. Kiger, P. S. Linsley, L. Lum, Y. Ma, B. Mathey-Prevot, D. E. Root, D. M. Sabatini, J. Taipale, N. Perrimon, and R. Bernards. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3777-779. [DOI] [PubMed] [Google Scholar]
- 15.Furusawa, T., H. Moribe, H. Kondoh, and Y. Higashi. 1999. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol. Cell. Biol. 198581-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallop, J. L., P. J. Butler, and H. T. McMahon. 2005. Endophilin and CtBP/BARS are not acyl transferases in endocytosis or Golgi fission. Nature 438675-678. [DOI] [PubMed] [Google Scholar]
- 17.Gassmann, R., A. Carvalho, A. J. Henzing, S. Ruchaud, D. F. Hudson, R. Honda, E. A. Nigg, D. L. Gerloff, and W. C. Earnshaw. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gizatullin, F., Y. Yao, V. Kung, M. W. Harding, M. Loda, and G. I. Shapiro. 2006. The Aurora kinase inhibitor VX-680 induces endoreduplication and apoptosis preferentially in cells with compromised p53-dependent postmitotic checkpoint function. Cancer Res. 667668-7677. [DOI] [PubMed] [Google Scholar]
- 19.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 1004568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106348-360. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo Carcedo, C., M. Bonazzi, S. Spano, G. Turacchio, A. Colanzi, A. Luini, and D. Corda. 2004. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science 30593-96. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 225296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, M., C. Uchida, Y. Takano, M. Kitagawa, and Y. Okano. 2004. Cell cycle-dependent regulation of the human aurora B promoter. Biochem. Biophys. Res. Commun. 316930-936. [DOI] [PubMed] [Google Scholar]
- 25.Kops, G. J., B. A. Weaver, and D. W. Cleveland. 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5773-785. [DOI] [PubMed] [Google Scholar]
- 26.Kuppuswamy, M., S. Vijayalingam, L. J. Zhao, Y. Zhou, T. Subramanian, J. Ryerse, and G. Chinnadurai. 2008. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol. Cell. Biol. 28269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanni, J. S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 181055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laoukili, J., M. R. Kooistra, A. Bras, J. Kauw, R. M. Kerkhoven, A. Morrison, H. Clevers, and R. H. Medema. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7126-136. [DOI] [PubMed] [Google Scholar]
- 29.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 222934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37251-282. [DOI] [PubMed] [Google Scholar]
- 31.Lindberg, H. K., X. Wang, H. Jarventaus, G. C. Falck, H. Norppa, and M. Fenech. 2007. Origin of nuclear buds and micronuclei in normal and folate-deprived human lymphocytes. Mutat. Res. 61733-45. [DOI] [PubMed] [Google Scholar]
- 32.Mackay, A. M., A. M. Ainsztein, D. M. Eckley, and W. C. Earnshaw. 1998. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140991-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiato, H., J. DeLuca, E. D. Salmon, and W. C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 1175461-5477. [DOI] [PubMed] [Google Scholar]
- 34.Mirnezami, A. H., S. J. Campbell, M. Darley, J. N. Primrose, P. W. Johnson, and J. P. Blaydes. 2003. Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription. Curr. Biol. 131234-1239. [DOI] [PubMed] [Google Scholar]
- 35.Nardini, M., S. Spano, C. Cericola, A. Pesce, A. Massaro, E. Millo, A. Luini, D. Corda, and M. Bolognesi. 2003. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 223122-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibu, Y., and M. S. Levine. 2001. CtBP-dependent activities of the short-range Giant repressor in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 986204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paliwal, S., S. Pande, R. C. Kovi, N. E. Sharpless, N. Bardeesy, and S. R. Grossman. 2006. Targeting of C-terminal binding protein (CtBP) by ARF results in p53-independent apoptosis. Mol. Cell. Biol. 262360-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinsky, B. A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15486-493. [DOI] [PubMed] [Google Scholar]
- 39.Polyak, K., T. Waldman, T. C. He, K. W. Kinzler, and B. Vogelstein. 1996. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 101945-1952. [DOI] [PubMed] [Google Scholar]
