Abstract
“Neutropenia” refers to deficient numbers of neutrophils, the most abundant type of white blood cell. Two main forms of inherited neutropenia are cyclic neutropenia, in which neutrophil counts oscillate with a 21-day frequency, and severe congenital neutropenia, in which static neutropenia may evolve at times into leukemia. Mutations of ELA2, encoding the protease neutrophil elastase, can cause both disorders. Among other genes, severe congenital neutropenia can also result from mutations affecting the transcriptional repressor Gfi1, one of whose genetic targets is ELA2, suggesting that the two act through similar mechanisms. In order to identify components of a common pathway regulating neutrophil production, we conducted yeast two-hybrid screens with Gfi1 and neutrophil elastase and detected a novel protein, PFAAP5 (also known as N4BP2L2), interacting with both. Expression of PFAAP5 allows neutrophil elastase to potentiate the repression of Gfi1 target genes, as determined by reporter assays, RNA interference, chromatin immunoprecipitation, and impairment of neutrophil differentiation in HSCs with PFAAP5 depletion, thus delineating a mechanism through which neutrophil elastase could regulate its own synthesis. Our findings are consistent with theoretical models of cyclic neutropenia proposing that its periodicity can be explained through disturbance of a feedback circuit in which mature neutrophils inhibit cell proliferation, thereby homeostatically regulating progenitor populations.
Neutrophils provide innate immunity against bacterial and fungal pathogens (13). Consequently, deficient numbers of neutrophils (neutropenia) increase vulnerability to infection. The two main forms of inherited neutropenia in humans (1, 12, 23, 34) are cyclic neutropenia and severe congenital neutropenia (SCN) (also known as “Kostmann syndrome”). Cyclic neutropenia is an autosomal dominant disorder in which the peripheral neutrophil count oscillates with a 21-day frequency. SCN is a genetically heterogeneous disorder comprised of static neutropenia and predisposition to myelodysplasia and acute myelogenous leukemia.
Heterozygous mutations in ELA2, the gene encoding the neutrophil granule serine protease neutrophil elastase, cause cyclic neutropenia (33) and are the most common cause of SCN (7, 17). SCN can also rarely result from dominantly acting mutations of the gene encoding the zinc finger-containing transcriptional repressor Gfi1 (55), whose deficiency in mice also produces neutropenia (30, 36). Other genes responsible for hereditary forms of neutropenia include HAX1, involved in apoptosis, among other functions (57), and recently found to be mutated in many cases of autosomal recessive SCN (41); AP3B1, encoding a subunit of the AP3 complex involved in posttranslational vesicular trafficking and whose autosomal recessive mutation leads to human Hermansky Pudlak syndrome type 2 (5) and canine cyclic neutropenia (8); certain alleles of the gene mutated in Wiskott-Aldrich syndrome (2), which encodes a protein assisting with organization of the actin cytoskeleton; G6PC3, encoding glucose-6 phosphatase, involved in gluconeogenic and glycogenolytic pathways in the endoplasmic reticulum and which was recently found to be mutated in a syndromic form of neutropenia (14); the gene encoding ras-associated protein RAB27A, the myosin gene MYO5, or the SLAC2A/melanophilin gene (12), mutations in which result in Griscelli hemophagocytic syndrome; and the CXCR4 gene, mutations in which cause the WHIM syndrome of myelokathexis (38).
Some of the factors responsible for hereditary neutropenia demonstrate molecular genetic interactions. ELA2 is among the genes transcriptionally repressed by Gfi1 (30, 55), and neutropenic mutations of Gfi1 consequently result in ELA2 overexpression. AP3B1 has a role in the translocation of neutrophil elastase from the Golgi apparatus to the granules of neutrophils, and disturbance of AP3B1 eventuates in intracellular mislocalization of neutrophil elastase (8). Transcriptional profiles of SCN patients with either ELA2 (37) or HAX1 (58) mutations show downregulation of ELA2. Thus, it is possible that the different neutropenia genes represent components of a pathway regulating neutrophil production.
In fact, the bizarre oscillations characteristic of cyclic neutropenia might be taken as evidence of a regulatory mechanism gone awry. Theoretical models of cyclic neutropenia have invoked a defective feedback circuit (28, 51) in which mature neutrophils somehow inhibit progenitor cells once the cell count has risen sufficiently and in which it can be imagined that pathologically excessive inhibition might dampen neutrophil production temporarily until mature neutrophils become depleted and the negative feedback signal abates, thereby establishing a cyclical pattern. Such a feedback circuit accords with the “chalone” hypothesis (56), independently predicting that neutrophils homeostatically regulate their own production by inhibiting their progenitors. Intriguingly, even prior to the determination of the role of neutrophil elastase in neutropenia, an attempt to biochemically identify the postulated chalone led to the isolation of neutrophil elastase as a molecule likely functioning in this capacity (46).
Current hypotheses for the molecular pathology underlying neutropenia center on the intracellular mislocalization (8, 32) and/or misfolding (24, 42) of mutant neutrophil elastase. Regardless of which is correct, a better understanding of the disease mechanism requires an explanation for the rhythmic phenomenon of cyclic neutropenia. With the goal of defining genetic pathways regulating neutrophil production, we sought to uncover factors interacting with neutrophil elastase and Gfi1. Remarkably, we discovered a novel protein, PFAAP5 (phosphonoformate immunoassociated protein 5, also known as N4BP2L2), which appears to make contact with both. Here, we characterize PFAAP5's association with neutrophil elastase and Gfi1 and investigate its potential participation in a feedback circuit in which neutrophil elastase regulates its own transcription.
MATERIALS AND METHODS
Yeast two-hybrid assay.
We used the Matchmaker two-hybrid system (Clontech). We have previously reported on the yeast two-hybrid screen performed with Gfi1 (20). For neutrophil elastase, we prepared fusions of the indicated sequences to the Gal4 DNA binding domain vector pGBKT7 and screened at high stringency (in media lacking adenine) against 106 clones from a human bone marrow cDNA library (Clontech) inserted into the Gal4 DNA activation domain vector pGADT7. Interacting plasmid clones were retransformed with the bait plasmid and control bait encoding a LexA-lamin C fusion, and the identities of bait-dependent positive interactors were determined by DNA sequencing.
Plasmids.
PFAAP5 cDNA was reverse transcribed from total human bone marrow RNA (and found to be identical to GenBank entry BC010643) and cloned into pCS2+ and pCS2+Myc-tag vectors (62). Expression constructs in pCS2+ and pCS2+Myc-tag vectors containing ELA2 (47) and GFI1 (21, 55) cDNAs and their derivatives have been previously described, with the exception of the glycosylation variant of ELA2, N95A/N144A, which was constructed by site-directed mutagenesis of full-length ELA2 (containing the carboxyl-terminal propeptide) in the same vector system.
Antibody production.
Three different affinity-purified chicken polyclonal antibodies were raised by Aves Laboratories against the PFAAP5 peptide C155 to E171, the neutrophil elastase carboxyl-terminal propeptide C225 to H238, and the internal epitope Q88 to D103.
Immunohistochemistry.
Immunolocalization of the neutrophil elastase N95A/N144A construct transiently transfected into RBL1 cells was performed as previously described (47), with the addition of counterstaining for LAMP1 (lysosomal membrane glycoprotein 1) with 5 μg/ml primary mouse monoclonal antibody LY1C6 (GeneTex) and 10 μg/ml secondary Alexa Fluor 555-conjugated donkey anti-mouse immunoglobulin G (Invitrogen). Imaging was done with a Zeiss 510 laser scanning confocal microscope with a 63× Plan-Apochromat objective and Zeiss LSM Image Browser software.
Coimmunoprecipitation.
NIH 3T3 cells (4 × 106 in four 10-cm plates) were transiently transfected using Lipofectamine Plus (Invitrogen) with 24 μg each of pCS2+ expression vectors for PFAAP5, Gfi1, and neutrophil elastase in three sets in which one of each of the expression vectors contained an amino-terminal Myc epitope tag and in an additional control set of experiments in which all three expression vectors lacked the epitope tag. Each of the four sets of experiments was repeated twice: once using a form of neutrophil elastase containing the carboxyl-terminal propeptide and once using a form of neutrophil elastase lacking this sequence. Forty hours following transient transfection, cells were harvested and lysed using the Complete Lysis-M kit (Roche). To reduce nonspecific binding, the lysates were brought to a volume of 1 ml in lysis buffer and incubated with 50 μl recombinant protein G-agarose beads (Invitrogen) at 4°C for 30 min with gentle agitation. The supernatants were split into three aliquots, brought to a volume of 1 ml using lysis buffer, and incubated with 25 μg anti-Myc 9E10 mouse monoclonal antibody (Roche) for 3 h at 4°C with gentle agitation. Recombinant protein G-agarose beads (50 μl) were added, and incubation was resumed overnight. The immunoprecipitates were collected and washed six times with 1 ml of phosphate-buffered saline and then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting, as previously described (20, 21), except that for detection of Gfi1 we used 3.33 ng/ml of mouse monoclonal antibody 2.5D.17 (Sigma) conjugated to horseradish peroxidase (using the Lightning-Link HRP conjugation kit; Innova Biosciences); for detection of neutrophil elastase containing the carboxyl-terminal propeptide and PFAAP5, we used 2 μg/ml and 0.5 μg/ml of the respective primary chicken antibody to carboxyl-terminal propeptide C225-H238, followed by a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-chicken secondary antibody (Aves Laboratories).