- 40.Poortinga, G., M. Watanabe, and S. M. Parkhurst. 1998. Drosophila CtBP: a hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 172067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 966683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieder, C. L., and H. Maiato. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7637-651. [DOI] [PubMed] [Google Scholar]
- 43.Ruchaud, S., M. Carmena, and W. C. Earnshaw. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8798-812. [DOI] [PubMed] [Google Scholar]
- 44.Sablina, A. A., G. V. Ilyinskaya, S. N. Rubtsova, L. S. Agapova, P. M. Chumakov, and B. P. Kopnin. 1998. Activation of p53-mediated cell cycle checkpoint in response to micronuclei formation. J. Cell Sci. 111977-984. [DOI] [PubMed] [Google Scholar]
- 45.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 9210467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher, J. M., A. Golden, and P. J. Donovan. 1998. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1431635-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, Y., J. Sawada, G. Sui, B. el Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422735-738. [DOI] [PubMed] [Google Scholar]
- 48.Spyer, M., and M. J. Allday. 2006. The transcriptional co-repressor C-terminal binding protein (CtBP) associates with centrosomes during mitosis. Cell Cycle 5530-537. [DOI] [PubMed] [Google Scholar]
- 49.Steigemann, P., C. Wurzenberger, M. H. Schmitz, M. Held, J. Guizetti, S. Maar, and D. W. Gerlich. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136473-484. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, F. E., H. Beamish, R. Warrener, and B. Gabrielli. 2008. Histone deacetylase inhibitors induce mitotic slippage. Oncogene 271345-1354. [DOI] [PubMed] [Google Scholar]
- 51.Terada, Y., M. Tatsuka, F. Suzuki, Y. Yasuda, S. Fujita, and M. Otsu. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Touitou, R., M. Hickabottom, G. Parker, T. Crook, and M. J. Allday. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 757749-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tripathi, M. K., S. Misra, S. V. Khedkar, N. Hamilton, C. Irvin-Wilson, C. Sharan, L. Sealy, and G. Chaudhuri. 2005. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J. Biol. Chem. 28017163-17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23683-690. [DOI] [PubMed] [Google Scholar]
- 55.Vader, G., R. H. Medema, and S. M. Lens. 2006. The chromosomal passenger complex: guiding Aurora-B through mitosis. J. Cell Biol. 173833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verger, A., K. G. Quinlan, L. A. Crofts, S. Spano, D. Corda, E. P. Kable, F. Braet, and M. Crossley. 2006. Mechanisms directing the nuclear localization of the CtBP family proteins. Mol. Cell. Biol. 264882-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker, J. A., D. R. Boreham, P. Unrau, and A. M. Duncan. 1996. Chromosome content and ultrastructure of radiation-induced micronuclei. Mutagenesis 11419-424. [DOI] [PubMed] [Google Scholar]
- 58.Wang, S. Y., M. Iordanov, and Q. Zhang. 2006. c-Jun NH2-terminal kinase promotes apoptosis by down-regulating the transcriptional co-repressor CtBP. J. Biol. Chem. [DOI] [PubMed]
- 59.Warrener, R., H. Beamish, A. Burgess, N. J. Waterhouse, N. Giles, D. Fairlie, and B. Gabrielli. 2003. Tumor cell-selective cytotoxicity by targeting cell cycle checkpoints. FASEB J. 171550-1552. [DOI] [PubMed] [Google Scholar]
- 60.Yang, J. S., L. Zhang, S. Y. Lee, H. Gad, A. Luini, and V. W. Hsu. 2006. Key components of the fission machinery are interchangeable. Nat. Cell Biol. 81376-1382. [DOI] [PubMed] [Google Scholar]
- 61.Zaffaroni, N., M. Pennati, and M. G. Daidone. 2005. Survivin as a target for new anticancer interventions. J. Cell Mol. Med. 9360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Q., S. Y. Wang, A. C. Nottke, J. V. Rocheleau, D. W. Piston, and R. H. Goodman. 2006. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc. Natl. Acad. Sci. USA 1039029-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Q., Y. Yoshimatsu, J. Hildebrand, S. M. Frisch, and R. H. Goodman. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115177-186. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, L. J., T. Subramanian, Y. Zhou, and G. Chinnadurai. 2006. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J. Biol. Chem. 2814183-4189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.