ChIP.
For chromatin immunoprecipitation (ChIP) assays, the set of combinations of transfected vectors was the same as for the coimmunoprecipitation experiments described above, except that the number of transfected NIH 3T3 cells and amount of DNA were halved. ChIP assays were performed with the Chromatin Immunoprecipitation Assay kit (Upstate), using 25 μg 9E10 as the immunoprecipitating antibody. Semiquantitative PCR was performed for each assay using gene-specific primer pairs. Each assay was performed in triplicate, with representative data shown.
Transcriptional reporter assays.
Transcriptional reporter assays were performed as previously described (55), with the following modifications. NIH 3T3 cells were transiently transfected using Lipofectamine Plus. The Gfi1-responsive pB30×2-TKCAT reporter construct (0.5 μg) (25) was cotransfected with 1 μg of the internal-control Renilla luciferase vector pRL-TK, the indicated quantity of each experimental plasmid, and, in order to keep the total amount of DNA constant across experimental conditions, either zero or varying quantities of pCS2 plus β-galactosidase. Cell lysates were harvested 40 h later. To ensure equal expression of the two forms of neutrophil elastase (with and without the carboxyl-terminal propeptide), a Western blot was performed using chicken polyclonal antibodies to the internal epitope Q88-D103 (processed as described above for Western blots utilizing the chicken antibody to the carboxyl-terminal propeptide), followed by reprobing with C-11 horseradish peroxidase-conjugated antibody to actin (Santa Cruz Biotechnology) as an internal loading control. Chloramphenicol acetyltransferase (CAT) activity was assayed with the CAT ELISA kit (Roche) and normalized to Renilla luciferase activity measured with the Dual-Luciferase assay (Promega). Experiments were performed in triplicate and assayed twice each, with reporting of the standard error of the mean between experiments. The following modifications were performed in assays evaluating the effects of reduced expression of PFAAP5: custom small interfering RNA (siRNA) directed against the mouse ortholog (GenBank accession number BC037393) of PFAAP5 (sense strand, ACAACAUUGUCUCGAAUUCTT; antisense strand, GAAUUCGAGACAAUGUUGUTT) and negative control no. 2 siRNA were supplied by Ambion, and siRNA (50 μmol) and 1 μg of each reporter (pRL-TK and pB30×2-TKCAT) were cotransfected with 10 ng of Gfi1 and/or 40 ng of neutrophil elastase (with or without the carboxyl-terminal propeptide) cDNA contained in pCS2+ expression vectors.
siRNA assays in HL-60 cells.
Predesigned Silencer siRNAs (100 μmol; Ambion) directed against human PFAAP5 (no. 22205), GFI1 (no. 16706), or ELA2 (no. 41659) were electroporated into HL-60 cells as previously described (21) and, after 48 h of quantitative real-time reverse transcription (RT)-PCR was performed, were normalized as before (21), using TaqMan (Applied Biosystems) gene expression assays (PFAAP5, Hs00208459_m1; GFI1, Hs00382207_m1; ELA2, Hs00357734_m1; AZU1, Hs00156049_m1).
Isolation of human hematopoietic stem cells (HSCs).
Human umbilical cord blood and human adult peripheral stem cells were obtained under a protocol approved by the Cincinnati Children's Hospital Medical Center Institutional Review Board. CD34+ cells were enriched to >90% purity by immunomagnetic bead selection (Miltenyi Biotec) and cryopreserved. Once thawed, the cells were cultured in Iscove's modified Dulbecco's medium containing 20% fetal bovine serum and supplemented with recombinant human stem cell factor, thrombopoietin, and Flt-3L (StemCell Technologies), all at 10 ng/ml.
Short hairpin RNA (shRNA) constructs and lentiviral transduction.
Lentiviral shPFAAP5 vector MISSION pLKO.1-shPFAAP5-puro constructs were obtained from Sigma. Yellow fluorescent protein (YFP) was subcloned into the KpnI/BamHI site of pLKO.1-shPFAAP5 vectors TRCN0000122131 and TRCN0000145053, replacing puromycin. shGFI1 constructs have been previously described (50). HEK 293T cells cultured in Dulbecco's modified Eagle medium with 10% fetal bovine serum were transfected by calcium phosphate with the lentiviral vectors, vesicular stomatitis virus glycoprotein envelope expression plasmid, and delta-8.9 packaging plasmid. Virus was collected and filtered through a 0.45-μm filter, pelleted by centrifugation, and then purified by a secondary centrifugation through a 20% sucrose cushion. For transduction, CD34+ cells were cultured in the presence of lentiviral supernatant supplemented with Polybrene overnight. Three days after transduction, the cells were sorted for YFP expression on a FACSAria cell sorter (BD Biosciences).
CFU assays.
Two thousand YFP+ sorted CD34+ CB cells were seeded per 35-mm plate in MethoCult H4100 medium (StemCell Technologies) supplemented with 20% BIT 9500 serum substitute (StemCell Technologies), 100 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/ml penicillin-streptomycin, and the human cytokines erythropoietin (6 U/ml), granulocyte colony-stimulating factor (10 ng/ml), interleukin-6 (20 ng/ml), interleukin-3 (20 ng/ml), and stem cell factor (20 ng/ml), obtained from StemCell Technologies. Cultures were plated in replicate, and colonies of more than 50 cells were scored after 14 days of incubation.
RESULTS
Two-hybrid screens with Gfi1 and neutrophil elastase each recover PFAAP5.
The yeast two-hybrid system is a tool for detecting protein-protein interactions. In order to identify proteins capable of interacting with Gfi1, we recently (20) performed a yeast two-hybrid screen of human bone marrow cDNA utilizing the zinc finger region of human Gfi1 as bait. Under high-stringency conditions (Table 1), we recovered just 28 interacting proteins (in the initial report [20], we characterized Gfi1's association with the PRDM5 transcription factor).
TABLE 1.
Yeast two-hybrid screen with Gfi1 zinc fingers
| Protein | No. of clonesa |
|---|---|
| Ubiquitin conjugating enzyme 9 (UBC9) | 13 (all overlap) |
| PIAS4 | 1 |
| Importin 8 | 1 |
| Ubiquitin A52 ribosomal fusion product | 4 (overlap) |
| PCNA | 1 |
| PRDM5 | 1 |
| Four and a half LIM domains 1 variant | 2 (overlap) |
| GLI-Kruppel family member HKR2 | 3 (overlap) |
| ZXDC | 2 (identical) |
| Small nuclear ribonucleoprotein D1 | 4 (2 identical, all overlap) |
| High-mobility group Box 2 | 4 (all overlap) |
| Poly(rC) binding protein 1 | 2 (overlap) |
| CD81 | 4 (3 identical, overlap) |
| Erythrocyte membrane protein band 4.2 | 1 |
| Synaptotagmin-like 1, isoform CRA_b | 1 |
| Translocase of inner mitochondrial membrane 50 homolog | 1 |
| Succinate dehydrogenase complex subunit B | 1 |
| Titin, isoform CRA_a | 1 |
| Fibrillin 2 | 1 |
| von Willebrand factor preproprotein | 1 |
| Eukaryotic translation initiation factor 3, subunit 7 zeta, 66/67kDa, isoform CRA_b | 1 |
| Ribosomal protein S20 | 1 |
| Eukaryotic translation initiation factor 3 subunit 7 | 1 |
| Hypothetical protein LOC54976 | 1 |
| PFAAP5 | 1 |
| Hypothetical protein XP_001133338 | 1 |
| Hypothetical protein similar to 60S ribosomal protein L29 | 1 |
| Trinucleotide repeat containing 5, isoform CRA_a | 3 (all overlap) |
Independent clones with overlapping sequences are indicated by “overlap”; identical clones are noted. cDNAs not producing open reading frames are not shown.
We later performed a yeast two-hybrid screen of the same library using human neutrophil elastase as bait. We initially found that expression of full-length ELA2 cDNA proved toxic to yeast (not shown), consistent with previously described fungicidal properties (54). We reasoned that the antimicrobial activity might be confined to a particular part of the protein. Noting that the tertiary structure of neutrophil elastase is comprised of two nearly equal-size beta-barrel structures joined at amino acid P110 (11), we constructed baits corresponding to each half of neutrophil elastase. The amino-terminal half of neutrophil elastase (I1 to P110) again proved toxic to yeast (not shown), but the half containing the more carboxyl terminal of the two beta-barrel domains (P110 to H238) could be stably cloned in yeast. Another consideration is that, in addition to “prepro” processing as part of signal peptide cleavage and zymogen activation occurring at the amino-terminal end of the protein, neutrophil elastase undergoes processing at its carboxyl terminus (26). It is typically purified as a “mature” form in which a 20-amino-acid-residue carboxyl-terminal propeptide is absent, but an “immature” form in which the carboxyl terminus remains intact (27) can also be detected. Consequently, we prepared two different baits from the distal half of neutrophil elastase, including a shortened one (P110 to Q218) terminating at the site of carboxyl-terminal cleavage in addition to the full-length form (P110 toH238) extending to the site of translation termination.
Under high-stringency conditions, screening of the bait (P110 to Q218) representing the mature form of neutrophil elastase yielded so many positives (Table 2) that it was not possible to characterize them all. There were in excess of 2,500 interacting clones; of these, we limited ourselves to determining the sequences of 200 and (because some proteins were recovered multiple times as overlapping or identical clones) identified 46 different proteins, including known substrates and other neutrophil granule enzymes. It is perhaps not surprising that neutrophil elastase demonstrates promiscuous interactions, because as a protease with broad specificity (9), it presumably recognizes many proteins as substrates.
TABLE 2.
Yeast two-hybrid screen with carboxyl domain of neutrophil elastase
| Protein | No. of clones
|
|
|---|---|---|
| − C-terminal | + C-terminal | |
| Iga alpha 1, chain C | 27 (21 unique, all overlap) | |
| Ig kappa light chain | 5 (2 overlap) | |
| Ig alpha2 | 3 (all overlap) | |
| Ig lambda chain | 2 (overlap) | |
| Collagen, alpha 1 | 3 (2 overlap) | |
| Collagen, alpha 2 | 3 (2 overlap) | |
| Collagen, alpha 2, type IV | 1 | |
| Complement C1q chain A | 2 (overlap) | |
| Complement C1q chain C | 1 | |
| Ficolin | 2 (overlap) | |
| α2-Macroglobulin | 2 (overlap) | |
| Plasminogen activator | 2 (overlap) | |
| Cytochrome oxidase II | 3 (overlap) | |
| Cytochrome oxidase III | 1 | |
| Cytochrome oxidase Va | 1 | |
| Cytochrome oxidase Vb | 1 | |
| Guanidinoacetate N-methyltransferase | 3 (2 identical, all overlap) | |
| NADH-ubiquinone oxidoreductase | 1 | |
| NADH dehydrogenase | 2 (no overlap) | |
| Succinate-ubiquinone oxidoreductase Ip | 1 | |
| Alcohol dehydrogenase | 1 | |
| Fructose 1,6-diphosphate | 2 (overlap) | |
| VPS29 | 1 | |
| Azurocidin | 1 | |
| Cathepsin D | 1 | |
| Chitotriosidase | 2 (overlap) | |
| Lactoferrin | 6 (all overlap) | |
| PFAAP5 | - | 1 |
| Deltex 2 | 1 | |
| Amino-terminal enhancer of split | 1 | |
| Acyl-coenzyme A:cholesterol acyltransferase | 3 (all overlap) | |
| ATP citrate lyase | - | 2 (identical) |
| PIGA | 1 | |
| Phospholipase A2, IV (cPLA2) | 1 | |
| Aminolevulinate δ-synthase 2 | 1 | |
| 5′-Aminolevulinate synthase | 1 | |
| Bile acid β-glucosidase | 1 | |
| Uroporphyrinogen decarboxylase | 1 | |
| Actin, β/γ | 9 (8 overlap) | |
| DNAJ | 1 | |
| HSP60 | 1 | |
| AN11 | 1 | |
| WD repeat protein 8 | 2 (identical) | |
| WD repeat domain 6 protein | 1 | |
| MMP9 | 1 | |
| Peripheral benzodiazepine receptor, alternate form | 2 (identical) | |
Ig, immunoglobulin.
What is surprising, however, is that screening of the bait (P110 to H238) representing the immature form of neutrophil elastase containing the intact carboxyl terminus yielded just two interacting proteins (ATP citrate lyase, an enzyme synthesizing fatty acids and contributing to cytokine-stimulated proliferation of hematopoietic cells [6], and PFAAP5, previously identified as an expressed sequence tag but whose predicted protein has remained uncharacterized) (Table 2), neither of which we recovered with, and neither of which was capable of interacting with (not shown), the bait lacking the carboxyl terminus. Remarkably, PFAAP5 was also among the few interacting proteins that we detected in the yeast two-hybrid screen employing Gfi1. We therefore focused further studies on PFAAP5 and hypothesized that it may have a role in regulating neutrophil production by establishment of a molecular link between neutrophil elastase and Gfi1, which in turn contributes to the transcriptional regulation of neutrophil elastase, as predicted for a feedback circuit.
PFAAP5 coimmunoprecipitates with neutrophil elastase and Gfi1 in transfected cells.
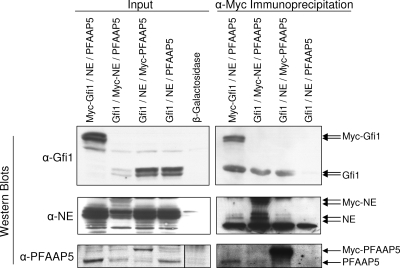
To corroborate interactions between these three proteins, we attempted to coimmunoprecipitate PFAAP5 with neutrophil elastase and/or Gfi1 from HL-60 promyelocytes. Unfortunately, the chicken antibodies that we generated to PFAAP5 and to the carboxyl terminus of neutrophil elastase did not prove suitable for immunoprecipitation, necessitating the use of ectopically expressed constructs containing an epitope tag. An additional consideration is that neutrophil elastase in HL-60 and other cell lines of myeloid derivation is predominantly found in the mature form lacking the carboxyl terminus (47) required for interaction with PFAAP5 in the yeast two-hybrid assay. We had previously observed that nonmyeloid cells, such as NIH 3T3 mouse fibroblasts, lack the ability to appropriately process the termini of transfected neutrophil elastase (47) and reasoned that in such cells the immature form of neutrophil elastase still containing an intact carboxyl terminus might be sufficiently stable to permit detection of potential intracellular interactions with PFAAP5 and/or Gfi1. We therefore performed coimmunoprecipitation in transfected NIH 3T3 cells. We prepared expression constructs for neutrophil elastase (containing the carboxyl-terminal propeptide), Gfi1, and PFAAP5, each with or without an amino-terminal Myc epitope tag, and then cotransfected each tagged version together with the other two untagged proteins (along with a negative control in which the Myc epitope tag was absent from all three proteins), performed immunoprecipitation with anti-Myc, and developed Western blots with antibodies against native forms of each of the three proteins (Fig. 1). In order to detect interactions specifically dependent on the carboxyl-terminal propeptide of neutrophil elastase, we developed an antibody to this sequence and employed it for the Western blots shown in Fig. 1 (middle row). The results showed that each Myc-tagged protein coimmunoprecipitated with each of the other two proteins. (Note that the Myc-tagged version of each protein demonstrated a higher molecular weight, as expected, but that for each Myc-tagged protein an additional band comigrated with the transfected native protein, suggesting that neutrophil elastase might have degraded the Myc tag in the immunoprecipitated fraction. Note also that coimmunoprecipitation was bidirectional, although immunoprecipitation against PFAAP5 appeared to more robustly pull down neutrophil elastase than vice versa.) As qualified under these experimental conditions involving overexpression of epitope-tagged constructs in a nonhematopoietic cell line, each of the three proteins can associate with at least one of the other two, but we cannot differentiate between all three proteins being in one complex versus two proteins together in each of the three possible pairwise combinations.
FIG. 1.
Coimmunoprecipitation of PFAAP5, Gfi1, and neutrophil elastase. NIH 3T3 cells were transfected with Gfi1, neutrophil elastase (NE), and PFAAP5 expression vectors (or a β-galactosidase-expressing control). In each set, the Myc epitope tag was present in one or none (fourth lane from left) of the expression vectors. Input lysates before (left blot) or after (right blot) immunoprecipitation with anti-Myc (α-Myc) were detected by Western blotting with anti-Gfi1 (top row), anti-neutrophil elastase carboxyl-terminal propeptide (middle row), and anti-PFAAP5 (bottom row). Transfection with a β-galactosidase-expressing control vector was included to demonstrate antibody specificity in input Western blots. (For the PFAAP5 input Western blot, the lanes are cropped from a single exposure of a continuous membrane, as indicated with a vertical line separating the rightmost lane.)
ChIP reveals occupancy of neutrophil elastase and PFAAP5 on a Gfi1-targeted promoter.
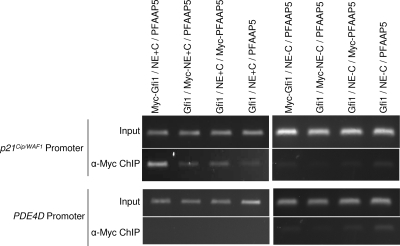
We have previously used ChIP to show that Gfi1 binds to the promoter of the cyclin-dependent kinase inhibitor gene p21Cip/WAF1 in association with histone-modifying proteins (21). We therefore determined if neutrophil elastase and PFAAP5 could also be immunoprecipitated in association with Gfi1 from chromatin corresponding to the p21Cip/WAF1 promoter (Fig. 2). We performed the experiments similarly to the coimmunoprecipitation experiments described above; we cotransfected NIH 3T3 cells with the Myc epitope-tagged version of each protein, along with the other two untagged proteins, but we completed two series: in one (Fig. 2, top left), neutrophil elastase contained the carboxyl-terminal propeptide required for PFAAP5 interaction, and in the other (Fig. 2, top right), neutrophil elastase lacked the carboxyl-terminal propeptide. We then immunoprecipitated sheared chromatin with anti-Myc and used PCR to evaluate the presence of bound p21Cip/WAF1 promoter fragments. All three proteins could be pulled down from the p21Cip/WAF1 promoter, but only in the presence of the form of neutrophil elastase containing the carboxyl-terminal propeptide required for PFAAP5 interaction. (Immunoprecipitation against Gfi1 provided the strongest signal, however, indicating that not all Gfi1 associates with the other two factors.) We also performed a control experiment in which none of the three constructs encoded an epitope-tagged version of the expressed protein (Fig. 2, fourth lane from left of each set of experiments) and included as a negative control the PDE4D promoter (Fig. 2, bottom), which we had previously shown is not a target to which Gfi1 can bind (21). We conclude that all three proteins are capable of forming a complex on the promoter of a gene targeted for repression by Gfi1 and that a specific interaction between neutrophil elastase containing an intact carboxyl terminus and PFAAP5 appears to be required for participation of the last two factors.
FIG. 2.
ChIP of PFAAP5, Gfi1, and neutrophil elastase from the p21Cip/WAF1 promoter. Vectors expressing neutrophil elastase with (NE+C) (first four lanes from left) or without (NE-C) (last four lanes) its carboxyl-terminal propeptide were cotransfected into NIH 3T3 cells, along with vectors expressing Gfi1 and PFAAP5. Each expressed protein was labeled one at a time with Myc epitope (lanes 1 to 3 and 5 to 7), or all were unlabeled (lanes 4 and 8). Ethidium-stained, agarose gel electrophoresis-separated PCR fragments of Gfi1 binding sites in the p21Cip/WAF1 promoter (top two rows) performed before (Input) or after (ChIP) immunoprecipitation with anti-Myc (α-Myc) are shown with negative control experiments for the PDE4D promoter (bottom two rows), which lacks Gfi1 binding sites.
PFAAP5 mediates neutrophil elastase and Gfi1 transcriptional repressor cooperation in reporter assays performed with siRNA.
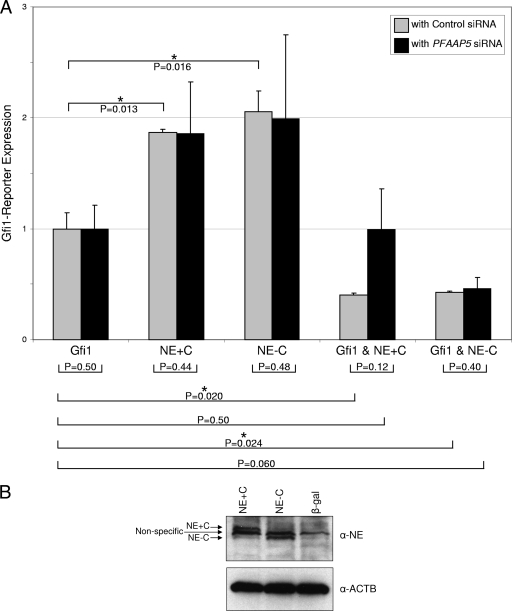
Without a coactivator, such as PRDM5 (20) or C/EBPɛ (39), Gfi1 functions as a transcriptional repressor. To determine if neutrophil elastase, in the presence or absence of PFAAP5, can influence Gfi1's transcriptional repressor activity, we performed transient-transfection reporter assays, along with RNA interference studies of PFAAP5, in NIH 3T3 cells. NIH 3T3 cells express endogenous PFAAP5 (as determined by RT-PCR [not shown]). We therefore transfected NIH 3T3 cells with neutrophil elastase and Gfi1 expression vectors, along with a Gfi1-responsive reporter plasmid plus either PFAAP5-specific or control siRNA (Fig. 3A). In the presence of control siRNA (Fig. 3A), which does not alter PFAAP5 expression levels or otherwise affect the reporter assay (as determined with initial assays [not shown]), Gfi1 demonstrated a basal level of repressor function (here normalized to a value of 1) (Fig. 3A, Gfi1). Without Gfi1, neutrophil elastase containing or lacking the carboxyl terminus showed no repressor activity (equivalent to control transfections [not shown]). However, Gfi1 cotransfected in the presence of neutrophil elastase yielded more potent repression than Gfi1 alone (regardless of whether neutrophil elastase contained or lacked the carboxyl terminus). To determine if the potentiation of Gfi1 repressor function by neutrophil elastase requires PFAAP5, we depleted its expression using siRNA (Fig. 3A). Reduction of PFAAP5 (as confirmed by RT-PCR[not shown]) affected the cooperative transcriptional repressor activity of Gfi1 only when in the presence of neutrophil elastase containing the carboxyl terminus, so that repressor activity was equivalent to that of Gfi1 acting alone. Western blotting (Fig. 3B) showed that the carboxyl-terminal propeptide does not influence the abundance of either form of neutrophil elastase used in this assay. We conclude that neutrophil elastase with or without its carboxyl terminus can enhance transcriptional repression by Gfi1 but that, as might be expected, only the form of neutrophil elastase containing the carboxyl terminus and capable of interacting with PFAAP5 requires PFAAP5 for this effect.
FIG. 3.
Potentiation of Gfi1 repression by neutrophil elastase in the presence of PFAAP5, as shown by reporter assay with RNA interference. (A) NIH 3T3 cells were transiently transfected with vectors expressing Gfi1 (10 ng) and/or neutrophil elastase (40 ng), with (NE+C) or without (NE-C) its carboxyl-terminal propeptide, along with a Gfi1-responsive CAT reporter, in the presence of siRNA directed against the endogenous mouse ortholog of PFAAP5 (black bars) or a control siRNA (gray bars). CAT activity was normalized to cotransfected luciferase, and the total quantity of transfected DNA was held constant by the addition of a β-galactosidase expression vector. (B) Western blot showing equal expression of NE+C and NE-C in NIH 3T3 cells under conditions equivalent to those in panel A. α-ACTB, anti-β-actin loading control. P values were determined by a two-tailed t test. **, P < 0.01; *, P < 0.05. The error bars indicate standard errors of the means.
Nuclear localization of neutrophil elastase enhances Gfi1 corepressor activity in reporter assays.
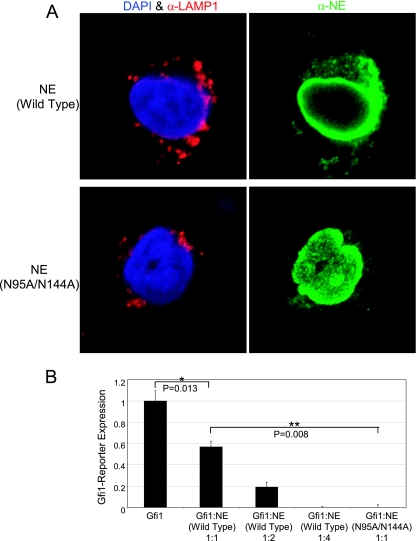
Gfi1, like other transcription factors, is found in the nucleus (25). PFAAP5 is also located in the nucleus (our results [not shown], as well as the EMBL GFP-cDNA localization database [http://gfp-cdna.embl.de/loc-html/Q92802.html]). Neutrophil elastase, on the other hand, primarily distributes to cytoplasmic granules, although it has been purified from elsewhere, including from the nucleus, as reviewed previously (34). We reasoned that if neutrophil elastase were acting in concert with Gfi1 and PFAAP5, then facilitating its trafficking to the nucleus should improve its potency in functioning as a corepressor. In other work investigating glycosylation, we found that replacement of two asparagine residues (N95 and 144A) in neutrophil elastase unexpectedly resulted in its increased concentration within the nucleus, compared to a granular and nuclear envelope distribution for the wild type, as shown by immunohistochemical localization in transiently transfected RBL1 cells (Fig. 4A). We took advantage of this incidental finding to determine if nuclear localization of neutrophil elastase influences its repressor activity. Using transient transfection of NIH 3T3 cells with a Gfi1-responsive reporter, as before, we found that, compared to a titration of wild-type neutrophil elastase (containing the carboxyl terminus) expression vector (Fig. 4B, first four bars), the nuclear-localizing form of neutrophil elastase (N95A/N144A, in which the two asparagine residues responsible for glycosylation are replaced with alanine in a full-length construct including the carboxyl terminus) demonstrated more than four-times-greater corepressor activity (Fig. 4B, right bar) in the presence of Gfi1. These results indicate that neutrophil elastase's ability to function as a corepressor improves when a greater concentration distributes to the nucleus, although we cannot exclude potential effects resulting from loss of glycosylation in the nuclear-localizing construct.
FIG. 4.
Enhancement of neutrophil elastase corepressor activity through nuclear localization. (A) Preferential localization of a neutrophil elastase N95A/N144A glycosylation variant in the nucleus. Shown is indirect immunofluorescence localization by confocal microscopy of neutrophil elastase (NE) (green) on the right, with DAPI (4′,6′-diamidino-2-phenylindole) (blue) nuclear and granular LAMP1 (red) counterstaining on the left column, for the wild type (top row) or a N95A/N144A glycosylation variant (bottom row) in transiently transfected RBL1 cells. (B) Transient-transfection reporter assay in NIH 3T3 cells showing better Gfi1 corepression with a nuclear-localizing glycosylation variant of neutrophil elastase. Cells were transfected with Gfi1-responsive CAT reporter or Gfi1 expression vector (45 ng), and the mass ratios of wild-type neutrophil elastase or neutrophil elastase N95A/N144A expression vector (both with the carboxyl-terminal propeptide intact) are indicated. CAT activity was normalized to cotransfected luciferase, and the total quantity of transfected DNA was held constant by the addition of a β-galactosidase expression vector. P values were determined by a two-tailed t test. **, P < 0.01; *, P < 0.05. The error bars indicate standard errors of the means.
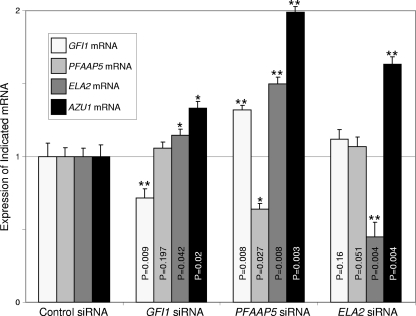
Silencing of neutrophil elastase, Gfi1, or PFAAP5 upregulates ELA2 and AZU1 in HL-60 promyelocytes.
To determine if the three factors neutrophil elastase, Gfi1, and PFAAP5 act together to promote transcriptional repression of neutrophil elastase, as well as Gfi1 (which autoinhibits its own transcription [18]), as predicted for a negative feedback circuit, we used RNA interference to evaluate the effect of reducing the expression of each factor one at a time. We therefore transfected HL-60 promyelocytes with siRNAs corresponding to either GFI1, PFAAP5, ELA2, or a control sequence. For the purpose of our experiments, designed to investigate a feedback loop hypothesis, we obviously could not use ELA2 expression levels to interrogate downstream transcriptional effects of ELA2 silencing. Therefore, as a surrogate “readout” for determining whether neutrophil elastase participates in the repression of its own transcription, we evaluated expression from the AZU1 gene (encoding azurocidin). AZU1 was previously shown to also be repressed by Gfi1 (19). Along with PRTN3 (encoding proteinase 3), ELA2 and AZU1 comprise a family of closely related genes encoding neutrophil granule proteins that likely arose by duplication in closely spaced intervals on the terminus of the short arm of human chromosome 19 and whose expression appears to be under the control of common transcriptional regulatory elements (59, 64). AZU1 expression should therefore parallel that of ELA2, but because we were not manipulating AZU1 transcript levels with siRNA (as with ELA2), AZU1 should therefore permit assessment of whether neutrophil elastase has autoinhibitory effects on its transcription. The experiment (Fig. 5) showed that siRNA directed against GFI1, PFAAP5, or ELA2 not only reduced the level of each targeted transcript (as intended), but elevated the expression of AZU1, with knockdown of PFAAP5 yielding the strongest effect. These results suggest that all three factors can act together to repress transcription from the ELA2-containing cluster of serine protease genes on chromosome 19.
FIG. 5.
Elevated expression of AZU1 following depletion of Gfi1, PFAAP5, or neutrophil elastase in HL-60 cells. HL-60 cells were transfected with siRNA corresponding to each of the indicated transcripts, and the expression of each gene was evaluated by real-time RT-PCR. P values were determined by a two-tailed t test. **, P < 0.01; *, P < 0.05. The error bars indicate standard deviations.
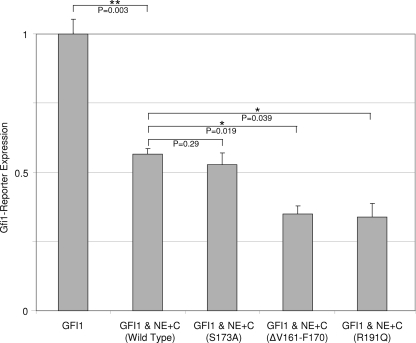
The transcriptional effects of neutrophil elastase are independent of proteolytic activity and are overdampened by cyclic neutropenia mutations.
To determine whether potentiation of transcriptional repression by Gfi1 is dependent upon neutrophil elastase's catalytic activity as a protease, we repeated the reporter assays utilizing an enzymatically inactive form of neutrophil elastase (S173A) in which the catalytic serine had been replaced with an alanine residue (47). The proteolytically inactive form of neutrophil elastase functioned equivalently to the wild type (Fig. 6), demonstrating that transcriptional repressor activity is independent of enzymolysis (and additionally excludes artifacts related to selective proteolysis of the CAT reporter). To evaluate whether mutations responsible for cyclic neutropenia disturb neutrophil elastase's transcriptional repressor activity, we repeated the reporter assays utilizing two different mutations (ΔV161-F170 and R191Q) that cause cyclic neutropenia (33). Each mutation enhances neutrophil elastase's transcriptional repressor activity in the presence of Gfi1. This observation is consistent with the feedback model as an explanation for disease: a mutant form of neutrophil elastase that overly inhibits its own synthesis, as well as other terminal effectors of granulocytic differentiation under the control of the Gfi1 transcriptional program, should lead to cyclical suppression of neutrophil formation.
FIG. 6.
Independence of neutrophil elastase (NE) transcriptional effects from protease activity and enhanced transcriptional dampening from cyclic neutropenia mutations. NIH 3T3 cells were transiently transfected with vectors expressing Gfi1 (10 ng) and neutrophil elastase (40 ng) containing its carboxyl-terminal propeptide, along with a Gfi1-responsive CAT reporter. Wild-type neutrophil elastase was compared to a proteolytically inactive non-disease-causing mutation (S173A) and two different mutations (ΔV161-F170 and R191Q) responsible for cyclic neutropenia. CAT activity was normalized to cotransfected luciferase. P values were determined by a two-tailed t test. **, P < 0.01; *, P < 0.05. The error bars indicate standard errors of the means.
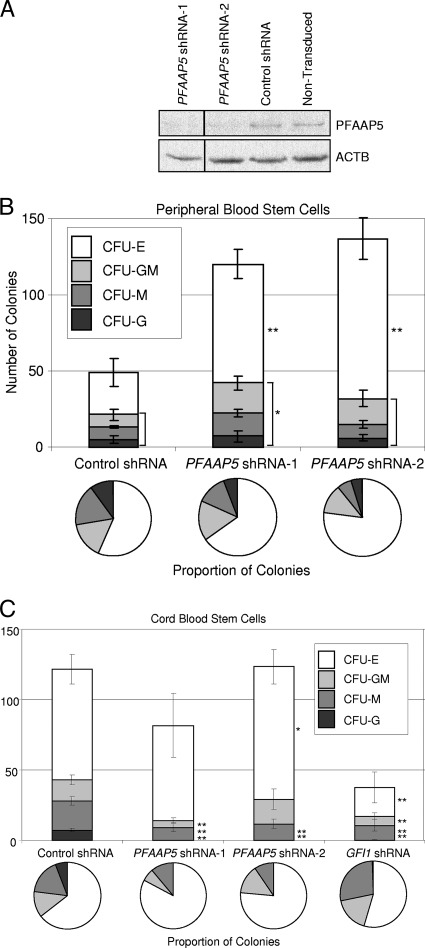
Silencing of PFAAP5 in HSCs promotes proliferation while reducing the proportion differentiating into myeloid cells.
To investigate PFAAP5's contributions to HSC differentiation, we purified human CD34+ hematopoietic stem cells and transduced them with lentiviral vectors expressing two different shRNAs targeting PFAAP5 (Fig. 7A). In vitro colony formation assays of adult peripheral blood stem cells revealed that depletion of PFAAP5 reduces the proportion differentiating into myeloid types of colonies (Fig. 7B). Colony formation assays of CD34+ human cord blood from a different individual revealed more pronounced effects on reducing or eliminating, respectively, macrophage and granulocyte colony formation (Fig. 7C). The differences in colony formation assays between the two sources of HSCs may be related to the different developmental stages from which they were derived (cord blood versus adult) or to interindividual differences related to the genetic backgrounds of the donors. Nevertheless, the positive control experiments with depletion of Gfi1 (Fig. 7C) demonstrated that, overall, loss of PFAAP5 has effects similar to those of the loss of Gfi1 (30, 36), except that, unlike for Gfi1, there is no reduction in erythrocyte colony formation with PFAAP5 depletion.
FIG. 7.
Effects of PFAAP5 depletion on HSC proliferation and myeloid differentiation. (A) Western blot showing reduced PFAAP5 expression in human CD34+ peripheral blood cells transduced with lentiviral vectors containing either of two different shRNAs targeting PFAAP5 compared to a nontargeting control shRNA and nontransduced cells. Expression of β-actin (ACTB) was used as a loading control. (The lanes are cropped from a single exposure of a continuous membrane, as indicated with a vertical line separating the leftmost lane.) (B) Colony formation assays performed on human CD34+ peripheral blood cells repeated in triplicate (control and shRNA-1) and quadruplicate (shRNA-2). CFU-E, CFU-erythroid; CFU-GM, CFU-granulocyte/macrophage; CFU-M, CFU macrophage; CFU-G, CFU-granulocyte. (C) Colony formation assays performed on human CD34+ cord blood cells repeated in sextuplicate. The error bars show standard deviations. P values were determined by a two-tailed t test. **, P < 0.01; *, P < 0.05.
DISCUSSION
How mutations in ELA2 and other genes disrupt hematopoiesis and cause neutropenia has remained unexplained. Perhaps the most striking—yet puzzling—clue is the cycling neutrophil count. Studies of circadian and other rhythms have led to the identification of transcriptionally coupled feedback loops responsible for regulating biological clocks (35, 43), and similarly, theoretical studies of cyclic neutropenia (28, 51) have also invoked disturbance of feedback circuits as a potential explanation for blood cell oscillation in this disease, although these models were proposed before the responsible genes were recognized. Another important finding derives from observations of individuals who are mosaic for ELA2 mutations (3), where the mutant and wild type are equally distributed among progenitor cell populations yet it is only the wild type that is found in circulating neutrophils, indicating that mutant neutrophil elastase acts cell autonomously to prevent the maturation of precursors.
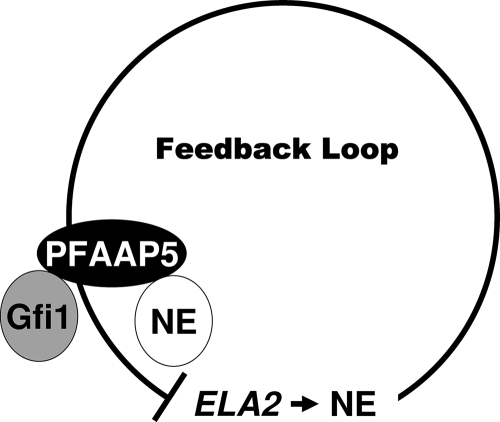
The possibility, then, that neutrophil elastase, whose expression is restricted to the progenitor cell population that it is capable of inhibiting (46), can communicate with Gfi1, a transcription factor governing both neutrophil elastase expression (19, 30, 55) and the production of neutrophils (29, 65), has immediate attraction, for it has the potential to “close” a postulated feedback circuit (Fig. 8). We therefore became intrigued by detecting PFAAP5 in yeast two-hybrid screens that otherwise identified few interacting partners for either neutrophil elastase (when in a form containing the carboxyl-terminal propeptide) or Gfi1.
FIG. 8.
Schematic of PFAAP5's role in the feedback loop. NE, neutrophil elastase.
PFAAP5 is the previously uncharacterized product of a transcript deposited in GenBank (accession no. AF530063) after it was found to be upregulated following treatment of Jurkat T lymphocytes with the herpesvirus drug phosphonoformate. Its predicted polypeptide structure (using the Robetta protein-modeling algorithm [40; http://robetta.bakerlab.org]) reveals a bipartite topology (not shown) compatible with our proposal for a role as an adapter capable of bridging interactions between two functionally disparate proteins. As noted, PFAAP5 is a nuclear protein, and it contains a predicted nuclear localization signal (K133 to K160, according to the PredictNLS algorithm [16; http://cubic.bioc.columbia.edu/predictNLS]) but is not expected to bind DNA (according to the LOCtree algorithm [52; http://cubic.bioc.columbia.edu/cgi/var/nair/loctree/query]). PFAAP5 lacks obvious similarity to other known proteins, except for the vicinity of its carboxyl terminus, which demonstrates homology to a portion of B3BP (BCL-3 binding protein) sharing a highly conserved ATPase domain (including a “Walker A” motif between R408 and K415 of PFAAP5), which contributes to B3BP interactions with the BCL-3 oncoprotein and p300/CBP histone acetyltransferase (63), offering further support that PFAAP5 participates in transcriptional regulation. PFAAP5 is most strongly expressed in the bone marrow in CD33+ promyelocytes and CD34+ HSCs and in the peripheral blood in monocytes (according to GNF Gene Expression Atlas 2 [60] as accessed from the Genome Browser [http://genome.ucsc.edu]), suggesting that it is important for neutrophil differentiation. PFAAP5 is also one of the most strongly downregulated transcripts following exposure to arsenic compounds (4), whose toxicity includes neutropenia (10) and whose therapeutic effect in the treatment of leukemia is to induce differentiation of promyelocytic blasts (61). PFAAP5 becomes phosphorylated at serine 199 in response to DNA damage, as recently determined (49) through a large-scale analysis of substrates for the protein kinases ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3 related).
Silencing of PFAAP5 expression in HSCs significantly impaired myeloid differentiation. In cord blood obtained from one human donor, RNA interference depletion of PFAAP5 completely abolished neutrophil production in in vitro colony formation assays. These results suggest that PFAAP5 may have an indispensable role in regulating neutrophil production.
PFAAP5 is a nuclear protein, as is Gfi1 (25). Neutrophil elastase, however, is predominately found in the granules of neutrophils. Nevertheless, other studies indicate that neutrophil elastase travels to the nucleus and is capable of influencing transcription. Before attaining oncogenic activity, the PML-RARα fusion transcription factor, generated by chromosomal translocation in promyelocytic leukemia, becomes cleaved, and biochemical and genetic experiments have demonstrated that neutrophil elastase proteolyses PML-RARα within the nucleus (44, 45). Neutrophil activation in response to pathogens results in dispersal of net-like structures comprised of chromatin embedded with neutrophil elastase (15, 22). Neutrophil elastase cleaves the telomere binding protein TRF2 within the nucleus during radiation-induced apoptosis of HL-60 promyelocytes (53). Our incidental finding that mutation of N-linked glycosylation sites promotes nuclear accumulation of neutrophil elastase (and thereby makes it a more potent transcriptional corepressor) also hints at cellular mechanisms facilitating its nuclear trafficking. How neutrophil elastase translocates to the nucleus is not known. The nuclear trafficking of neutrophil elastase may occur through a posttranslational pathway distinct from its granule-trafficking pathway.
There is a precedent for proteases “moonlighting” as transcription factors. VPE is a cysteine protease of plant vacuoles (arguably equivalent to neutrophil granules) and is involved in tissue differentiation and apoptosis, but it can also function as a transcription factor (48). VPE mutations affecting its proteolytic activity do not interfere with its transcription factor capabilities, indicating that these two functions are separate. Similarly, for neutrophil elastase, we found that mutationally inactivating the catalytic serine required for proteolysis did not influence its ability to function as a transcriptional repressor.
PFAAP5 may have a role in promoting the maturation of the carboxyl terminus of neutrophil elastase. The RNA interference studies showed that PFAAP5 is required to help neutrophil elastase act as a corepressor with Gfi1 in HL-60 promyelocytes, where neutrophil elastase is processed to remove the carboxyl propeptide, and therefore should no longer interact with PFAAP5. On the other hand, a “preprocessed” version of neutrophil elastase, in which the carboxyl propeptide was deleted, functioned as a corepressor just as well as the full-length version of neutrophil elastase containing an intact propeptide in NIH 3T3 fibroblasts, and its activity remained unchanged when PFAAP5 was knocked down by RNA interference. We reconcile these observations by positing that PFAAP5 is required for neutrophil elastase's transcriptional activity prior to the removal of the carboxyl terminus and that only once the propeptide is removed does neutrophil elastase become transcriptionally active.
We also examined the effects of mutations in neutrophil elastase that are responsible for cyclic neutropenia. Two different mutations (ΔV161-F170 and R191Q) each enhanced neutrophil elastase's ability to potentiate Gfi1 transcriptional repressor activity. This observation is in agreement with the proposed model, in which pathogenic overdampening of the feedback circuit would be expected to inhibit production of neutrophil elastase, as well as other transcriptional functions that are under the control of Gfi1. With respect to SCN, it may be significant that many of the mutations of neutrophil elastase that cause SCN truncate the protein just prior to the carboxyl-terminal propeptide (34) and therefore lack the region of neutrophil elastase that appears to be required for interaction with PFAAP5. Thus, mutations affecting neutrophil elastase's carboxyl terminus might indirectly disrupt Gfi1 function via PFAAP5 and thereby also impair transcriptional programs under Gfi1's control—including neutrophil differentiation and the cell cycle (29, 31, 65).
Acknowledgments
This study was supported by NIH grants R01HL79507 and R01DK58161 (to M.S.H.), R01HL079574 and R01CA105152 (to H.L.G.), and F30AG030316 (to S.J.S.). S.J.S. was additionally supported by the University of Washington Medical Scientist Training Program (NIH T32GM007266, Poncin Fund, and ARCS Fellowship). M.E.B.R. was supported by NIH grant T32HD07463.
We appreciatively acknowledge Chung-Ying Huang's contributions to early phases of this work.
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Ancliff, P. J. 2003. Congenital neutropenia. Blood Rev. 17209-216. [DOI] [PubMed] [Google Scholar]
- 2.Ancliff, P. J., M. P. Blundell, G. O. Cory, Y. Calle, A. Worth, H. Kempski, S. Burns, G. E. Jones, J. Sinclair, C. Kinnon, I. M. Hann, R. E. Gale, D. C. Linch, and A. J. Thrasher. 2006. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood 1082182-2189. [DOI] [PubMed] [Google Scholar]
- 3.Ancliff, P. J., R. E. Gale, M. J. Watts, R. Liesner, I. M. Hann, S. Strobel, and D. C. Linch. 2002. Paternal mosaicism proves the pathogenic nature of mutations in neutrophil elastase in severe congenital neutropenia. Blood 100707-709. [DOI] [PubMed] [Google Scholar]
- 4.Argos, M., M. G. Kibriya, F. Parvez, F. Jasmine, M. Rakibuz-Zaman, and H. Ahsan. 2006. Gene expression profiles in peripheral lymphocytes by arsenic exposure and skin lesion status in a Bangladeshi population. Cancer Epidemiol. Biomarkers Prev. 151367-1375. [DOI] [PubMed] [Google Scholar]
- 5.Badolato, R., and S. Parolini. 2007. Novel insights from adaptor protein 3 complex deficiency. J. Allergy Clin. Immunol. 120735-741. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, D. E., G. Hatzivassiliou, F. Zhao, C. Andreadis, and C. B. Thompson. 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 246314-6322. [DOI] [PubMed] [Google Scholar]
- 7.Bellanne-Chantelot, C., S. Clauin, T. Leblanc, B. Cassinat, F. Rodrigues-Lima, S. Beaufils, C. Vaury, M. Barkaoui, O. Fenneteau, M. Maier-Redelsperger, C. Chomienne, and J. Donadieu. 2004. Mutations in the ELA2 gene correlate with more severe expression of neutropenia: a study of 81 patients from the French Neutropenia Register. Blood 1034119-4125. [DOI] [PubMed] [Google Scholar]
- 8.Benson, K. F., F. Q. Li, R. E. Person, D. Albani, Z. Duan, J. Wechsler, K. Meade-White, K. Williams, G. M. Acland, G. Niemeyer, C. D. Lothrop, and M. Horwitz. 2003. Mutations associated with neutropenia in dogs and humans disrupt intracellular transport of neutrophil elastase. Nat. Genet. 3590-96. [DOI] [PubMed] [Google Scholar]
- 9.Bieth, J. G. 1998. Leukocyte elastase, p. 54-60. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, San Diego, CA.
- 10.Binet, F., H. Cavalli, E. Moisan, and D. Girard. 2006. Arsenic trioxide (AT) is a novel human neutrophil pro-apoptotic agent: effects of catalase on AT-induced apoptosis, degradation of cytoskeletal proteins and de novo protein synthesis. Br. J. Haematol. 132349-358. [DOI] [PubMed] [Google Scholar]
- 11.Bode, W., E. Meyer, Jr., and J. C. Powers. 1989. Human leukocyte and porcine pancreatic elastase: X-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry 281951-1963. [DOI] [PubMed] [Google Scholar]
- 12.Bohn, G., K. Welte, and C. Klein. 2007. Severe congenital neutropenia: new genes explain an old disease. Curr. Opin. Rheumatol. 19644-650. [DOI] [PubMed] [Google Scholar]
- 13.Borregaard, N., K. Theilgaard-Monch, J. B. Cowland, M. Stahle, and O. E. Sorensen. 2005. Neutrophils and keratinocytes in innate immunity—cooperative actions to provide antimicrobial defense at the right time and place. J. Leukoc. Biol. 77439-443. [DOI] [PubMed] [Google Scholar]
- 14.Boztug, K., G. Appaswamy, A. Ashikov, A. A. Schaffer, U. Salzer, J. Diestelhorst, M. Germeshausen, G. Brandes, J. Lee-Gossler, F. Noyan, A. K. Gatzke, M. Minkov, J. Greil, C. Kratz, T. Petropoulou, I. Pellier, C. Bellanne-Chantelot, N. Rezaei, K. Monkemoller, N. Irani-Hakimeh, H. Bakker, R. Gerardy-Schahn, C. Zeidler, B. Grimbacher, K. Welte, and C. Klein. 2009. A syndrome with congenital neutropenia and mutations in G6PC3. N. Engl. J. Med. 36032-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkmann, V., U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch, and A. Zychlinksy. 2004. Neutrophil extracellular traps kill bacteria. Science 3031532-1535. [DOI] [PubMed] [Google Scholar]
- 16.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dale, D. C., R. E. Person, A. A. Bolyard, A. G. Aprikyan, C. Bos, M. A. Bonilla, L. A. Boxer, G. Kannourakis, C. Zeidler, K. Welte, K. F. Benson, and M. Horwitz. 2000. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood 962317-2322. [PubMed] [Google Scholar]
- 18.Doan, L. L., S. D. Porter, Z. Duan, M. M. Flubacher, D. Montoya, P. N. Tsichlis, M. Horwitz, C. B. Gilks, and H. L. Grimes. 2004. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 322508-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan, Z., and M. Horwitz. 2003. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl. Acad. Sci. USA 1005932-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan, Z., R. E. Person, H. H. Lee, S. Huang, J. Donadieu, R. Badolato, H. L. Grimes, T. Papayannopoulou, and M. S. Horwitz. 2007. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol. Cell. Biol. 276889-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan, Z., A. Zarebski, D. Montoya-Durango, H. L. Grimes, and M. Horwitz. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 2510338-10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, T. A., U. Abed, C. Goosmann, R. Hurwitz, I. Schulze, V. Wahn, Y. Weinrauch, V. Brinkmann, and A. Zychlinsky. 2007. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenda, D. S., and D. C. Link. 2006. Mechanisms of disordered granulopoiesis in congenital neutropenia. Curr. Top. Dev. Biol. 74133-176. [DOI] [PubMed] [Google Scholar]
- 24.Grenda, D. S., M. Murakami, J. Ghatak, J. Xia, L. A. Boxer, D. Dale, M. C. Dinauer, and D. C. Link. 2007. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood 1104179-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 166263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gullberg, U., N. Bengtsson, E. Bulow, D. Garwicz, A. Lindmark, and I. Olsson. 1999. Processing and targeting of granule proteins in human neutrophils. J. Immunol. Methods 232201-210. [DOI] [PubMed] [Google Scholar]
- 27.Gullberg, U., A. Lindmark, G. Lindgren, A. M. Persson, E. Nilsson, and I. Olsson. 1995. Carboxyl-terminal prodomain-deleted human leukocyte elastase and cathepsin G are efficiently targeted to granules and enzymatically activated in the rat basophilic/mast cell line RBL. J. Biol. Chem. 27012912-12918. [DOI] [PubMed] [Google Scholar]
- 28.Haurie, C., D. C. Dale, and M. C. Mackey. 1998. Cyclical neutropenia and other periodic hematological disorders: a review of mechanisms and mathematical models. Blood 922629-2640. [PubMed] [Google Scholar]
- 29.Hock, H., M. J. Hamblen, H. M. Rooke, J. W. Schindler, S. Saleque, Y. Fujiwara, and S. H. Orkin. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 4311002-1007. [DOI] [PubMed] [Google Scholar]
- 30.Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson, S. Cameron, and S. H. Orkin. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18109-120. [DOI] [PubMed] [Google Scholar]
- 31.Horman, S. R., C. S. Velu, A. Chaubey, T. Bourdeau, J. Zhu, W. E. Paul, B. Gebelein, and H. L. Grimes. 2009. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood 1135466-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horwitz, M., K. F. Benson, Z. Duan, F. Q. Li, and R. E. Person. 2004. Hereditary neutropenia: dogs explain human neutrophil elastase mutations. Trends Mol. Med. 10163-170. [DOI] [PubMed] [Google Scholar]
- 33.Horwitz, M., K. F. Benson, R. E. Person, A. G. Aprikyan, and D. C. Dale. 1999. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat. Genet. 23433-436. [DOI] [PubMed] [Google Scholar]
- 34.Horwitz, M. S., Z. Duan, B. Korkmaz, H. H. Lee, M. E. Mealiffe, and S. J. Salipante. 2007. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood 1091817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunt, T., and P. Sassone-Corsi. 2007. Riding tandem: circadian clocks and the cell cycle. Cell 129461-464. [DOI] [PubMed] [Google Scholar]
- 36.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30295-300. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi, H., M. Kobayashi, K. Nakamura, N. Konishi, S. Miyagawa, T. Sato, H. Toyoda, Y. Komada, S. Kojima, Y. Todoroki, K. Ueda, and O. Katoh. 2003. Dysregulation of transcriptions in primary granule constituents during myeloid proliferation and differentiation in patients with severe congenital neutropenia. J. Leukoc. Biol. 73225-234. [DOI] [PubMed] [Google Scholar]
- 38.Kawai, T., and H. L. Malech. 2009. WHIM syndrome: congenital immune deficiency disease. Curr. Opin. Hematol. 1620-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanna-Gupta, A., H. Sun, T. Zibello, H. M. Lee, R. Dahl, L. A. Boxer, and N. Berliner. 2007. Growth factor independence 1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene inactivating mutation in the C/EBPɛ gene. Blood 1094181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, D. E., D. Chivian, L. Malmstrom, and D. Baker. 2005. Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins 61(Suppl. 7)193-200. [DOI] [PubMed] [Google Scholar]
- 41.Klein, C., M. Grudzien, G. Appaswamy, M. Germeshausen, I. Sandrock, A. A. Schaffer, C. Rathinam, K. Boztug, B. Schwinzer, N. Rezaei, G. Bohn, M. Melin, G. Carlsson, B. Fadeel, N. Dahl, J. Palmblad, J. I. Henter, C. Zeidler, B. Grimbacher, and K. Welte. 2007. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease). Nat. Genet. 3986-92. [DOI] [PubMed] [Google Scholar]
- 42.Kollner, I., B. Sodeik, S. Schreek, H. Heyn, N. von Neuhoff, M. Germeshausen, C. Zeidler, M. Kruger, B. Schlegelberger, K. Welte, and C. Beger. 2006. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood 108493-500. [DOI] [PubMed] [Google Scholar]
- 43.Lakin-Thomas, P. L. 2006. Transcriptional feedback oscillators: maybe, maybe not. J. Biol. Rhythms 2183-92. [DOI] [PubMed] [Google Scholar]
- 44.Lane, A. A., and T. J. Ley. 2003. Neutrophil elastase cleaves PML-RARα and is important for the development of acute promyelocytic leukemia in mice. Cell 115305-318. [DOI] [PubMed] [Google Scholar]
- 45.Lane, A. A., and T. J. Ley. 2005. Neutrophil elastase is important for PML-retinoic acid receptor alpha activities in early myeloid cells. Mol. Cell. Biol. 2523-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leitch, H. A., and J. G. Levy. 1994. Reversal of camal-mediated alterations of normal and leukemic in-vitro myelopoiesis using inhibitors of proteolytic activity. Leukemia 8605-611. [PubMed] [Google Scholar]
- 47.Li, F. Q., and M. Horwitz. 2001. Characterization of mutant neutrophil elastase in severe congenital neutropenia. J. Biol. Chem. 27614230-14241. [DOI] [PubMed] [Google Scholar]
- 48.Matarasso, N., S. Schuster, and A. Avni. 2005. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell 171205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuoka, S., B. A. Ballif, A. Smogorzewska, E. R. McDonald III, K. E. Hurov, J. Luo, C. E. Bakalarski, Z. Zhao, N. Solimini, Y. Lerenthal, Y. Shiloh, S. P. Gygi, and S. J. Elledge. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 3161160-1166. [DOI] [PubMed] [Google Scholar]
- 50.Montoya-Durango, D. E., C. S. Velu, A. Kazanjian, M. E. Rojas, C. M. Jay, G. D. Longmore, and H. L. Grimes. 2008. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J. Biol. Chem. 28332056-32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morley, A. 1979. Cyclic hemopoiesis and feedback control. Blood Cells 5283-296. [PubMed] [Google Scholar]
- 52.Nair, R., and B. Rost. 2005. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 34885-100. [DOI] [PubMed] [Google Scholar]
- 53.Nakagami, Y., M. Ito, T. Hara, and T. Inoue. 2002. Loss of TRF2 by radiation-induced apoptosis in HL60 cells. Radiat. Med. 20121-129. [PubMed] [Google Scholar]
- 54.Newman, S. L., L. Gootee, J. E. Gabay, and M. E. Selsted. 2000. Identification of constituents of human neutrophil azurophil granules that mediate fungistasis against Histoplasma capsulatum. Infect. Immun. 685668-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Person, R. E., F. Q. Li, Z. Duan, K. F. Benson, J. Wechsler, H. A. Papadaki, G. Eliopoulos, C. Kaufman, S. J. Bertolone, B. Nakamoto, T. Papayannopoulou, H. L. Grimes, and M. Horwitz. 2003. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rytomaa, T. 1973. Role of chalone in granulopoiesis. Br. J. Haematoly. 24141-146. [DOI] [PubMed] [Google Scholar]
- 57.Schaffer, A. A., and C. Klein. 2007. Genetic heterogeneity in severe congenital neutropenia: how many aberrant pathways can kill a neutrophil? Curr. Opin. Allergy Clin. Immunol. 7481-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skokowa, J., J.-P. Fobiwe, D. Lan, M. Germeshausen, and K. Welte. 2007. Defective expression of neutrophil serine proteases in myeloid progenitors of congenital neutropenia patients carrying either ELA2 or HAX1 mutations. Blood 109A3299. [Google Scholar]
- 59.Sturrock, A., K. F. Franklin, S. Wu, and J. R. Hoidal. 1998. Characterization and localization of the genes for mouse proteinase-3 (Prtn3) and neutrophil elastase (Ela2). Cytogenet. Cell Genet. 83104-108. [DOI] [PubMed] [Google Scholar]
- 60.Su, A. I., M. P. Cooke, K. A. Ching, Y. Hakak, J. R. Walker, T. Wiltshire, A. P. Orth, R. G. Vega, L. M. Sapinoso, A. Moqrich, A. Patapoutian, G. M. Hampton, P. G. Schultz, and J. B. Hogenesch. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 994465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tallman, M. S. 2008. The expanding role of arsenic in acute promyelocytic leukemia. Semin. Hematol. 45S25-S29. [DOI] [PubMed] [Google Scholar]
- 62.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 81434-1447. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe, N., S. Wachi, and T. Fujita. 2003. Identification and characterization of BCL-3-binding protein: implications for transcription and DNA repair or recombination. J. Biol. Chem. 27826102-26110. [DOI] [PubMed] [Google Scholar]
- 64.Wong, E. T., D. E. Jenne, M. Zimmer, S. D. Porter, and C. B. Gilks. 1999. Changes in chromatin organization at the neutrophil elastase locus associated with myeloid cell differentiation. Blood 943730-3736. [PubMed] [Google Scholar]
- 65.Zeng, H., R. Yucel, C. Kosan, L. Klein-Hitpass, and T. Moroy. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 234116-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]